Abstract
Ongoing ecological research is concerned with analysing climate-induced changes in species distribution. For this purpose, the projection must have high-quality bioclimatic variables from historical and future climatic periods for the projection. To date, there are many global bioclimatic variables on this topic. Nevertheless, a consistent dataset with identical model variables from historic and projected periods is rare. We present 26 bioclimatic variables that are calculated based on a large ensemble consisting of 70 bias-adjusted GCM-RCM simulations for 1971–2098. Both, the historic and the projection periods were calculated using the same models to ensure consistency between the periods. The variables are validated against E-OBS observations from which we calculated the same bioclimatic variables. For projection periods we chose 20 year ranges between 2021–2098. Here, we offer two versions of them: (1) variables separated into RCP 2.6, 4.5 and 8.5, including percentiles among the realisations and within the RCPs; and (2) variables per realisation separately. We then extracted the temporal 5th, 50th and 95th percentile per period as representing values.
Similar content being viewed by others
Background & Summary
Current ecological research deals with the subject of analysing climate-induced changes in species distribution. In recent years the research interest concerning various ecological topics greatly increased1,2,3. Common knowledge is that the distribution of vegetation highly depends on the climate conditions4,5. To quantify this climate induced forcing, direct gradients in the form of bioclimatic variables are widely used as predictors as they have physiological relevance to species6. A vast amount of global bioclimatic variables are already available with various spatial and temporal resolutions, based on different source data. Data from historic periods are often generated using direct observations such as station data in conjunction with indirect observations, for instance remote sensing data (WorldClim7,8) or based on remote sensing data alone (MERRAclim9). Others were derived from the before mentioned WorldClim in combination with CRU10 data (CliMond11,12) or are either based on ERA-Interim13 in combination with station data (CHELSA14) or ERA5 reanalysis data15 (BIOCLIMATE16,17). A very different approach is used to derive past (Holocene), present and future projections (ecoClimate18) through simulation data from CMIP5 (Coupled Modeling Intercomparison Projects)19 and PMIP3 (Paleoclimate Modeling Intercomparison Projects)20. Similarly, simulation data from GCMs (Global Circulation Models) are utilised to derive past conditions for the late Quaternary21 (last 120,000 years).
Nearly all data sources mentioned provide historic data but also future projections (WorldClim, CliMond, BIOCLIMATE, ecoClimate). However, solely ecoClimate uses identical source data for all time periods. Especially WoldClim data7,8 can be considered as standard predictor variables for species distribution models. The data basis for WoldClim’s future and historic periods are different. For the historic period 1970 to 2000 it is gridded station data in conjunction with remote sensing products. For the future projections it is CMIP622 GCMs (General Circulation Models or Global Circulation Models). This can introduce a climate model bias, which can be minimised through bias-adjustment in order to correct the statistics of the projections during a historic time period. Nevertheless, the provided data of the historic time periods differ in methodology from the projection data. This introduces additional bias as both datasets exhibit a different characteristic which will be passed on to following applications. The CMIP6 GCMs carry a spatial resolution of 20 km to 260 km23 depending on the model used. On the other hand, CMIP5 simulations with RCM for Europe in the framework of EURO-CORDEX24,25,26 offer a resolution of ~12.5 km and are to date regional climate projections with the highest resolution in large ensembles.
We present 26 bioclimatic variables with an European extent for the historic 1971 to 2000 and the projection period 2021 to 2098, both based on regionalised CMIP5 simulation data from EURO-CORDEX and ReKlies-DE27 from within the Helmholtz-Initiative Climate Adaptation and Mitigation (HI-CAM) initiative28. The input climate variables are bias-adjusted using E-OBS data (version v20.0e)29 and spatially disaggregated. The extracted historic bioclimatic data are validated against equivalent E-OBS based variables. Our variables allow training and projection on the same simulation data and thus same characteristics, which lessens the bias for further applications such as species distribution modelling.
