Abstract
The relationship between diabetes mellitus (DM) and Alzheimer’s disease (AD) is so strong that scientists called it “brain diabetes”. According to several studies, the critical factor in this relationship is brain insulin resistance. Due to the rapid global spread of both diseases, overcoming this cross-talk has a significant impact on societies. Long non-coding RNAs (lncRNAs), on the other hand, have a substantial impact on complex diseases due to their ability to influence gene expression via a variety of mechanisms. Consequently, the regulation of lncRNA expression in chronic diseases permits the development of innovative therapeutic techniques. However, developing a new drug requires considerable time and money. Recently repurposing existing drugs has gained popularity due to the use of low-risk compounds, which may result in cost and time savings. in this study, we identified drug repurposing candidates capable of controlling the expression of common lncRNAs in the cross-talk between DM and AD. We also utilized drugs that interfered with this cross-talk. To do this, high degree common lncRNAs were extracted from microRNA-lncRNA bipartite network. The drugs that interact with the specified lncRNAs were then collected from multiple data sources. These drugs, referred to as set D, were classified in to positive (D+) and negative (D−) groups based on their effects on the expression of the interacting lncRNAs. A feature selection algorithm was used to select six important features for D. Using a random forest classifier, these features were capable of classifying D+ and D− with an accuracy of 82.5%. Finally, the same six features were extracted for the most recently Food and Drug Administration (FDA) approved drugs in order to identify those with the highest likelihood of belonging to D+ or D−. The most significant FDA-approved positive drugs, chromium nicotinate and tapentadol, were presented as repurposing candidates, while cefepime and dihydro-alpha-ergocryptine were recommended as significant adverse drugs. Moreover, two natural compounds, curcumin and quercetin, were recommended to prevent this cross-talk. According to the previous studies, less attention has been paid to the role of lncRNAs in this cross-talk. Our research not only did identify important lncRNAs, but it also suggested potential repurposed drugs to control them.
Similar content being viewed by others
Introduction
Recent studies1,2 have linked insulin resistance (IR) and impaired insulin signaling to the cross-talk between diabetes mellitus (DM) and Alzheimer’s disease (AD). Many scientists believe that Alzheimer's disease is both a neurological and neuroendocrine disorder. Therefore, Type 3 Diabetes (T3D) is the most appropriate term to describe this complicated disease3. Other cellular and molecular factors involved in the cross-talk between DM and AD are oxidative stress, obesity, inflammation, dysregulation of Apolipoprotein E (APOE), Insulin Degradation Enzyme (IDE), Glucose Transporter type 4 (GLUT4), and Acetylcholinesterase (AChE)4. Nowadays, computational-based methods are very popular because they save time and money, so experts are using computational methods to introduce important genes, proteins, drugs etc.
To overcome this cross-talk, various computational-based studies have been performed so far. Mittal et al. extracted differentially expressed genes from Type 2 diabetes mellitus (T2DM) and Alzheimer’s patients. Then, they constructed a protein–protein interaction network using the proteins' first neighbors. Pathway enrichment analysis was also performed, and notable proteins as well as significant pathways were introduced5.
Using Biological Expression Language, Karki et al.6 proposed six shared pathways between T2DM and AD. They also discussed the dangers of certain anti-diabetic drugs in the progression of AD.
Hu et al.7 employed data from the ROSMAP Project to introduce common pathways between T2DM and AD. They analyzed multi-omic data using an inference method.
In addition to the importance of proteins, recent research suggests that non-coding RNAs (ncRNAs) play an important role in the progression of complex diseases8,9,10,11. The reason is that they are encoded by the majority of the human genome. Furthermore, approximately two-thirds of the experimentally detected microRNAs (miRNAs) are expressed in the brain and are active participants in its functions12. Long non-coding RNAs (lncRNAs) have also been shown to play a biomarker role in the detection of IR, inflammation, and DM, as well as the progression of AD13,14.
Ghiam et al.15 investigated the role of the ncRNAs as potential biomarkers in the cross-talk between DM and AD.
In addition to the biomarker role of ncRNAs in early disease detection, they may also be used as pharmacological targets in the treatment of a variety of disorders16. On the other hand, a new drug takes a long time to complete all phases of drug design (at least 10 years). This procedure is both time-consuming and expensive. To address the aforementioned problems, computer-aided drug design (CADD) has been proposed as a method for predicting new drugs for further study. Methods of drug repositioning (also known as drug repurposing) have been utilized to identify new therapeutic roles for existing drugs. For this purpose, Shakil used molecular docking to investigate the effects of two antidiabetic drugs, Ertugliflozin and Sotagliflozin, and their targets, Sodium glucose cotransporter 2 and AChE, on AD. Finally, he emphasized the sotagliflozin structure for future Type 3 diabetes treatment17.
This study aims to introduce (reuse) Food and Drug Administration (FDA) approved drugs for regulating the expression of common lncRNAs in the cross-talk between DM and AD. Additionally, drugs with side effects were reported.
The following section discusses data collection, machine learning, and network-based methods. Positive drugs (candidates for repurposing) and negative drugs (those with adverse effects) were proposed in the results section, and we will elaborate on them in the discussion section.
Materials and methods
Materials
Collection of data
Insulin is required for normal cognitive functions of the hippocampus. According to scientists, the primary factor in the cross-talk between DM and AD is insulin resistance in the brain18. Consequently, significant mRNAs were extracted from DM, AD, and IR. The set of common mRNAs was selected as follows:
where DMp, ADp and IRp denote the sets of curated proteins associated with DM, AD and IR in the Disgenet database respectively.
