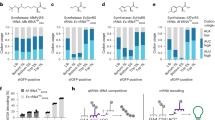
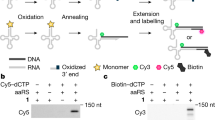
Abstract
A critical component of genetic code expansion applications is an aminoacyl-tRNA synthetase (RS)/tRNA pair that faithfully encodes a noncanonical amino acid (ncAA) in response to a specific codon. Here we detail a procedure to select an ncAA-specific RS from a publicly available 3.2-million-member Methanomethylophilus alvus pyrrolysyl-RS (MaPylRS) active site mutant library. Four main parts of the procedure are: (1) preparing the library for use and creating needed cell lines; (2) life and death selections that, respectively, select for functional RSs and select against RSs that incorporate canonical amino acids; (3) three fluorescence-based status checks that provide information about the efficiency and fidelity of the surviving RSs in incorporating the target ncAA; and (4) characterizing top hits to find the best ones for use in applications. The resulting RS/tRNA pairs can be used in either bacterial or eukaryotic cells to study proteins of interest. Additionally, the stability of the MaPylRSs makes them useful in cell-free ncAA-protein expression and amenable to structural and other in vitro characterizations. This Protocol is usable by those with basic molecular biology expertise and features a reliable positive control scheme for selections, status checks at different stages to interpret the level of success and a robust procedure to characterize newly engineered tRNA–RS pairs. Users of this Protocol can expect to select ncAA-specific RS/tRNA pairs from the library within about 30–50 d depending on preparation needs.
Key points
-
This Protocol outlines the steps to isolate aminoacyl-tRNA synthetase/tRNA pairs that incorporate noncanonical amino acids into proteins. RSs are selected from a publicly available 3.2-million-member MaPylRS active site mutant library and involves a series of life and death selections and fluorescence-based enrichment and evaluation.
-
Aminoacyl-tRNA synthetase hits with high fidelity and efficiency in incorporating the desired noncanonical amino acids are suitable for use in prokaryotic and eukaryotic cells and in cell-free protein expression systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




















Similar content being viewed by others
Data availability
The data used to illustrate procedural outcomes of this protocol are available as Source data within the article or reported by supporting primary research papers23. Additional relevant data may be available upon request to the authors. Source data are provided with this paper.
References
Shandell, M. A., Tan, Z. & Cornish, V. W. Genetic code expansion: a brief history and perspective. Biochemistry 60, 3455–3469 (2021).
de la Torre, D. & Chin, J. W. Reprogramming the genetic code. Nat. Rev. Genet. 22, 169–184 (2021).
Jann, C., Giofré, S., Bhattacharjee, R. & Lemke, E. A. Cracking the code: reprogramming the genetic script in prokaryotes and eukaryotes to harness the power of noncanonical amino acids. Chem. Rev. 124, 10281–10362 (2024).
Koch, N. G. & Budisa, N. Evolution of pyrrolysyl-tRNA synthetase: from methanogenesis to genetic code expansion. Chem. Rev. 124, 9580–9608 (2024).
Perdiguero, A. N. & Liang, A. D. Practical approaches to genetic code expansion with aminoacyl-tRNA synthetase/tRNA pairs. CHIMIA 78, 22–31 (2024).
Xie, J. & Schultz, P. G. An expanding genetic code. Methods 36, 227–238 (2005).
Xie, J. & Schultz, P. G. A chemical toolkit for proteins—an expanded genetic code. Nat. Rev. Mol. Cell Biol. 7, 775–782 (2006).
Krahn, N., Tharp, J. M., Crnković, A. & Söll, D. Engineering aminoacyl-tRNA synthetases for use in synthetic biology. Enzymes 48, 351–395 (2020).
Beattie, A. T., Dunkelmann, D. L. & Chin, J. W. Quintuply orthogonal pyrrolysyl-tRNA synthetase/tRNAPyl pairs. Nat. Chem. 15, 948–959 (2023).