Methods
Data source
Regional Climate simulations serve as input for the calculation of bioclimatic variables. These climate simulations were bias-corrected and disaggregated within the Helmholtz-Initiative (HI-CAM)28 and consist of regionalised CMIP5 simulations from EURO-CORDEX and ReKlies-DE27. They encompass data based on 11 Regional Climate Model (RCM) downscaling simulations as output from 10 different Global Climate Models (GCM). These simulations consist of 88 realisations that can be assigned to 3 different Representative Concentration Pathways (RCP) 2.6, 4.5 and 8.5 defined by Intergovernmental Panel on Climate Change (IPCC)30. Realisations with GCM CNRM-CM5 and/or RCM HIRHAM5 involved were omitted according to errata31 and (https://www.euro-cordex.net/078730/index.php.en, last access: 20th June 2024) at the time of simulation creation. This leads to omitting 18 realisations and therefore leading to 70 valid realisations, (final realisations listed under Table 1). Thereof 18 realisations are counted as RCP 2.6, 13 realisations as RCP 4.5 and 39 realisations as RCP 8.5.
The simulation data have a spatial resolution of 0.11° (12.5 km) and are bias-adjusted using the trend-preserving ISIMIP3BASD methodology32. An ensemble version (v20.0e)29 of gridded E-OBS data33 with a different gridding method and improved uncertainty estimate served as reference data for the bias-adjustment. During bias-adjustment the realisation data (HI-CAM) are aligned with the statistics of the reference data. The time period considered here is 1971 to 2000. They include daily minimum, maximum and average temperature as well as daily precipitation. The E-OBS data33 were derived based on climate stations across Europe and tested thoroughly for their quality29,34. The underlying climate station data vary in their temporal and spatial coverage across Europe and carry some inhomogeneities. Consequently, the gridded E-OBS data also exhibit inhomogeneities34. E-OBS data users of any version have to be aware of these limitations. The ensemble version v20.0e was remapped from their original resolution 0.25° (25 km) to 0.11° using a conservative remapping method to fit the realisation data (HI-CAM) resolution.
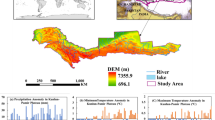
The bias-adjusted realisation data are then spatially disaggregated from 0.11° to 0.03125° (3 km) using External Drift Kriging (v2.0.0)35 with elevation36 as external drift factor. Realisation variables with daily temporal resolution considered for the bioclimatic calculation are as follows: daily average air temperature (tas) [°C]37, maximum air temperature (tasmax) [°C]38, minimum air temperature (tasmin) [°C]39 and precipitation (pr) [mm d−1]40. UFZ mHM-Hydrological model output41,42,43 daily potential evapotranspiration (pet) [mm d−1]44 and weekly soil water content for 6 different soil layers (swc) [mm] serve as additional input. The realisation data and mHM model output are available in the WGS84 reference frame (EPSG: 4326) with a spatial resolution of 0.03125° for the extent of the European continent excluding Turkey, Iceland and Russia (lat: 35° lon: −11°, lat: 71.5° lon: 41°) and temporal range from 1971 to 2098, Fig. 1. We chose to use biogeographical regions of Europe (EBR)45 to facilitate the presentation of bioclimatic variables in the following.
HI-CAM data availability for Europe. Dividing the area in eight biogeographical regions of Europe (EBR)45 facilitates the presentation of bioclimatic variables in selected subsequent figures.
Calculation of bioclimatic variables
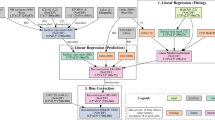
Figure 2 illustrates the process of bioclimatic variable computation using capital letters within the following subsections. A total of 26 bioclimatic variables were selected following these of ANUCLIM46 and WorldClim variables7,8 as well as literature review, see Table 2. The variables tas, tasmax, tasmin, pr, pet and swc serve as input to the calculation. For swc the sum over all soil layers per time step per realisation was calculated beforehand. We utilised anuclim and atmos indicators from Python package xclim47 as well as algorithms of our own for the calculation. We calculated the bioclimatic variables for each realisation with a temporal sampling period of 1 year for the time range 1971 to 2000 serving as historic and 2021 to 2098 serving as projection time period. This accounts for 26 bioclimatic variables for 70 realisations [A]. The following descriptions apply pixel-wise to each bioclimatic variable.