The information of miRNAs and lncRNAs were collected from miRTarBase19, LncBase20, and Starbase21 databases. We attempted to select the most reliable mRNA–miRNA, miRNA–lncRNA, and lncRNA–drug interactions with experimental evidence. To do so, we looked for mRNA–miRNA and miRNA–lncRNA interactions in the Starbase database with CLIP-Data ≥ 3 (strict selection), mRNA–miRNA interactions in the miRTarBase database with strong evidence, and miRNA–lncRNA interactions in the LncBase database with high confidence. These interactions are chosen to be functional with the best binding capabilities.
The lncRNA-drug information was gathered from the D-lnc22 and NancoRNA23 databases. Finally, the information of the latest FDA approved drugs with the structural files were downloaded from DrugBank24.
Method
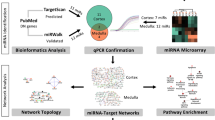
We extracted common mRNAs in DM, AD, and IR from the curated Disgenet database in the first step. After retrieving miRNAs and lncRNAs from experimentally detected databases for the selected mRNAs, high degree miRNAs and lncRNAs were extracted from mRNA–miRNA and miRNA–lncRNA networks, respectively. Following that, for the significant lncRNAs, drug-lncRNA interactions were extracted from various datasets, and drugs were classified into positive and negative groups based on their effects on the regulation of the selected lncRNAs. To build a model for selecting FDA-approved drugs for repurposing, one-dimensional and two-dimensional (1D-2D) features of the intended drugs were extracted, and the feature selection algorithm was used to select features that could best separate positive and negative sets. On the basis of the selected features, the random forest classifier was then constructed, and performance metrics were reported. Finally, using a constructed random forest model, the probability of each FDA-approved drug belonging to positive or negative sets was predicted, and the most relevant drugs to each set were introduced by defining a threshold. Those in the positive group were proposed as potential candidates for repurposing to regulate important lncRNAs, whereas those in the negative group were proposed as adverse drugs in the cross-talk between DM and AD. Figure 1 shows the workflow of the study.
In the following, each step has been described in detail.
Identification of significant miRNAs based on the mRNA-miRNA network
A mRNA–miRNA bipartite network was built in the first state. The nodes in one part of this network represent a set of selected mRNAs, while the nodes in the other part represent miRNAs which have interacted with at least one of the selected mRNAs retrieved from databases. Assume that P = {p1, p2, …, px} and M = {m1, m2, …, ml} represent the set of mRNAs and miRNAs, respectively. If miRNA mj interacts with mRNA pi, there is an edge between pi and mj.
High degree miRNAs were chosen as significant in the constructed network. These miRNAs are denoted by M′ = {m′1, m′2, m′3, …, m′s}.
Identification of significant lncRNAs based on the miRNA-lncRNA network
In this state, the miRNA-lncRNA bipartite network was constructed. Nodes in one part are the set of significant miRNAs (M′) and nodes in the other part are the set of lncRNAs received from databases which have interacted with at least one of the selected miRNAs. Assume that L = {l1, l2, …, lr} is the set of mentioned lncRNAs. If lncRNA lj interacts with miRNA m'i, there is an edge between m'i and lj. High degree lncRNAs were chosen as significant in this network. These lncRNAs are denoted by L′ = {l′1, l′2, l′3, …, l′t}.
Drug collection based on the lncRNA-drug network
At first, the regulation (upregulation or downregulation) effect of members of L′ in both DM and AD were diagnosed. Let D = {d1, d2, d3, …, dz} denoted the set of drugs which interacted with at least one of the elements in L′. Suppose that di interacts with both l′i and l′j which are upregulated and downregulated respectively in both DM and AD. It means that di inhibits both of them therefore, the effect of di on l′i is positive (therapeutic effect) but on l′j is negative (exacerbation effect). In this case we remove di from D. Accordingly, drugs with contradictory effect were omitted. Let D′ = {d′1, d′2, d′3, …, d′n} are denoted the set of remaining drugs. Now we constructed the lncRNA-drug bipartite network which nodes in one part are L′ and the other part are D′. There is an edge between l′i and d′j if drug d′j interacts with lncRNA l′i. Next, based on the therapeutic effect or side effect of D′ on the regulation of members in L′, D′ was divided in to D+ = {d′1, …, d′j} which cure the intended diseases and D− = {d′j+1, …, d′n} which severely increase the negative effects of the intended diseases. For instance, suppose that l′i is upregulated in both DM and AD and d′i inhibits it therefore, d′i will be categorized in D+. On the other hand, suppose that l′j is upregulated too but d′k causes overexpression of this lncRNA (inverse effect) so, by using this drug, the negative effects of both DM and AD will increase. As a result, d′k will be categorized in D−.
The regulation (upregulation or downregulation) effect of L′ members in both DM and AD was initially identified. D = {d1, d2, d3, …, dz} represents the set of drugs that interacted with at least one element in L′.
Suppose that di interacts with l′i and l′j, which are respectively upregulated and downregulated in both DM and AD. It indicates that di inhibits both l′i and l′j therefore, the effect of di on l′i is beneficial (therapeutic effect), while its effect on l′j is detrimental (exacerbation effect). In this instance, we eliminate di from D. Thus, drugs with contradictory effects were excluded. Let D′ = {d′1, d′2, d′3, …, d′n} represents the remaining drugs. Now we have constructed the lncRNA-drug bipartite network, with L′ nodes in one part and D′ nodes in the other. If d′j interacts with lncRNA l′i, there is an edge between l′i and d′j. Next, based on the therapeutic effect or side effect of D′ on the regulation of members in L′, D′ was divided into D+ = {d′1, …, d′j} which cure the intended diseases and D− = {d′j+1, …, d′n} which significantly intensify the negative effects of the intended diseases. For example, if l′i is upregulated in both DM and AD and d′i inhibits it, then d′i will be classified as D+. Alternatively, if l′j is also upregulated but d′k causes overexpression of this lncRNA, then the use of this drug will exacerbate the negative effects of both DM and AD. Consequently, d′k will be classified as D−.