Neumann, H., Slusarczyk, A. L. & Chin, J. W. De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J. Am. Chem. Soc. 132, 2142–2144 (2010).
Cervettini, D. et al. Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase–tRNA pairs. Nat. Biotechnol. 38, 989–999 (2020).
Spinck, M., Guppy, A. & Chin, J. W. Automated orthogonal tRNA generation. Nat. Chem. Biol. 21, 657–667 (2025).
Wang, L., Zhang, Z., Brock, A. & Schultz, P. G. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl Acad. Sci. USA 100, 56–61 (2003).
Chin, J. W., Martin, A. B., King, D. S., Wang, L. & Schultz, P. G. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl Acad. Sci. USA 99, 11020–11024 (2002).
Chin, J. W. et al. Addition of p-Azido-l-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 124, 9026–9027 (2002).
Wang, L., Brock, A., Herberich, B. & Schultz, P. G. Expanding the genetic code of Escherichia coli. Science 292, 498–500 (2001).
Dumas, A., Lercher, L., Spicer, C. D. & Davis, B. G. Designing logical codon reassignment—expanding the chemistry in biology. Chem. Sci. 6, 50–69 (2014).
Lino, B. R., Williams, S. J., Castor, M. E. & Van Deventer, J. A. Reaching new heights in genetic code manipulation with high throughput screening. Chem. Rev. 124, 12145–12175 (2024).
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Ann. Rev. Biochem. 79, 413–444 (2010).
Nguyen, D. P. et al. Genetic encoding of photocaged cysteine allows photoactivation of TEV protease in live mammalian cells. J. Am. Chem. Soc. 136, 2240–2243 (2014).
Cropp, T. A., Anderson, J. C. & Chin, J. W. Reprogramming the amino-acid substrate specificity of orthogonal aminoacyl-tRNA synthetases to expand the genetic code of eukaryotic cells. Nat. Protoc. 2, 2590–2600 (2007).
Grasso, K. T. et al. A facile platform to engineer Escherichia coli tyrosyl-tRNA synthetase adds new chemistries to the eukaryotic genetic code, including a phosphotyrosine mimic. ACS Cent. Sci. 8, 483–492 (2022).
Avila-Crump, S. et al. Generating efficient Methanomethylophilus alvus pyrrolysyl-trna synthetases for structurally diverse non-canonical amino acids. ACS Chem. Biol. 17, 3458–3469 (2022).
Galles, G. D. et al. Tuning phenylalanine fluorination to assess aromatic contributions to protein function and stability in cells. Nat. Commun. 14, 59 (2023).
Porter, J. J. et al. Genetically encoded protein tyrosine nitration in mammalian cells. ACS Chem. Biol. 14, 1328–1336 (2019).
Jang, H. S., Jana, S., Blizzard, R. J., Meeuwsen, J. C. & Mehl, R. A. Access to faster eukaryotic cell labeling with encoded tetrazine amino acids. J. Am. Chem. Soc. 7245–7249 (2020).
Seki, E., Yanagisawa, T., Kuratani, M., Sakamoto, K. & Yokoyama, S. Fully productive cell-free genetic code expansion by structure-based engineering of Methanomethylophilus alvus pyrrolysyl-tRNA synthetase. ACS Synth. Biol. 9, 718–732 (2020).
Meineke, B., Heimgärtner, J., Lafranchi, L. & Elsässer, S. J. Methanomethylophilus alvus Mx1201 provides basis for mutual orthogonal pyrrolysyl tRNA/aminoacyl-tRNA synthetase pairs in mammalian cells. ACS Chem. Biol. 13, 3087–3096 (2018).
Beránek, V., Willis, J. C. W. & Chin, J. W. An evolved Methanomethylophilus alvus pyrrolysyl-tRNA synthetase/tRNA pair is highly active and orthogonal in mammalian cells. Biochemistry 58, 387–390 (2019).