The process of bioclimatic variable calculation for historical 1971 to 2000 and projection period 2021 to 2098 outlined by a flowchart. Capital letters indicate the steps and are referred to in the following descriptions. The total of 70 HI-CAM28 EURO-CORDEX24,25,26 CMIP519 climate realisations serve as input [A]. We calculated the historical period based on the median of all realisations [B] and validated them against E-OBS version (v20.0e)29 [C]. In addition, the realisations can be assigned to three different RCPs (Representative Concentration Pathways) − 2.6, 4.5, 8.5 [D]. Here, we extracted the 5th, 50th and 95th percentile within each RCP [E] to account for the variance within. From all periods (historic and projection) we have then extracted the temporal median [F].
Calculation of ensemble statistics
The time range 1971 to 2000 serves as historic time period due to data availability not before 1971. Over the course of the period all realisations run quite similar [B]. We extracted the temporal 5th, 50th and 95th percentile per realisation to receive three values representing the time period [F]. This serves as input to our data validation [C].
For the projection time period, we selected 4 time periods with a similar range to the WorldClim8 data, which are common predictor variables in species distribution modelling studies. The periods are as follows 2021 to 2040, 2041 to 2060, 2061 to 2080 and 2079 to 2098 due to data availability until 2098 and to keep the length of 20 years consistent. Here we extracted two versions for the bioclimatic variables.
-
(1)
As we can assign each realisation to an RCP [D], Table 1, we extracted the 50th but also 5th and 95th percentile from all realisations belonging to the same RCP [E] to obtain the variance within. The annual 50th percentile development of selected variables per EBR is shown in Figs. 3, 4, 5 and 6. In addition, Figs. 7, 8, 9 and 10 reveal the annual course of the 5th, 50th and 95th percentile for the continental EBR as an example. Subsequently, we extracted the temporal 5th, 50th and 95th percentile for each RCP and period similar to the historic data [F]. This results in 4 periods and 3 RCPs each accompanied by 9 percentiles (within RCP and temporal 5th, 50th, 95th).
Fig. 3 Fig. 4 Fig. 5 Fig. 6 Fig. 7 Fig. 8 Fig. 9 -
(2)
In addition, we have extracted the temporal 5th, 50th and 95th percentile for each period (realisation and projection). This results in 4 periods and 3 percentiles, for all 70 realisations and allows the user to chose only realisations of interest for further analysis. An example of the temporal course of BioVar1 (Mean Temperature) for the realisation (met) 46 is included in Fig. 11.
Fig. 11 Temporal course over all 5 time steps (historic and projections) of BioVar1 - Mean Temperature - for realisation (met) 46, which is defined within RCP 4.5. Subfigure (a) shows the 5th, (b) the 50th and (c) the 95th temporal percentile per time step. The blue line within the legend indicates the areal mean.
All described calculations were performed using Python package xarray48. We re-projected the final data products to ETRS89 Lambert Azimuthal Equal Area (EPSG: 3035) with resampling method nearest neighbour using rioxarray49 and set the spatial resolution to 3 km.
Data Records
The dataset can be obtained from a Zenodo repository50 which is comprised of ten tar.gz files.
Each file inside the tar.gz contains all 26 bioclimatic variables with order and names according to Table 2 column Variable name. The filenames for the realisations are described as follows:
bioVars_1971–2000_met.tar.gz → Projections per realisations for period 1971–2000
bioVars_2021–2040_met.tar.gz → Projections per realisations for period 2021–2040
bioVars_2041–2060_met.tar.gz → Projections per realisations for period 2041–2060
bioVars_2061–2080_met.tar.gz → Projections per realisations for period 2061–2080
bioVars_2079–2098_met.tar.gz → Projections per realisations for period 2079–2098
Regarding the reference and projection periods the filenames inside the tar.gz files are
biovars_met01_1971–2000_5.tif
with 01 as realisation number, 1971–2000 as period and 5 as temporal percentile within the period.
For bioVars separated into RCPs the filenames are described as follows:
bioVars_2021–2040_rcp.tar.gz → Projections per RCP for period 2021–2040
bioVars_2041–2060_rcp.tar.gz → Projections per RCP for period 2041–2060
bioVars_2061–2080_rcp.tar.gz → Projections per RCP for period 2061–2080
bioVars_2079–2098_rcp.tar.gz → Projections per RCP for period 2079–2098
Regarding the RCP for the projection periods the filenames inside the tar.gz files are
biovars_rcp2.6_50perc_2061–2080_5.tif
with 2.6 as RCP, 50 as percentile within the RCP, 2061–2080 as period and 5 as temporal percentile within the period.
