Abstract
The aim of the study was sequencing of the mitogenome of Hygrobates turcicus Pešić, Esen & Dabert, 2017 to expand knowledge of the polymorphism and cryptic or pseudocryptic diversity within Hydrachnidia. The samples originated from Bulgaria, Vidima River near Debnewo, 42°56′41.4′′N, 24°48′44.6′′E, depth 0.4 m, stones on the bottom, water flow 0.71 m/s, temperature 10 °C, pH 8.53, oxygen 110%, conductivity 279 µS/cm, hardness 121 CaO mg/l; 11 males, 27 females, 2 deutonymphs 12.x.2019 leg. Zawal, Michoński & Bańkowska; one male and one female dissected and slides mounted. The study was carried out using the following methods: DNA extraction, sequencing, assembly and annotation, comparison with other populations of H. turcicus, and multigene phylogeny. As a result of the study, it was determined that the mitogenome is 15,006 bp long and encodes for 13 proteins, 2 rRNAs, and 22 tRNAs. The genome is colinear with those of H. longiporus and H. taniguchii, the difference in size originating from a non-coding region located between protein-coding genes ND4L and ND3. Five genes have alternative start-codon, and four display premature termination. The multigene phylogeny obtained using all mitochondrial protein-coding genes unambiguously associates H. turcicus with the cluster formed by H. longiporus and H. taniguchii.
Similar content being viewed by others
Introduction
Water mites (Hydrachnidia) are very diverse and species rich group of macroinvertebrates. They occupy almost all freshwater environments. An updated version of Limnofauna Europaea (www.watermite.org) shows the improvement of the knowledge on European water mite biodiversity. From the 1062 species listed in 1978 year, 28% have been synonymized or excluded because of their uncertain status (species incertae), while at the same time over 200 species were added1. There is still a clear gap in alpha-taxonomy and knowledge upon phenotypic polymorphism and cryptic diversity within Hydrachnidia. Recent publications2,3,4,5,6,7,8,9,10 indicate the presence of many unrecognized species, especially in the southern part of Europe, which can be distinguished by molecular methods.
The publication of the three parts of an identification key11,12,13 initiated a new trend in researchers on European water mites, facilitating or enabling ecological and biological research. The release of these keys was preceded by numerous revisions of individual genera14,15,16 based on morphological data. However, the development of molecular barcoding17 quickly suggested that alpha-taxonomy is still weakly recognised and there are many species-complex containing cryptic or pseudo-cryptic species. An important work on integrative taxonomy and phylogeny of water mites was published by Dabert et al.18. Their analyses, based on nuclear ribosomal genes such as 18S, 28S and fragments of the mitochondrial gene of the cytochrome c oxidase subunit 1 (cox1), provided evidence about the relationships within the group. Recent taxonomical studies2,3,4,5,6,7,8 indicated a lack of knowledge about phenotypic polymorphism and cryptic or pseudo-cryptic diversity within Hydrachnidia. Therefore, more genetic data are needed. One of the ways to achieve this is to sequence complete mitochondrial genomes. There is an important, rapidly growing literature dedicated to the sequencing of complete mitogenomes, but despite this, up to now only 12 mitogenomes of Hydrachnidia have been made available and published19,20,21,22,23.
To participate in this effort, we have undertaken the sequencing of the complete mitogenome of Hygrobates turcicus Pešić, Esen & Dabert, 2017, a species belonging to genera extensively studied by DNA-barcoding2,3,4,5,6,9,10. We compared the characteristics of this mitogenome with those of Hygrobates longiporus Thor, 1898 and Hygrobates taniguchii Imamura, 1954, and used the data to perform a multigene phylogeny.
Results
Mitogenome of H. turcicus
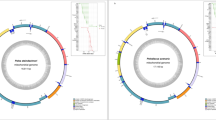
The mitogenome of H. turcicus (GenBank accession number OM336267) is 15,006 bp long (Fig. 1) (Table 1). It contains 13 conserved protein-coding genes, two rRNAs, and 22 tRNAs. It is colinear with the mitogenomes of H. longiporus and H. taniguchii, but its size is ca. 1300 bp longer (Table 1). The extra-length originates mostly from a non-coding region, which is located between protein-coding genes ND3 and ND4L. A comparison of the length, start and stop codon of the protein-coding genes of Hygrobates spp. is presented in Table 2. Two genes of H. turcicus present a premature termination, namely cox2 and cox3. In the case of cox3, this is common to the three species of Hygrobates spp. The distribution of the start codons is 7 ATG, 3 ATT, 2 ATA and 1 TTG. This TTG start codon was found in the ND5 gene, which discriminates H. turcicus from H. longiporus and H. taniguchii. A canonical start codon could not be identified in this case.
Comparison with other populations of H. turcicus
The percentages of identities between the available cox1 genes of H. turcicus are listed in Table 3. These percentages range between 99.52 and 99.84%, with the highest percentage (99.84%) being found 10 times out of 16.
Multigene phylogeny
The phylogenetic tree inferred from concatenated mitochondrial protein-coding genes displays high bootstrap values, ranging from 98 to 100%. The snail parasites Riccardoella spp. appear as an outgroup. Phylogenetic analyses clustered the analysed Parasitengona species into 6 maximally supported clades, three of them (1. Sperchon plumeifer and Mideopsis roztoczensis; 2. Unionicola parkeri and U. foili; 3. H. longiporus, H. taniguchii and H. turcicus) corresponding to the water mites. H. turcicus was recovered with high statistical support (98%) as a sister branch to H. longiporus and H. taniguchii (Fig. 2).