Willis, J. C. W. & Chin, J. W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 10, 831–837 (2018).
Gan, Q. & Fan, C. Orthogonal translation for site-specific installation of post-translational modifications. Chem. Rev. 124, 2805–2838 (2024).
Bednar, R. M. et al. Genetic incorporation of two mutually orthogonal bioorthogonal amino acids that enable efficient protein dual-labeling in cells. ACS Chem. Biol. 16, 2612–2622 (2021).
Meineke, B., Heimgärtner, J., Eirich, J., Landreh, M. & Elsässer, S. J. Site-specific incorporation of two ncaas for two-color bioorthogonal labeling and crosslinking of proteins on live mammalian cells. Cell Rep. 31, 107811 (2020).
Nikić, I. & Lemke, E. A. Genetic code expansion enabled site-specific dual-color protein labeling: superresolution microscopy and beyond. Curr. Opin. Chem. Biol. 28, 164–173 (2015).
Van Fossen, E. M. et al. Nanobody assemblies with fully flexible topology enabled by genetically encoded tetrazine amino acids. Sci. Adv. 8, eabm6909 (2022).
Roy, G. et al. Development of a high yielding expression platform for the introduction of non-natural amino acids in protein sequences. MAbs 12, 1684749 (2020).
Ma, J. S. Y. et al. Versatile strategy for controlling the specificity and activity of engineered T cells. Proc. Natl Acad. Sci. USA 113, E450–E458 (2016).
Li, Q. et al. Developing covalent protein drugs via proximity-enabled reactive therapeutics. Cell 182, 85–97.e16 (2020).
Switzer, H. J. et al. Employing non-canonical amino acids towards the immobilization of a hyperthermophilic enzyme to increase protein stability. RSC Adv 13, 8496–8501 (2023).
Bednar, R. M. et al. Immobilization of proteins with controlled load and orientation. ACS Appl. Mater. Interfaces 11, 36391–36398 (2019).
Robertson, W. E. et al. Sense codon reassignment enables viral resistance and encoded polymer synthesis. Science 372, 1057–1062 (2021).
Koch, N. G., Baumann, T. & Budisa, N. Efficient unnatural protein production by pyrrolysyl-trna synthetase with genetically fused solubility tags. Front. Bioeng. 9, 807438 (2021).
Cao, L., Liu, J., Ghelichkhani, F., Rozovsky, S. & Wang, L. Genetic incorporation of ϵ-N-benzoyllysine by engineering Methanomethylophilus alvus pyrrolysyl-tRNA synthetase. ChemBioChem 22, 2530–2534 (2021).
Fricke, R. et al. Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-α-amino acids in vitro and in vivo. Nat. Chem. 15, 960–971 (2023).
Gottfried-Lee, I., Perona, J. J., Karplus, P. A., Mehl, R. A. & Cooley, R. B. Structures of Methanomethylophilus alvus pyrrolysine tRNA-synthetases support the need for de novo selections when altering the substrate specificity. ACS Chem. Biol. 17, 3470–3477 (2022).
Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Genetically encoding N-ε-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 (2008).
Italia, J. S., Latour, C., Wrobel, C. J. J. & Chatterjee, A. Resurrecting the bacterial tyrosyl-tRNA synthetase/tRNA pair for expanding the genetic code of both E. coli and eukaryotes. Cell Chem. Biol. 25, 1304–1312.e5 (2018).
Chin, J. W. et al. An expanded eukaryotic genetic code. Science 301, 964–967 (2003).
Wu, N., Deiters, A., Cropp, T. A., King, D. & Schultz, P. G. A genetically encoded photocaged amino acid. J. Am. Chem. Soc. 126, 14306–14307 (2004).
Italia, J. S. et al. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat. Chem. Biol. 13, 446–450 (2017).
Kim, Y., Cho, S., Kim, J. -C. & Park, H.-S. tRNA engineering strategies for genetic code expansion. Front. Genet. 15, 1373250 (2024).