In addition, the validation results are part of the Zenodo repository50. The folders eobs and worldclim each contain scatter plots of all possible validation variables (folder plots) as well as statistics (folder stats) for R2 and RMSE.
Technical Validation
We applied the bioclimatic calculation including the temporal 5th, 50th and 95th percentile on the E-OBS data to compare them with our historical bioclimatic variables based on HI-CAM data. Similarly, these E-OBS bioclimatic variables were reprojected to ETRS89 Lambert Azimuthal Equal Area (EPSG: 3035) using nearest neighbour as resampling method and the spatial resolution was set to 3 km. As the variables potential evaporation and soil water content are not available from E-OBS, we omitted bioclimatic variables that rely on these variables, which are Mean Soil Water Content and Water Deficit. We compared our historic variables with E-OBS variables based on the period percentiles resolution (selection of scatter plots, Figs. 12, 13). A quantitative analysis was run on the period percentiles resolution. In this quantitative analysis, we randomly sampled 10 pixel values per entity which is variable, temporal percentile and EBR from E-OBS as well as variable, realisation, temporal percentile and EBR from HI-CAM bioclimatic variables. For each of these entities we calculated the R2 and RMSE, see tables in Zenodo repository50. This process was iterated 20 times and statistics were averaged among them. We see highest R2 values for bioclimatic variables without precipitation involved (e.g. Mean Temperature - BioVar1, Diurnal Temperature Range - BioVar2, Mean Temperature of the Warmest Quarter - BioVar10, Mean Temperature of the Coldest Quarter - BioVar11 and Growing Degree Days above 5 °C - BioVar21 - please see Zenodo repository50 for these plots). Two rather extreme event bioclimatic variables Dry Days - BioVar23 and Max Consecutive Dry Days - BioVar24 - please see Zenodo repository50 for these plots) show lowest agreement across all EBRs. The three smallest EBRs (Arctic, Pannonian and Steppic) show lower agreement with E-OBS variables than other EBRs. The origin of this lower agreement might evolve from lower station density in these areas (Spain, the Balkans and Northern Scandinavia) that were used for generating the E-OBS data34. The lower density in Spain, which is largely part of EBR Mediterranean, affects the agreement less as it does in the low station density EBRs that are much smaller (Pannonian and Steppic in the Balkans as well as Arctic in Northern Scandinavia).
Scatterplots of E-OBS version (v20.0e)29 vs. historical bioclimatic BioVar13 (Index of Aridity) for the time period 1971 to 2000 and each realisation separately. The comparison was iterated 20 times for 10 randomly sampled pixel per EBR45. These plots include 5 randomly sampled pixel per realisation from within the comparison data pool. Subplots indicate the temporal (a) 5th, (b) 50th and (c) 95th percentile.
Scatterplots of E-OBS version (v20.0e)29 vs. historical bioclimatic BioVar20 (Last Spring Frost) for the time period 1971 to 2000 and each realisation separately. The comparison was iterated 20 times for 10 randomly sampled pixel per EBR45. These plots include 5 randomly sampled pixel per realisation from within the comparison data pool. Subplots indicate the temporal (a) 5th, (b) 50th and (c) 95th percentile.
As the bias-correction of the HICAM variables was done using the E-OBS v20.029 data, we are confident, that a change to the newest E-OBS v30.029 data for the validation might not improve the data to a significant amount. E-OBS v20.0 data were used in recent publications and proofs to be a well accepted standard (among other51,52). The newest E-OBS version (v30.0)29 offers a longer time-frame, more variables at higher spatial resolution as well as an enlarged station density in comparison to E-OBS v20.029,53. Merely, the enlarged station density might improve our bioclimatic variables from the process of bias-correction. However, our study was designed and conducted using E-OBS v20.029 for bias-correction, ensuring consistency in data analysis, validation and results interpretation. Changing to E-OBS v30.029 might introduce discrepancies in results due to changes in data handling, resolution or underlying corrections.
To rule out lower qualitative biolimatic variables due to lower station density in E-OBS v20.029 and overoptimistic validation using the same data for bias-correction as well as validation, we enlarged the validation using WorldClim data version 2.18. We utilised the data with 2.5 minutes spatial resolution, re-projected and resampled these data to EPSG:3035 and 3 km to match our bioclimatic variables. As WorldClim8 offers the temporal mean per time period, we used our 50th temporal percentile data for validation. We were able to validate 17 of our 26 bioclimatic variables, that are available from WorldClim8 as well.