Features of drugs
For drugs in D′, we converted sdf files to mol files for using Open Babel25, a tool for converting chemical file formats. PaDEL26, a software for calculating molecular descriptors and fingerprints, was also employed to extract 1D–2D features based on the structural properties of D′ members. Only those features that best classified D′ into D+ and D- were selected. The algorithm for feature selection is as follows:
-
1.
Eliminating constant features: Features that have the same value for all drugs in D' were omitted because they had no impact on the classification process.
-
2.
Removing redundant features: To improve performance, highly correlated features should be removed. To achieve this, we computed the Pearson correlation coefficient using the "findcorrelation" function from the "caret" package in R, with a cutoff of 0.9. This function takes the absolute values of pair-wise correlations into account. The function eliminates the variable with the highest mean absolute correlation when two variables have a high correlation.
-
3.
Discarding features with low entropy: To find efficient variables for the model, entropy was calculated for each feature using the entropy function in R, and features with entropy less than θ were removed.
-
4.
Removing features with a p-value greater than 0.05: For the remaining attributes, the t-student test was used to select features with significant differences (p value < 0.05) between D+ and D−.
Finally, significant attributes F = {f1, f2, f3, …, fw} were chosen
Model designing
A random forest classifier was constructed using features extracted from the previous state.
Out of bagging error (OOB) was calculated using Python's random forest library to determine the optimal number of trees. OOB is a type of random forest mean prediction error in which trees without the intended sample in their bootstrap are utilized. The model was then evaluated using five-fold cross validation, and performance metrics were reported.
Choosing FDA-approved drugs that are relevant to members of D′
In the same manner as in previous states, PaDEL was used to extract 1D-2D features for FDA-approved drugs, and the same set of extracted features, F, was selected. Let DFDA = {df1, df2, df3, …, dfv} denote drugs with selected features. Then, for DFDA, outlier detection was utilized to select the most pertinent FDA-approved drugs for D′.
The procedure for detecting outliers is as follows:
-
1.
Number of atoms: very small drugs with nATOM ≤ µ was removed from the study since they have no impact on the classification process.
-
2.
Applicability of domain: the warning leverage for each compound was calculated using the formula: hi = xiT (XT X)−1 xi
where:
-
3.
xi is the query molecule's description-row vector
-
4.
X is the k*n matrix (k descriptor values for each one of the n training molecules)
-
5.
h* (warning leverage) is usually set to 3p/n, where p is the number of model variables (|F|) plus one and n is the number of training compounds
Outliers were identified as drugs with a hi value greater than the warning leverage and were removed from the dataset.
-
6.
One class classification: although relevant drugs were selected from previous steps by removing outliers, to further optimize the running time of the classification algorithm, it is necessary to select highly relevant drugs to D′. To achieve this, for the remaining drugs in the DFDA, a single class classification method employing Support Vector Machine (SVM) classifier with radial basis27 was utilized, and other outliers were excluded.
Finally, the most relevant compounds to D' were chosen from all FDA-approved drugs. These drugs are denoted by D′FDA = {df′1, df′2, df′3, …, df′q}. Now, this set was fed into the newly constructed random forest model, and for each drug the probability of belonging to D+ or D− was calculated. The drugs with probabilities of 0.95 or higher were selected. Those with a positive effect (belongs to D+) were suggested as repurposing candidates and those with a negative effect (belongs to D−) were introduced as warning drugs that could simultaneously increase the risk of DM and AD.
Results
Network analysis
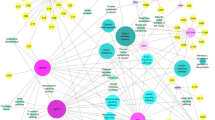
Based on the data collection section above, 9 mRNAs shared by DM, AD, and IR were extracted from the curated Disgenet database (Table 1, first column). There are 412 miRNAs which have interacted with at least one of these mRNAs. Then we constructed the mRNA-miRNA bipartite network and significant miRNAs were extracted. In this network, no miRNA interacts with the INS and LEP proteins, so these two proteins were omitted from our investigation. Then, miRNAs with the highest degree (degree 4) were chosen as significant. By this strategy, 7 miRNAs were identified (Table 1, second column). Similarly, we constructed the miRNA-lncRNA bipartite network. This network contains 1700 lncRNAs, only three of them have the highest degree of 7, and eight of them have degree 6. These 11 lncRNAs were deemed significant (Table 1, forth column). These degrees are chosen so that approximately 10 miRNAs and 10 lncRNAs are considered as significant. For instance, if we considered degree 3 for the first network, we would select more than 40 miRNAs, and if we considered degree 5 for lncRNAs, we would select more than 30 lncRNAs. Supplementary Tables S1 and S2 list all mRNA-miRNA and miRNA-lncRNA interactions in these two networks. However, the final tripartite network was built using 7 selected mRNAs, 7 selected miRNAs, and 11 selected lncRNAs. Results have been shown in Fig. 2 and Table 1.”
Final mRNA-miRNA-lncRNA network. The first layer (purple nodes) consists of 7 shared mRNAs between DM and AD that were extracted from curated Disgenet database. The second layer (blue nodes) consists of 7 high degree miRNAs obtained from mRNA-miRNA bipartite network and the last layer (yellow nodes) consists of 11 high degree lncRNAs achieved from miRNA-lncRNA bipartite network.
Significant mRNAs, miRNAs, and lncRNAs are summarized in Table 1. No interacting miRNAs have been reported in the existing databases for INS and LEP among the 9 mRNAs. The hub miRNAs and lncRNAs are shown in columns 2 and 4, and the associated degrees of the intended miRNAs and lncRNAs are shown in columns 3 and 5.