Park, H.-S. et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–1154 (2011).
Guo, J., Melançon, C. E. III, Lee, H. S., Groff, D. & Schultz, P. G. Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angewandte Chemie Int. Edn 48, 9148–9151 (2009).
Stieglitz, J. T. & Van Deventer, J. A. High-throughput aminoacyl-tRNA synthetase engineering for genetic code expansion in yeast. ACS Synth. Biol. 11, 2284–2299 (2022).
Kim, J.-C., Kim, Y., Cho, S. & Park, H.-S. Noncanonical amino acid incorporation in animals and animal cells. Chem. Rev. 124, 12463–12497 (2024).
Liu, D. R. & Schultz, P. G. Progress toward the evolution of an organism with an expanded genetic code. Proc Natl Acad Sci USA 96, 4780–4785 (1999).
Santoro, S. W., Wang, L., Herberich, B., King, D. S. & Schultz, P. G. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 20, 1044–1048 (2002).
Santoro, S. W. & Schultz, P. G. in Directed Enzyme Evolution: Screening and Selection Methods (eds Arnold, F. H. & Georgiou, G.) 291–312 (Humana, 2003).
Schmidt, M. J. & Summerer, D. Directed evolution of orthogonal pyrrolysyl-tRNA synthetases in Escherichia coli for the genetic encoding of noncanonical amino acids. Methods Mol. Biol. 1728, 97–111 (2018).
Furuhata, Y. et al. Directed evolution of aminoacyl-tRNA synthetases through in vivo hypermutation. Nat. Commun. 16, 4832 (2025).
Dunkelmann, D. L. et al. Adding α,α-disubstituted and β-linked monomers to the genetic code of an organism. Nature 625, 603–610 (2024).
Soni, C. et al. A translation-independent directed evolution strategy to engineer aminoacyl-tRNA synthetases. ACS Cent. Sci. 10, 1211–1220 (2024).
Ficaretta, E. D. et al. Optimized directed evolution of E. coli leucyl-tRNA synthetase adds many noncanonical amino acids into the eukaryotic genetic code including ornithine and N-ϵ-acetyl-methyllysine. Angewandte Chemie Int. Edn 64, e202423172 (2025).
Galles, G. D., Infield, D. T., Mehl, R. A. & Ahern, C. A. Selection and validation of orthogonal tRNA/synthetase pairs for the encoding of unnatural amino acids across kingdoms. Methods Enzymol. 654, 3–18 (2021).
Koch, N. G. & Budisa, N. in Genetically Incorporated Non-Canonical Amino Acids Methods in Molecular Biology, vol 2676 (eds. Tsai, Y. H. & Elsässer, S. J.) https://doi.org/10.1007/978-1-0716-3251-2_1 (2025).
Nikić, I. et al. Debugging eukaryotic genetic code expansion for site-specific click-PAINT super-resolution microscopy. Angew Chem. Int. Ed. Engl. 55, 16172–16176 (2016).
Cho, C.-C., Blankenship, L. R., Ma, X., Xu, S. & Liu, W. The pyrrolysyl-tRNA synthetase activity can be improved by a P188 mutation that stabilizes the full-length enzyme. J. Mol. Biol. 434, 167453 (2022).
Yanagisawa, T., Ishii, R., Fukunaga, R., Nureki, O. & Yokoyama, S. Crystallization and preliminary X-ray crystallographic analysis of the catalytic domain of pyrrolysyl-tRNA synthetase from the methanogenic archaeon Methanosarcina Mazei. Acta Cryst. F 62, 1031–1033 (2006).
Taylor, C. J. et al. Engineering mutually orthogonal PylRS/tRNA pairs for dual encoding of functional histidine analogues. Protein Sci. 32, e4640 (2023).
Kille, S. et al. Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth. Biol. 2, 83–92 (2013).