Similar to the validation using E-OBS29 data, we randomly sampled 20 pixel values per variable and EBR from WorldClim8 as well as variable, realisation and EBR from HI-CAM bioclimatic variables. The amount was increased to account for slightly shifted pixel locations between both inputs. Again, this process was iterated 20 times and statistics were averaged among them.
The results for BioVar1 (Mean Temperature) and BioVar4 (Temperature Seasonality), see Fig. 14, as well as BioVar12 (Precipitation Sum) and BioVar15 (Precipitation Seasonality), Fig. 15, remain very promising, see statistics in the Zenodo repository50. Some variables from WorldClim version 2.18 show a slightly different methodology of extraction (Temperature Seasonality – BioVar4, Diurnal Temperature Range - BioVar2), which results in higher RMSE values, see plots and statistics in the Zenodo repository50. Partly unfavourable R2 values (BioVar2) e.g. for Boreal EBR might evolve from the fact, that WorldClim8 variables are averaged for the time period 1970–2000 whereas for BioVars the 50th percentile is utilised for the time period 1971–2000. Especially for extreme variables with extreme seasonal climatic differences (e.g. EBR Boreal) this can lead to unfavourable R2 results.
Scatterplots of WorldClim version (2.1)8 vs. historical bioclimatic variables for the time period 1971 to 2000 for each realisation separately. The comparison was iterated 20 times for 20 randomly sampled pixel per EBR45. These plots include 5 randomly sampled pixel per realisation from within the comparison data pool. Subplots indicate the (a) BioVar1 (Mean Temperature) and (b) BioVar4 (Temperature Seasonality). The axes limits vary in result of different calculation of WorldClim and historical Temperature Seasonality.
Scatterplots of WorldClim version (2.1)8 vs. historical bioclimatic variables for the time period 1971 to 2000 for each realisation separately. The comparison was iterated 20 times for 20 randomly sampled pixel per EBR45. These plots include 5 randomly sampled pixel per realisation from within the comparison data pool. Subplots indicate the (a) BioVar12 (Precipitation Sum) and (b) BioVar15 (Precipitation Seasonality). The axes limits vary in result of different calculation of WorldClim and historical Precipitation Seasonality.
Usage Notes
The bioclimatic data are provided here in three parts. (1) The historic time period, which is derived for the time period 1971 to 2000 per realisation. Whereas for the projection period (2) the realisations are then separated into three RCPs and combined within. Here the combining encompasses the 5th, 50th, 95th percentile which allows to account for the variance among the realisations within the RCP. If only specific realisations are to be taken into account (3) we provide all realisations separately. All three parts are supplied as the temporal 5th, 50th, 95th percentile for predefined periods as described in the Method section. We are encouraged to define similar temporal periods (historic as well as projection time periods) as in WorldClim8. Providing the historical and the projection periods derived from the same simulation data enables the usage of these bioclimatic variables with low shifting bias among. Use cases for these variables might be species distribution modelling. The variables from the historic period act as predictor variables during the model training process and the variables from the projection periods (RCP or singular realisations) serve as predictor variables for the model projection. A principal component analysis (PCA) or a correlation analysis of all variables within the historic period reveals a high correlation of multiple variables and should be considered for further use of the data. Despite the utilisation of E-OBS v20.029 (released October 2019) instead of E-OBS v30.029 (released September 2024) for bias-correction and validation, we are confident about the quality of our bioclimatic variables. Nevertheless, areas of more sparse station coverage between both E-OBS versions might introduce some slight uncertainties in our bioclimatic variables in those regions.
Code availability
The code, we generated the bioclimatic variables with, can be accessed under the zenodo repository54
Python packages and versions are listed within the requirements.txt file.
References
Dutra Silva, L., Bento Elias, R. & Silva, L. Modelling invasive alien plant distribution: A literature review of concepts and bibliometric analysis. Environmental Modelling & Software 145, 105203, https://doi.org/10.1016/J.ENVSOFT.2021.105203 (2021).
Elith, J. & Leathwick, J. R. Species Distribution Models: Ecological Explanation and Prediction Across Space and Time. Annual Review of Ecology, Evolution, and Systematics 40, 677–697, https://doi.org/10.1146/ANNUREV.ECOLSYS.110308.120159 (2009).