To validate the role of obtained mRNAs in the cross-talk between DM and AD, their dysregulation’s type was investigated. The literature review indicates that HMOX1, NOS3, PPARG, and TNF are upregulated in both diseases, while INSR, SLC2A4, and SOD2 are downregulated. The formation of neurofibrillary tangles is caused by the upregulation of HMOX1, which increases tau hyperphosphorylation and results in neurofibrillary tangle formation28. Overexpression of PPARG causes amyloid beta (Aβ) neurotoxicity by targeting glycogen synthase kinase-3beta (GSK3β) in the Wnt/Beta-Catenin pathway. The upregulation of NOS3 and TNF may increase apoptosis29 and inflammation30 in the memory region of the brains of patients with DM and AD, respectively. Conversely, downregulation of INSR reduces insulin's sensitivity to its receptors, resulting in insulin resistance in the brain31. SLC2A4 (GLUT4) is the brain's glucose transporter. Any dysregulation of the expression of this important protein may impair neuronal cells' ability to absorb glucose32. SOD2 downregulation also affects mitochondrial activity in the neuronal cells33. Summary of the results are presented in Table 2.
In this table, 7 mRNAs in the cross-talk between DM and AD (achieved from curated Disgenet database) and their mechanisms in the relationship between the two intended diseases are introduced using literature review.
For the 7 significant miRNAs identified by the miRNA-lncRNA bipartite network, we also conduct a literature review to confirm their role in the cross-talk between DM and AD.
In both diseases, hsa-miR-106b-5p, hsa-miR-17-5p, hsa-miR-20a-5p, hsa-miR-93-5p, and hsa-miR-200c-3p have been downregulated, whereas hsa-miR-20b-5p has been upregulated. Downregulation of hsa-miR-106b-5p, hsa-miR-17-5p, has-miR-20a-5p and hsa-miR-93-5p as well as overexpression of hsa-miR-20a-5p may alter the expression of amyloid precursor protein (APP) and subsequently influence Aβ, which is responsible for hyperphosphorylation of tau protein. In addition, altering the expression of hsa-miR-200c-3p may increase the expression of GSK3β, which results in hyperphosphorylation of tau34. Moreover, hsa-miR-3184-5p is a novel miRNA in the cross-talk between DM and AD, which is proposed in this study for future experimental investigation. Table 3 shows the summary of these results.
This table describes the mechanisms of 7 significant miRNAs in the cross-talk between DM and AD. Only miR-3184-5p introduced in this study as a novel miRNA in this cross-talk which needs additional experimental research.
Similarly, we investigated the role of obtained lncRNAs in DM and AD cross-talk through literature review. Recent studies suggest that NEAT1 and XIST, two important biomarkers in DM and AD, are upregulated in both diseases, which not only activate the Akt/mTOR signaling pathway but also dysregulate the expression of miR-124, resulting in an increase in BACE1 concentration and Aβ plaque deposition in the brain35,36,37. H19, another significant lncRNA, is upregulated in DM and AD, resulting in hippocampal neuron apoptosis38. Meanwhile, low levels of MALAT1 (downregulation) have been found in Alzheimer’s patients compared to controls, reducing the lncRNA’s anti-apoptotic effect39. Abdulle et al. also demonstrated the significance of MALAT1 as a target in diabetic-related diseases40. Additionally, HCG18 and BPLN3P are upregulated, which increases the risk of insulin resistance and type 1 diabetes, respectively41,42. The remaining obtained lncRNAs are novel in this cross-talk which are proposed in this study for future experimental analysis. Table 4 displays the summary of these results.
This table explains the role of the obtained lncRNAs in the cross-talk between DM and AD.
Among 11 significant lncRNAs, 5 of them are novel, proposed in this study for future experimental analysis.
Additionally, 40 drugs were extracted from lncRNA-drug bipartite network where 24 drugs classified as D− and 16 drugs as D+. These drugs interact with NEAT1, XIST, H19 and MALAT1. For example, Panobinostat increases NEAT1 and H19 expression in SK-NEP-1 and G401 cells43 According to Table 4, the upregulation of the targeted lncRNAs may cause cross-talk between DM and AD. Consequently, the use of this drug may increase the risk of both DM and AD. As a result, it is classified as D−. In contrast, Adriamycin inhibits NEAT1 expression in stomach cancer. In addition, it can increase MALAT1 expression in non-small cell lung cancer. It may therefore have a therapeutic effect on the cross-talk between DM and AD and is classified as D+.
The remaining 7 lncRNAs did not interact with any drugs in the existing lncRNA-drug databases. Table 5 displays the results of the negative or positive effect of all the obtained drugs on the expression of the 4 selected lncRNAs.
The drugs in the first column (D−) and the third column (D+) have a negative and positive effect on the common significant lncRNAs in DM and AD respectively. Furthermore, the interaction of lncRNAs with drugs is reported in columns 2 and 4, respectively.
Feature description
PaDEL was utilized to extract 1444 features from the 40 drugs that were chosen from the lncRNA-drug bipartite network. There were 293 omitted constant features. Then, 745 highly correlated features with cutoff = 0.9 were also removed (Fig. 3A). Low entropy features (θ < 1.5) that had no effect on classifying D+ and D− were removed as well (Fig. 3B).
The feature selection process. (A) The correlation matrix of 1444 features achieved from PaDEL has been displayed. The values range from − 1 to 1, with − 1 denoting a negative correlation and 1 denoting a positive correlation. Features that were highly correlated (cutoff = 0.9) were removed. (B) Features with low entropy (below the red dashed line) were omitted.
Consequently, 293 features remained. The t-student test was then performed to identify the most significant features, and 6 of them with a p value less than 0.05 that best classified D+ and D− were selected. The final drug-feature matrix, which includes 40 drugs and 6 significant features, is presented in Table S3 of the Supplementary materials.
Random forest classifier
A random forest classifier based on the drug-feature matrix was used to construct a model for further analysis. The optimal number of trees was determined to be approximately 100 based on the tradeoff between OOB error and accuracy. Figure 4 depicts the findings.