Stokes, A. L. et al. Enhancing the utility of unnatural amino acid synthetases by manipulating broad substrate specificity. Mol. Biosyst. 5, 1032–1038 (2009).
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Wang, W. et al. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat. Neurosci. 10, 1063–1072 (2007).
Wang, Q. & Wang, L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J. Am. Chem. Soc. 130, 6066–6067 (2008).
Seitchik, J. L. et al. Genetically encoded tetrazine amino acid directs rapid site-specific in vivo bioorthogonal ligation with trans-cyclooctenes. J. Am. Chem. Soc. 134, 2898–2901 (2012).
Tang, L. et al. Rational design for high bioorthogonal fluorogenicity of tetrazine-encoded green fluorescent proteins. Nat. Sci. 2, e20220028 (2022).
Studier, F. W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Tang, L. et al. Construction of ‘small-intelligent’ focused mutagenesis libraries using well-designed combinatorial degenerate primers. BioTechniques 52, 149–158 (2012).
Miyake-Stoner, S. J. et al. Generating permissive site-specific unnatural aminoacyl-tRNA synthetases. Biochemistry 49, 1667–1677 (2010).
Cooley, R. B., Karplus, P. A. & Mehl, R. A. Gleaning unexpected fruits from hard-won synthetases: probing principles of permissivity in non-canonical amino acid-tRNA synthetases. Chembiochem 15, 1810–1819 (2014).
Shilova, O., Kotelnikova, P., Proshkina, G., Shramova, E. & Deyev, S. Barnase–Barstar pair: contemporary application in cancer research and nanotechnology. Molecules 26, 6785 (2021).
Wang, L. & Schultz, P. G. A general approach for the generation of orthogonal tRNAs. Chem. Biol. 8, 883–890 (2001).
Cooley, R. B. et al. Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry 53, 1916–1924 (2014).
Sungwienwong, I. et al. Improving target amino acid selectivity in a permissive aminoacyl tRNA synthetase through counter-selection. Org. Biomol. Chem. 15, 3603–3610 (2017).
Bednar, R. M., Karplus, P. A. & Mehl, R. A. Site-specific dual encoding and labeling of proteins via genetic code expansion. Cell Chem. Biol. 30, 343–361 (2023).
Young, D. D. et al. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 50, 1894–1900 (2011).
Miyake-Stoner, S. J. et al. Probing protein folding using site-specifically encoded unnatural amino acids as FRET donors with tryptophan. Biochemistry 48, 5953–5962 (2009).
Eddins, A. J. et al. Truncation-free genetic code expansion with tetrazine amino acids for quantitative protein ligations. Bioconjugate Chem. 34, 2243–2254 (2023).
Ko, W., Kumar, R., Kim, S. & Lee, H. S. Construction of bacterial cells with an active transport system for unnatural amino acids. ACS Synth. Biol. 8, 1195–1203 (2019).
Luo, X. et al. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat. Chem. Biol. 13, 845–849 (2017).
Fan, C., Ip, K. & Söll, D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 590, 3040–3047 (2016).
Worst, E. G. et al. Cell-free expression with the toxic amino acid canavanine. Bioorg. Med. Chem. Lett. 25, 3658–3660 (2015).
DeLey Cox, V. E., Cole, M. F. & Gaucher, E. A. Incorporation of modified amino acids by engineered elongation factors with expanded substrate capabilities. ACS Synth. Biol. 8, 287–296 (2019).
Hohsaka, T., Kajihara, D., Ashizuka, Y., Murakami, H. & Sisido, M. Efficient incorporation of nonnatural amino acids with large aromatic groups into streptavidin in in vitro protein synthesizing systems. J. Am. Chem. Soc. 121, 34–40 (1999).
Chen, S. et al. Incorporation of phosphorylated tyrosine into proteins: in vitro translation and study of phosphorylated IκB-α and its interaction with NF-κB. J. Am. Chem. Soc. 139, 14098–14108 (2017).