Zimmermann, N. E., Edwards, T. C., Graham, C. H., Pearman, P. B. & Svenning, J. C. New trends in species distribution modelling. Ecography 33, 985–989, https://doi.org/10.1111/J.1600-0587.2010.06953.X (2010).
von Humboldt, A. & Bonpland, A. Essai Sur La Géographie Des Plantes: Accompagné D’Un Tableau Physique Des Régions Équinoxiales. In Jackson, S. T. & Romanowski, S. (eds.) Essay on the geography of plants, 274 (Levrault, Schoell et Compagnie., 1805).
Woodward, F. I. & Williams, B. G. Climate and plant distribution at global and local scales. Vegetatio 69, 189–197, https://doi.org/10.1007/BF00038700 (1987).
Guisan, A. & Zimmermann, N. E. Predictive habitat distribution models in ecology. Ecological Modelling 135, 147–186, https://doi.org/10.1016/S0304-3800(00)00354-9 (2000).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25, 1965–1978, https://doi.org/10.1002/JOC.1276 (2005).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37, 4302–4315, https://doi.org/10.1002/JOC.5086 (2017).
Vega, G. C., Pertierra, L. R. & Olalla-Tárraga, M. Á. MERRAclim, a high-resolution global dataset of remotely sensed bioclimatic variables for ecological modelling. Scientific Data 4, 1–12, https://doi.org/10.1038/sdata.2017.78 (2017).
New, M., Hulme, M. & Jones, P. Representing Twentieth-Century Space–Time Climate Variability. Part I: Development of a 1961–90 Mean Monthly Terrestrial Climatology. Journal of Climate 12, 829–856, 10.1175/1520-0442(1999)012<0829:RTCSTC>2.0.CO;2 (1999).
Kriticos, D. J. et al. CliMond: global high-resolution historical and future scenario climate surfaces for bioclimatic modelling. Methods in Ecology and Evolution 3, 53–64, https://doi.org/10.1111/J.2041-210X.2011.00134.X (2012).
Kriticos, D. J., Jarošik, V. & Ota, N. Extending the suite of bioclim variables: a proposed registry system and case study using principal components analysis. Methods in Ecology and Evolution 5, 956–960, https://doi.org/10.1111/2041-210X.12244 (2014).
Dee, D. P. et al. The ERA-Interim reanalysis: configuration and performance of the data assimilation system. Quarterly Journal of the Royal Meteorological Society 137, 553–597, https://doi.org/10.1002/qj.828 (2011).
Karger, D. N. et al. Climatologies at high resolution for the earth’s land surface areas. Scientific Data 4, 1–20, https://doi.org/10.1038/sdata.2017.122 (2017).
Hersbach, H. et al. The ERA5 global reanalysis. Quarterly Journal of the Royal Meteorological Society 146, 1999–2049, https://doi.org/10.1002/QJ.3803 (2020).
Wouters, H., Berckmans, J., Maes, R., Vanuytrecht, E. & De Ridder, K. Global bioclimatic indicators from 1950 to 2100 derived from climate projections, https://doi.org/10.24381/cds.a37fecb7 (2021).
Wouters, H., Berckmans, J., Maes, R., Vanuytrecht, E. & De Ridder, K. Downscaled bioclimatic indicators for selected regions from 1950 to 2100 derived from climate projections (2021).
Lima-Ribeiro, M. S. et al. EcoClimate: a database of climate data from multiple models for past, present, and future for macroecologists and biogeographers. Biodiversity Informatics 10, 1–21, https://doi.org/10.17161/BI.V10I0.4955 (2015).
Taylor, K. E., Stouffer, R. J. & Meehl, G. A. An overview of CMIP5 and the experiment design. Bulletin of the American Meteorological Society 93, 485–498, https://doi.org/10.1175/BAMS-D-11-00094.1 (2012).
Braconnot, P. et al. Evaluation of climate models using palaeoclimatic data. Nature Climate Change 2, 417–424, https://doi.org/10.1038/NCLIMATE1456 (2012).
Beyer, R. M., Krapp, M. & Manica, A. High-resolution terrestrial climate, bioclimate and vegetation for the last 120,000 years. Scientific Data 7, 1–9, https://doi.org/10.1038/s41597-020-0552-1 (2020).