Furthermore, fivefold cross validation (cv) was used to evaluate the constructed random forest classifier, and several performance measures were reported. During the evaluation 80% of the data was selected for the training dataset and 20% for the test dataset. As shown in Tables 6 and 7, the accuracy of each fold in a fivefold cv ranges between 0.75 and 0.875%, and the overall accuracy is 0.825. In addition, the precision and recall values for D+ and D− range from 0.75 to 1. The F1 measures for negative and positive sets are 0.89 and 0.86, respectively. Results have been shown in Tables 6 and 7.
Selecting relevant FDA approved drugs
The structural files of 2474 FDA-approved drugs (DFDA) were downloaded from DrugBank, and 1D-2D features were extracted from PaDEL. Drugs with a nATOM (number of atoms) less than 7 were eliminated (Fig. 5A). The 6 significant features obtained in the previous step were chosen for the remaining 2317 drugs. However, for 13 drugs, the value of some selected features was unavailable, so they were eliminated. The total number of drugs was thus reduced to 2304. To detect outliers, first, the applicability of the domain was determined, and then the warning leverage was computed as follows:
This figure depicts the DFDA's outlier detection process. (A) Those with a small number of atoms (nATOM = 7) were initially removed from the dataset (drugs below the red dashed line). (B) The warning leverage was then calculated using the applicability of domain, and outlier drugs were excluded (drugs above the red dashed line).
Each drug with a warning leverage equal to or exceeding the aforementioned threshold was omitted (Fig. 5B). Consequently, 94 drugs were eliminated.
The remaining 2210 drugs were classified using a one-class classification SVM method, and 1221 drugs (D′FDA) were selected as the most relevant to at least one of the 40 drugs.
Then, D′FDA was given as input into the constructed random forest model, and the probability of each drug belonging to the D+ or D− was calculated. The drugs with a probability of 0.95 or higher were deemed the most relevant. Due to their positive effect on the regulation of lncRNAs associated with DM and AD, certain D+ drugs have been proposed as repositioning candidates. Those in D− have been classified as adverse drugs as they have a negative effect on the regulation of corresponding lncRNAs. Table 8 lists all selected FDA-approved drugs discovered in this study.
The final FDA-approved negative and positive drugs are listed in this table. Positive drugs are suggested for repurposing. Negative drugs, on the other hand, have been introduced as warning drugs that may disrupt the expression of common lncRNAs and significantly increase the negative effects of both diseases simultaneously.
Discussion
In this work, using network and machine learning approaches, candidate drugs for repurposing were introduced based on common mRNAs, miRNAs, and lncRNAs associated with the cross-talk between DM and AD. Drugs that may exacerbate the negative effects of both diseases were also discussed. The first 11 significant lncRNAs, as listed in Table 1, are taken into account. Only four of these lncRNAs have been associated with drug interactions in lncRNA-drug databases: NEAT1, XIST, H19, and MALAT1. The interactions with the remaining seven lncRNAs have not yet been identified. Therefore, this study focuses on the four lncRNAs listed above in order to identify suitable drugs for the repositioning objective.
Scientists believe that inhibiting NEAT1 and XIST reduces Aβ plaques in patients' brains by regulating the expression of interacted ncRNAs and proteins in specific pathways. Therefore, silencing of the aforementioned lncRNAs may represent a novel potential therapeutic target in the treatment of both diseases44,45. Furthermore, researchers show that inhibiting H19 prevents apoptosis in hippocampal neurons in both diseases46. Additionally, experiments indicate that overexpression of MALAT1 increases the lncRNA's neuroprotective function in the brains of patients and decreases neuronal injury in the hippocampus47.
The discovery of drugs that can silence NEAT1, XIST, or H19 while also inducing MALAT1 overexpression could be considered a new treatment for DM and AD cross-talk.
Conversely, drugs that have a negative effect on the targeted lncRNAs must be avoided. According to Table 8, there are two categories of FDA-approved drugs. Positive and negative drugs are listed, respectively, in the first and third columns. Positive drugs have been recommended for diabetic and Alzheimer's patients. Estradiol, for instance, is effective in preventing Aβ plaques and tau hyperphosphorylation in the brains of Alzheimer's patients, in addition to reducing the risk of diabetes48,49. It has been demonstrated that drugs in the chromium family have a curable effect on DM, AD, and IR, as they not only improve cognitive function in older adults but also regulate glucose excretion in patients with DM and IR50. Consequently, it may be one of the most significant candidate drugs in this cross-talk. Tapentadol, a drug used to treat pain in the elderly, is another positive drug. According to researchers51 it may improve the behavior of elderly dementia patients. Additionally, it has been proposed as a novel therapy for diabetic neuropathy52. Niacin is a B vitamin that facilitates the transformation of food into energy. According to research on the effect of niacin on AD, scientists believe that including niacin in one's diet can aid in disease prevention.53. Volunteers who received less niacin were up to 70 percent more likely to develop the disease than those who received more. However, it has the potential to increase glucose levels in diabetic patients, so dosage must be closely monitored54. Sibutramine is used for weight loss. Additionally, it reduces LDL cholesterol, thereby reducing the cardiovascular risk in diabetic patients55. Moreover, since high cholesterol levels may increase the risk of AD56, it may be a new drug capable of curing Alzheimer’s patients, particularly by controlling APOE.
Curcumin, a bright yellow chemical produced by curcuma, and quercetin, a plant pigment found in grapefruit, onion, and other fruits and vegetables, are two positive natural compounds. According to numerous studies, curcumin plays an important role in reducing Aβ plaques and inhibiting tau aggregation, as well as controlling blood sugar and managing diabetes complications. In addition, quercetin reduces oxidative stress by regulating signaling pathways, as well as lowering hyperglycemia and improving IR. In AD, it also has neuroprotective properties.