Mira, P., Yeh, P. & Hall, B. G. Estimating microbial population data from optical density. PLoS ONE 17, e0276040 (2022).
Andrews, B. T., Schoenfish, A. R., Roy, M., Waldo, G. & Jennings, P. A. The rough energy landscape of superfolder GFP is linked to the chromophore. J. Mol. Biol. 373, 476–490 (2007).
Hirel, P. H., Schmitter, M. J., Dessen, P., Fayat, G. & Blanquet, S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl Acad. Sci. USA 86, 8247–8251 (1989).
Lundqvist, M. et al. Chromophore pre-maturation for improved speed and sensitivity of split-GFP monitoring of protein secretion. Sci. Rep. 9, 310 (2019).
Banerjee, S. & Mazumdar, S. Electrospray ionization mass spectrometry: a technique to access the information beyond the molecular weight of the analyte. Int. J. Anal. Chem. 2012, 282574 (2012).
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Mukai, T. et al. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci. Rep. 5, 9699 (2015).
Beyer, J. N. et al. Overcoming near-cognate suppression in a release factor 1-deficient host with an improved nitro-tyrosine tRNA synthetase. J. Mol. Biol. 432, 4690–4704 (2020).
Mehl, R. A. et al. Generation of a bacterium with a 21 amino acid genetic code. J. Am. Chem. Soc. 125, 935–939 (2003).
Discovery, C. et al. Chai-1: decoding the molecular interactions of life. Preprint at bioRxiv https://doi.org/10.1101/2024.10.10.615955 (2024).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Krishna, R. et al. Generalized biomolecular modeling and design with RoseTTAFold All-Atom. Science 384, eadl2528 (2024).
Acknowledgements
This work was supported by the GCE4All Biomedical Technology Optimization and Dissemination Center supported by National Institute of General Medical Science (grant no. RM1-GM144227 to R.A.M.). Graphpad Prism 10.1.1 was used to generate the figures to illustrate efficiency/fidelity and UP50 data.
Author information
Authors and Affiliations
Contributions
R.C. and R.M. conceived the protocol. N.A., Y.G., R.C. and R.M. developed the protocol, designed the experiments and analyzed the data. N.A. and Y.G. performed the experiments. N.A., Y.G., R.B. and R.C. generated figures. R.C. contributed data. N.A., Y.G., R.B., P.A.K., R.C. and R.M. wrote, provided input into and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interest.
Peer review
Peer review information
Nature Protocols thanks Abhishek Chatterjee, Nediljko Budisa and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Avila-Crump et al. ACS Chem Biol. 17, 3458–3469 (2022): https://doi.org/10.1021/acschembio.2c00639
Galles et al. Nat Commun. 14, 59 (2023): https://doi.org/10.1038/s41467-022-35761-w
Gottfried-Lee et al. ACS Chem Biol. 17, 3470–3477 (2022): https://doi.org/10.1021/acschembio.2c00640
Supplementary information
Supplementary Data
Sequences for Supplementary Tables 11 and 12.
Source data
Source Data Figs. 5, 9, 13, 14, 15, 16, 17, 18 and 20
Tab:Figure5 raw FACS data, Tab:Figure9 raw fluorescence and cell density, Tab:Figure13 raw FACS data, Tab:Figure14 raw FACS data, Tab:Figure15 nRFU data, Tab:Figure16 raw MS data, Tab:Figure17 raw fluorescence and cell density, Tab:Figure18 raw fluorescence and cell density, Tab:Figure20 nRFU data.
Source Data Fig. 11b
Uncropped gel.
Source Data Fig. 12b
Uncropped gel.
Source Data Figure 16b
Uncropped gel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alexander, N.D., Gangarde, Y.M., Bednar, R.M. et al. Selecting aminoacyl-tRNA synthetase/tRNA pairs for efficient genetic encoding of noncanonical amino acids into proteins. Nat Protoc (2025). https://doi.org/10.1038/s41596-025-01241-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41596-025-01241-w