Eyring, V. et al. Overview of the Coupled Model Intercomparison Project Phase 6 (CMIP6) experimental design and organization. Geoscientific Model Development 9, 1937–1958, https://doi.org/10.5194/GMD-9-1937-2016 (2016).
IPCC. Climate Change 2021 – The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge University Press, Cambridge, New York, 2023).
Jones, C., Giorgi, F. & Asrar, G. The Coordinated Regional Downscaling Experiment: CORDEX; An international downscaling link to CMIP5. CLIVAR Exchanges 16, 34–40 (2011).
Jacob, D. et al. EURO-CORDEX: New high-resolution climate change projections for European impact research. Regional Environmental Change 14, 563–578, https://doi.org/10.1007/s10113-013-0499-2 (2014).
Vautard, R. et al. Evaluation of the Large EURO-CORDEX Regional Climate Model Ensemble. Journal of Geophysical Research: Atmospheres 126, e2019JD032344, https://doi.org/10.1029/2019JD032344 (2021).
Hübener, H. et al. ReKliEs-De Ergebnisbericht, https://doi.org/10.2312/WDCC/ReKliEsDe_Ergebnisbericht (2017).
Samaniego, L. et al. High-resolution bias-adjusted and disaggregated climate simulation ensemble. Tech. Rep. (2022).
Cornes, R. C., van der Schrier, G., van den Besselaar, E. J. & Jones, P. D. An Ensemble Version of the E-OBS Temperature and Precipitation Data Sets. Journal of Geophysical Research: Atmospheres 123, 9391–9409, https://doi.org/10.1029/2017JD028200 (2018).
IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge University Press, Cambridge, New York, 2013).
Evin, G., Somot, S. & Hingray, B. Balanced estimate and uncertainty assessment of European climate change using the large EURO-CORDEX regional climate model ensemble. Earth System Dynamics 12, 1543–1569, https://doi.org/10.5194/ESD-12-1543-2021 (2021).
Lange, S. Trend-preserving bias adjustment and statistical downscaling with isimip3basd (v1.0). Geoscientific Model Development 12, 3055–3070, https://doi.org/10.5194/gmd-12-3055-2019 (2019).
Haylock, M. R. et al. A European daily high-resolution gridded data set of surface temperature and precipitation for 1950–2006. Journal of Geophysical Research: Atmospheres 113, 20119, https://doi.org/10.1029/2008JD010201 (2008).
Hofstra, N., Haylock, M., New, M. & Jones, P. D. Testing E-OBS European high-resolution gridded data set of daily precipitation and surface temperature. Journal of Geophysical Research: Atmospheres 114, 21101, https://doi.org/10.1029/2009JD011799 (2009).
Samaniego, L., Kumar, R. & Jackisch, C. Predictions in a data-sparse region using a regionalized grid-based hydrologic model driven by remotely sensed data. Hydrology Research 42, 338–355, https://doi.org/10.2166/NH.2011.156 (2011).
U.S. Geological Survey. USGS EROS Archive - Digital Elevation - Global Multi-resolution Terrain Elevation Data 2010 (GMTED2010) (2017).
HICAM tavg, https://tinyurl.com/HICAM-tavg.
HICAM tmax, https://tinyurl.com/HICAM-tmax.
HICAM tmin, https://tinyurl.com/HICAM-tmin.
HICAM pre, https://tinyurl.com/HICAM-pre.
Samaniego, L., Kumar, R. & Attinger, S. Multiscale parameter regionalization of a grid-based hydrologic model at the mesoscale. Water Resources Research 46, 5523, https://doi.org/10.1029/2008WR007327 (2010).
Kumar, R., Samaniego, L. & Attinger, S. Implications of distributed hydrologic model parameterization on water fluxes at multiple scales and locations. Water Resources Research 49, 360–379, https://doi.org/10.1029/2012WR012195 (2013).
Thober, S. et al. The multiscale routing model mRM v1.0: simple river routing at resolutions from 1 to 50 km. Geoscientific Model Development 12, 2501–2521, https://doi.org/10.5194/GMD-12-2501-2019 (2019).
HICAM pet, https://tinyurl.com/HICAM-pet.
European Environment Agency. Biogeographical regions (2016).