On the other hand, negative drugs have not been suggested for patients with DM and AD.
Diamorphine, for instance, increases tau hyperphosphorylation in AD patients by targeting the FYN protein57. The FDA has not approved zaleplon, a sleeping pill, for Alzheimer's patients because this class drugs have been linked to an increased risk of dementia in the elderly58. The anti-inflammatory drug, sulfasalazine, is used to treat Crohn’s disease and rheumatoid arthritis. Despite being an anti-inflammatory, chou et al. discovered that it may increase the risk of dementia in patients with rheumatoid arthritis. Hypoglycemia has been added to the list of side effects of sulfasalazine in T2DM patients59,60. Geodon is the brand name for the antipsychotic drug ziprasidone, which is used to treat schizophrenia and bipolar disorder. Experimental studies indicate that it increases the risk of death in Alzheimer's patients, so it is neither recommended nor approved for dementia patients61. Furthermore, it increases the risk of hyperglycemia and diabetes. Cefepime, an antibiotic used to treat bacterial infections, has been linked to disruption of brain function; therefore, Alzheimer's patients should only take it on a doctor's advice.
In DM or AD, drugs such as cocarboxylase, glipizide, glimepiride, and the ergotamine family are being investigated. They are, however, classified as negative drugs in our study because they have an adverse effect on the regulation of common lncRNAs, and further experimental research into their impact on DM and AD is recommended.
There is insufficient information regarding the associations between the remaining drugs and the intended diseases; therefore, more research is required.
In addition to the obtained drugs, we investigated the impact of well-known drugs on the expression of the significant mRNAs, miRNAs, and lncRNAs that are currently used for the treatment of DM and AD from DrugBank database. There are 52 and 11 DM and AD drugs, respectively. In our results, the anti-diabetic drug Rosiglitazone is categorized as a negative drug. Based on our findings, the use of this drug may increase the risk of AD by elevating NEAT1 expression (see Tables 4, 5). Therefore, this drug may be an important candidate for future experimental research on the role of anti-diabetic drugs in the progression of AD.
However, there is no other overlap between drugs that we introduced in this study and the remaining 62 DM or AD drugs in the DrugBank database.
In future work, we will increase the reliability of our study by evaluating the expression of introduced lncRNAs using human samples. Then, we intend to conduct a laboratory experiment at the Royan Institute for Stem Cells to test introduced candidate drugs on the obtained lncRNAs using mouse samples. On the other hand, we proposed that dysregulation of certain mRNAs and miRNAs causes cross-talk between DM and AD. Since the focus of this paper was on lncRNA-drug repurposing, one of the future studies could concentrate on drug repurposing based on introduced mRNAs and miRNAs.
This study can be expanded to include any other diseases with a correlation, such as DM and fatty liver or AD and Parkinson's disease. The pipeline remains unchanged; only the extracted data changes. Consequently, this work can serve as a guideline for similar work on two other diseases that interact with each other.
Conclusion
Drug repurposing is one of the most important methods for developing new drugs from existing ones for the treatment of diseases. This paper proposed a network and machine learning-based method that not only suggests drugs to inhibit DM and AD cross-talk, but also introduces drugs that exacerbate the negative effects of the aforementioned diseases.
For instance, the drugs chromium nicotinate and tapentadol, which are used to treat IR and pain, respectively, may regulate the expression of NEAT1, XIST, MALAT1, and H19 in the brains of diabetic and Alzheimer's patients. Consequently, these two drugs can be considered a novel treatment for this cross-talk. In addition, drugs such as ziprasidone and sulfasalazine, which are used to treat crohn's disease and schizophrenia, may increase the risk of both DM and AD. The list of repurposing candidate drugs and adverse drugs for which we wish to conduct experimental research to confirm our findings is presented in Table 8. Two of the drugs in this table, curcumin and quercetin, are natural compounds that can regulate the expression of H19, XIST, and MALAT1. Therefore, long-term use of these natural products may reduce the risk of DM and AD.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Abbreviations
- IR:
-
Insulin resistance
- DM:
-
Diabetes mellitus
- AD:
-
Alzheimer’s disease
- T3D:
-
Type 3 diabetes
- APOE:
-
Apolipoprotein E
- IDE:
-
Insulin degradation enzyme
- GLUT4:
-
Glucose transporter type 4
- AChE:
-
Acetylcholinesterase
- T2DM:
-
Type 2 diabetes mellitus
- NcRNA:
-
Non-coding RNA
- MiRNA:
-
MicroRNA
- LncRNA:
-
Long non-coding RNAs
- CADD:
-
Computer aided drug design
- FDA:
-
Food and Drug Administration
- OOB:
-
Out of bagging error
- SVM:
-
Support vector machine
- CV:
-
Cross validation
- Aβ:
-
Amyloid beta
References
Baglietto-Vargas, D., Shi, J., Yaeger, D. M., Ager, R. & LaFerla, F. M. Diabetes and Alzheimer’s disease crosstalk. Neurosci. Biobehav. Rev. 64, 272–287 (2016).
McNay, E. C. & Pearson-Leary, J. GluT4: A central player in hippocampal memory and brain insulin resistance. Exp. Neuro.l 323, 113076 (2020).
Kandimalla, R., Thirumala, V. & Reddy, P. H. Is Alzheimer’s disease a Type 3 diabetes? A critical appraisal. Biochim. Biophys. Acta 1863(5), 1078–1089 (2017).
Mittal, K. & Katare, D. P. Shared links between type 2 diabetes mellitus and Alzheimer’s disease: A review. Diabetes Metab. Syndr. 10(2 Suppl 1), S144-149 (2016).
Mittal, K., Mani, R. J. & Katare, D. P. Type 3 diabetes: Cross talk between differentially regulated proteins of type 2 diabetes mellitus and Alzheimer’s disease. Sci. Rep. 6(1), 25589 (2016).
Karki, R., Kodamullil, A. T. & Hofmann-Apitius, M. Comorbidity analysis between Alzheimer’s disease and type 2 diabetes mellitus (T2DM) based on shared pathways and the role of T2DM drugs. J. Alzheimer’s Dis. JAD 60(2), 721–731 (2017).
Hu, Z. et al. Shared causal paths underlying Alzheimer’s dementia and type 2 diabetes. Sci. Rep. 10(1), 4107 (2020).
DiStefano, J. K. The emerging role of long noncoding RNAs in human disease. Methods Mol. Biol. (Clifton, NJ) 1706, 91–110 (2018).
Lekka, E. & Hall, J. Noncoding RNAs in disease. FEBS Lett 592(17), 2884–2900 (2018).
Li, Y. & Kowdley, K. V. MicroRNAs in common human diseases. Genomics Proteomics Bioinform. 10(5), 246–253 (2012).
Natarajan, S. K., Smith, M. A., Wehrkamp, C. J., Mohr, A. M. & Mott, J. L. MicroRNA function in human diseases. Med. Epigenet. 1(1), 106–115 (2013).
Idda, M. L., Munk, R., Abdelmohsen, K. & Gorospe, M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA 9(2), e1463 (2018).
He, X. et al. LncRNAs: Key players and novel insights into diabetes mellitus. Oncotarget 8(41), 71325–71341 (2017).
Zhang, Y. et al. The role of non-coding RNAs in Alzheimer’s disease: from regulated mechanism to therapeutic targets and diagnostic biomarkers. Front. Aging Neurosci. 13, 654978 (2021).
Ghiam, S., Eslahchi, C., Shahpasand, K., Habibi-Rezaei, M. & Gharaghani, S. Exploring the role of non-coding RNAs as potential candidate biomarkers in the cross-talk between diabetes mellitus and Alzheimer’s disease. Front. Aging Neurosci. 14, 955461 (2022).
Matsui, M. & Corey, D. R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16(3), 167–179 (2017).
Shakil, S. Molecular interaction of anti-diabetic drugs with acetylcholinesterase and sodium glucose co-transporter 2. J. Cell. Biochem. 118(11), 3855–3865 (2017).
McNay, E. C. & Recknagel, A. K. Brain insulin signaling: A key component of cognitive processes and a potential basis for cognitive impairment in type 2 diabetes. Neurobiol. Learn. Mem. 96(3), 432–442 (2011).
Huang, H.-Y. et al. MiRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 48(D1), D148–D154 (2019).
Karagkouni, D. et al. DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 48(D1), D101–D110 (2019).
Yang, J. H. et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 39(Database issue), D202-209 (2011).
Jiang, W. et al. D-lnc: A comprehensive database and analytical platform to dissect the modification of drugs on lncRNA expression. RNA Biol. 16(11), 1586–1591 (2019).
Li, L. et al. NoncoRNA: A database of experimentally supported non-coding RNAs and drug targets in cancer. J. Hematol. Oncol. 13(1), 15 (2020).
DrugBank 5.0: a major update to the DrugBank database for 2018. https://doi.org/10.1093/nar/gkx1037.
Openbabel. http://openbabel.org/.
Yap, C. W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32, 1466–1474 (2011).
Bounsiar, A. & Madden, M. G. Kernels for One-class support vector machines. In: 2014 International Conference on Information Science & Applications (ICISA): 6–9 May 2014. 1–4. (2014).
Schipper, H. M. Heme oxygenase-1 in Alzheimer disease: A tribute to Moussa Youdim. J. Neural Transm. 118(3), 381–387 (2011).
Vallée, A. & Lecarpentier, Y. Alzheimer disease: Crosstalk between the canonical Wnt/Beta-catenin pathway and PPARs alpha and gamma. Front. Neurosci. 10, 459 (2016).
Park, K. S. et al. PPAR-gene expression is elevated in skeletal muscle of obese and type II diabetic subjects. Diabetes 46, 1230–1234 (1997).
Gabbouj, S. et al. Altered insulin signaling in Alzheimer’s disease brain—special emphasis on PI3K-Akt pathway. Front. Neurosci. 13, 629 (2019).
Kyrtata, N., Emsley, H., Sparasci, O., Parkes, L. & Dickie, B. A systematic review of glucose transport alterations in Alzheimer’s disease. Front. Neurosci. 15, 626636 (2021).
Massaad, C., Washington, T., Pautler, R. & Klann, E. Overexpression of SOD-2 reduces hippocampal superoxide and prevents memory deficits in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 106, 13576–13581 (2009).
Liu, S. et al. MicroRNAs in Alzheimer’s disease: Potential diagnostic markers and therapeutic targets. Biomed. Pharmacother. 148, 112681 (2022).
Chandrakumar, S., Prabu, P., Mohan, V. & Balasubramanyam, M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genomics 12, 1–9 (2018).
Huang, Z., Zhao, J., Wang, W., Zhou, J. & Zhang, J. Depletion of LncRNA NEAT1 rescues mitochondrial dysfunction through NEDD4L-dependent PINK1 degradation in animal models of Alzheimer’s disease. Front. Cell. Neurosci. 14, 28 (2020).
Yue, D. et al. Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 44, 630–636 (2019).
Yu, J. L., Li, C., Che, L. H., Zhao, Y. H. & Guo, Y. B. Downregulation of long noncoding RNA H19 rescues hippocampal neurons from apoptosis and oxidative stress by inhibiting IGF2 methylation in mice with streptozotocin-induced diabetes mellitus. J. Cell. Physiol. 234(7), 10655–10670 (2019).
Ma, P. et al. Long non-coding RNA MALAT1 inhibits neuron apoptosis and neuroinflammation while stimulates neurite outgrowth and its correlation with MiR-125b mediates PTGS2, CDK5 and FOXQ1 in Alzheimer’s disease. Curr. Alzheimer Res. 16(7), 596–612 (2019).