ANU Fenner School of Environment & Society. ANUCLIM.
Bourgault, P. et al. xclim: xarray-based climate data analytics, https://doi.org/10.5281/zenodo.10292044 (2023).
Hoyer, S. et al. xarray, https://doi.org/10.5281/zenodo.10023467 (2023).
Snow, A. D. et al. corteva/rioxarray: 0.15.0 Release, https://doi.org/10.5281/zenodo.8247542 (2023).
Reichmuth, A. et al. BioVars - bio-climatic datasets for Europe based on a large regional climate ensemble for periods between 1971 to 2098 (1.1.0). [Data set]. Zenodo., https://doi.org/10.5281/zenodo.14624171 (2025).
Aalbers, E. E., van Meijgaard, E., Lenderink, G., de Vries, H. & van den Hurk, B. J. J. M. The 2018 west-central European drought projected in a warmer climate: how much drier can it get. Natural Hazards and Earth System Sciences 23, 1921–1946, https://doi.org/10.5194/nhess-23-1921-2023 (2023).
Bouaziz, L. J. E. et al. Ecosystem adaptation to climate change: the sensitivity of hydrological predictions to time-dynamic model parameters. Hydrology and Earth System Sciences 26, 1295–1318, https://doi.org/10.5194/hess-26-1295-2022 (2022).
European Climate Assessment & Dataset E-OBS gridded dataset, https://www.ecad.eu/download/ensembles/download.php (2024).
Reichmuth, A. Calculation of bioVars - bio-climatic variables based on HI-CAM CMIP5 climate simulations (2.1). Zenodo., https://doi.org/10.5281/zenodo.14629225 (2025).
Conrad, V. Usual formulas of continentality and their limits of validity. Eos, Transactions American Geophysical Union 27, 663–664, https://doi.org/10.1029/TR027I005P00663 (1946).
Ohlemüller, R., Gritti, E. S., Sykes, M. T. & Thomas, C. D. Towards European climate risk surfaces: the extent and distribution of analogous and non-analogous climates 1931-2100. Global Ecology and Biogeography 15, 395–405, https://doi.org/10.1111/j.1466-822x.2006.00245.x (2006).
Acknowledgements
This work was funded by the Helmholtz knowledge transfer projects fund [grant number WT-0207]. The Helmholtz-Climate-Initiative (HI-CAM) is funded by the Helmholtz Associations Initiative and Networking Fund. The EURO-CORDEX initiative (https://euro-cordex.net) is a voluntary effort of many of the leading and most active institutions in the field of regional climate research in Europe. We acknowledge the EURO-CORDEX community for making their RCM simulation results publicly available. We also acknowledge the ReKliEs-De project (http://reklies.hlnug.de), funded by BMBF, for making their RCM simulation results publicly available. We express our gratitude for valuable cooperation during HI-CAM project with Kevin Sieck (GERICS - Climate Service Center Germany) and Thomas Remke (formerly GERICS). Also we thank the UFZ high performance computing system EVE and their administrators, in particular Ben Langenberg, Toni Harzendorf, Christian Krause and Conrad Ostertag for their support in scientific computation. We also acknowledge the E-OBS dataset from the EU-FP6 project UERRA (https://www.uerra.euhttps://www.uerra.eu) and the Copernicus Climate Change Service, and the data providers in the ECA&D project (https://www.ecad.euhttps://www.ecad.eu).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Anne Reichmuth: Conceptualisation, Methodology, Software, Formal Analysis, Validation, Visualisation, Writing – Original Draft Preparation. Oldrich Rakovec: Software, Methodology, Formal Analysis, Editing. Friedrich Boeing: Software, Methodology, Formal Analysis, Writing - Review & Editing. Sebastian Müller: Software. Luis Samaniego: Supervision, Conceptualisation. Andreas Marx: Methodology, Formal Analysis, Writing - Review & Editing. Hanna Komischke: Validation, Visualisation. Andreas Schmidt: Visualisation. Daniel Doktor: Funding Acquisition, Supervision, Project administration, Writing - Review & Editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Reichmuth, A., Rakovec, O., Boeing, F. et al. BioVars - A bioclimatic dataset for Europe based on a large regional climate ensemble for periods in 1971–2098. Sci Data 12, 217 (2025). https://doi.org/10.1038/s41597-025-04507-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04507-w