Abdulle, L. E. et al. MALAT1 as a diagnostic and therapeutic target in diabetes-related complications: A promising long-noncoding RNA. Int. J. Med. Sci. 16(4), 548–555 (2019).
Geng, G., Zhang, Z. & Cheng, L. Identification of a multi-long noncoding RNA signature for the diagnosis of type 1 diabetes mellitus. Front. Bioeng. Biotechnol. 8, 553 (2020).
Xia, Y., Zhang, Y. & Wang, H. Upregulated lncRNA HCG18 in patients with non-alcoholic fatty liver disease and its regulatory effect on insulin resistance. Diabetes Metab. Syndr. Obes. 14, 4747–4756 (2021).
Yan-Fang, T. et al. Molecular mechanism of the cell death induced by the histone deacetylase pan inhibitor LBH589 (panobinostat) in wilms tumor cells. PLoS ONE 10(7), e0126566 (2015).
Huang, S. et al. Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J. Cell. Physiol. 234(7), 11200–11207 (2019).
Yue, D. et al. Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 44(2), 630–636 (2020).
Zhang, Y. Y. et al. Silenced lncRNA H19 and up-regulated microRNA-129 accelerates viability and restrains apoptosis of PC12 cells induced by Aβ(25–35) in a cellular model of Alzheimer’s disease. Cell Cycle (Georgetown, Tex) 20(1), 112–125 (2021).
Li, L., Xu, Y., Zhao, M. & Gao, Z. Neuro-protective roles of long non-coding RNA MALAT1 in Alzheimer’s disease with the involvement of the microRNA-30b/CNR1 network and the following PI3K/AKT activation. Exp. Mol. Pathol. 117, 104545 (2020).
Mauvais-Jarvis, F. Is estradiol a biomarker of type 2 diabetes risk in postmenopausal women?. Diabetes 66(3), 568–570 (2017).
Wharton, W. et al. Short-term hormone therapy with transdermal estradiol improves cognition for postmenopausal women with Alzheimer’s disease: Results of a randomized controlled trial. J. Alzheimer’s Dis. JAD 26(3), 495–505 (2011).
Krikorian, R., Eliassen, J. C., Boespflug, E. L., Nash, T. A. & Shidler, M. D. Improved cognitive-cerebral function in older adults with chromium supplementation. Nutr. Neurosci. 13(3), 116–122 (2010).
Nunziata, E., De Siati, F. & Palleschi, L. Effectiveness and tolerability of tapentadol in very elderly patients with assessment of cognitive-behavioral aspects. Geriatr. Care 3(2), 973–982 (2017).
Bueno, A. G. M. J. M. & German, C. I. Tapentadol, a new horizon in the treatment of diabetic peripheral painful neuropathy. Acta Med. 16(1), 41–46 (2018).
Morris, M. C. et al. Dietary niacin and the risk of incident Alzheimer’s disease and of cognitive decline. J. Neurol. Neurosurg. Psychiatry 75(8), 1093–1099 (2004).
Chen, L. et al. Niacin-induced hyperglycemia is partially mediated via niacin receptor GPR109a in pancreatic islets. Mol. Cell. Endocrinol. 404, 56–66 (2015).
Weeke, P. et al. The weight lowering effect of sibutramine and its impact on serum lipids in cardiovascular high risk patients with and without type 2 diabetes mellitus—an analysis from the SCOUT lead-in period. BMC Endocr. Disord. 10(1), 3 (2010).
Sáiz-Vazquez, O., Puente-Martínez, A., Ubillos-Landa, S., Pacheco-Bonrostro, J. & Santabárbara, J. Cholesterol and Alzheimer’s disease risk: A meta-meta-analysis. Brain Sci. 10(6), 386 (2020).
Egervari, G. et al. Chromatin accessibility mapping of the striatum identifies tyrosine kinase FYN as a therapeutic target for heroin use disorder. Nat. Commun. 11(1), 4634 (2020).
Shih, H. I. et al. An increased risk of reversible dementia may occur after zolpidem derivative use in the elderly population: A population-based case-control study. Medicine 94(17), e809 (2015).
Chou, R. C., Kane, M., Ghimire, S., Gautam, S. & Gui, J. Treatment for rheumatoid arthritis and risk of Alzheimer’s disease: A nested case-control analysis. CNS Drugs 30(11), 1111–1120 (2016).
Stamatiades, G. A., Echouffo-Tcheugui, J. B. & Garber, J. R. Sulfasalazine-induced hypoglycemia in a patient with type 2 diabetes and end-stage renal disease. AACE Clin. Case Rep. 4(6), e493–e496 (2018).
Singh, S. & Wooltorton, E. Increased mortality among elderly patients with dementia using atypical antipsychotics. CMAJ Can. Med. Assoc. J. 173(3), 252 (2005).
Acknowledgements
The financial support of the Research Council of the University of Tehran is acknowledged.
Author information
Authors and Affiliations
Contributions
S.G. designed and performed experiments, analyzed data and wrote the paper. C.E., S.G., K.S. and M.H.-R. supervised the research. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ghiam, S., Eslahchi, C., Shahpasand, K. et al. Identification of repurposed drugs targeting significant long non-coding RNAs in the cross-talk between diabetes mellitus and Alzheimer’s disease. Sci Rep 12, 18332 (2022). https://doi.org/10.1038/s41598-022-22822-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-022-22822-9
This article is cited by
-
A novel approach to enhance glioblastoma multiforme treatment efficacy: non-coding RNA targeted therapy and adjuvant approaches
Clinical Epigenetics (2025)
-
DeepDrug as an expert guided and AI driven drug repurposing methodology for selecting the lead combination of drugs for Alzheimer’s disease
Scientific Reports (2025)