Cladogram illustrating the phylogenetic relationships for Hygrobates turcicus based on complete mitochondrial genome sequences. Mitochondrial genome rearrangement events are mapped on the branches of the best scoring maximum likelihood tree generated with RAxML-NG. Each node has 100% bootstrap support value.
Discussion
H. turcicus is a species distinguished recently on the basis of DNA-barcoding from the group of H. fluviatilis-complex species. It was described from Turkey and next mentioned from Bulgaria3,5. It is closely related with H. ulii Pesic, Saboori, Zawal & Dabert, 2019, and together with H. balcanicus Pesic, 2020, it is a separate clade in relation to the other species of H. fluviatilis-complex5,6. It is worth noting that based on cox1 comparisons (Table 3), the specimen sequenced in the current study didn’t exhibit a higher conservation with the other specimen from Bulgaria (GenBank: MN520308), more precisely from the Strymon river, than with 9 others specimens originating from Turkey. This might prove to be a limitation for accurate biogeographical studies that need to be considered when using single gene barcoding.
Molecular barcoding has proven useful to unveil the genetic diversity between closely related species, if not cryptic or semi-cryptic species of water mites, especially when obtained through the sequencing of the cox1 gene. It is usually sufficient to perform molecular phylogenies within these species. For more distantly related species, e.g. belonging to different families, more conserved nuclear genes such as the small subunit of the ribosomal RNA gene are sometimes preferred10,18,24. What we would like to emphasize in our work is that complete mitogenomes can prove to be also useful. The amount of data retrieved from the concatenation of all protein-coding genes led to the obtention of a phylogenetic tree with optimal support at the nodes. The results obtained in this study combine geographically distant but taxonomically related species (H. longiporus, H. taniguchii and H. turcicus) into one clade, thereby establishing a sister group for the clade comprising the genus Unionicola, which belongs to the same superfamily (Hygrobatoidea) and contrasting the rest of the water mite species (Sperchon clupeifer and Mideospis roztoczensis). At the same time, all species of water mites constitute one group, a sister group of species belonging to Trombidia (genus Leptotrombium). This illustrates the great usefulness of complete mitogenomes for the recognition of relationship between geographically and taxonomically distant taxa.
Based on our results and subsequent comparisons with the works of other authors22, we could also notice some differences among the genus Hygrobates for what concerns the start and stop codons of their mitochondrial genes. In the near future, it would be interesting to sequence mitogenomes of other species closely related to H. turcicus, to see how much these features are conserved.
Finally, studies such as ours will be helpful in the near future for members of the community who work on biomonitoring based on metabarcoding or environmental DNA. We might cite the recent article from Blattner et al.25, which includes Hygrobates norvegicus among other bioindicator species. This study was based on the amplification of several mitochondrial genes. Sequencing complete mitogenomes of duly identified specimen of water mites will help documenting the databases for later uses in similar studies.
Materials and Methods
Biological material
The H. turcicus samples were collected from stones on the bottom of Bulgaria, Vidima River near Debnewo (42°56′41.4′′N, 24°48′44.6′′E, depth 0.4 m, water flow 0.71 m/s, temperature 10 oC, pH 8.53, oxygen 110%, conductivity 279 µS/cm, hardness 121 CaO mg/l), including 11 males, 27 females, 2 deutonymphs. Collected in 12.x.2019 by Zawal, Michoński & Bańkowska. One male and one female were dissected and slide-mounted for morphological identification.
DNA extraction, sequencing, assembly and annotation
Water mites were collected by hand netting. Specimen were sorted out, initially identified and preserved in 96% ethanol, which is a method generally used in genetic research material5. Up to 50 specimens identified as H. turcicus were pooled together, and their DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany) as described previously23. Exoskeletons were retrieved after DNA extraction and mounted in Hoyer’s medium. Sequencing was performed at the Beijing Genomics Institute in Shenzhen, China, on a DNBSEQ platform in accordance with the company's procedure. A total of ca. 40 million clean 150 bp paired-end reads were obtained and assembled using SPAdes 3.14.026 using a k-mer of 125. The contig corresponding to the mitogenome was extracted, and the Consed27 package was used to verify its extremities. Annotations were done with the help of MITOS28 and manually curated.
Comparison with other populations of H. turcicus
The cox1 gene of H. turcicus was aligned with other 16 other sequences downloaded from GenBank and trimmed to a final size of 624 bp. The trimmed sequences were aligned on Clustal Omega online (ebi.ac.uk/Tools/msa/clustalo) to calculate the percentages of identities. We also computed the overall mean of genetic distances based on the Kimura 2-parameter model using MEGA 7.0. Standard error estimate(s) were obtained using bootstrap (1000 replicates).
Multigene phylogeny
We aligned the 13 complete mitochondrial genomes with MAFFT version 7.51029, using Riccardoella tokyoensis and Riccardoella reaumuri as outgroup terminals. We conducted maximum likelihood (ML) analyses using RAxML-NG30 under three different strategies. (1) One of the IR regions was removed from all mitochondrial genomes to reduce overrepresentation of duplicated sequences before we ran RAxML-NG on the unpartitioned alignment under GTR + I + G substitution model as a single partition; (2) The same data was partitioned by gene, exon, intron and intergenic spacer regions, allowing separate base frequencies, α-shape parameters, and evolutionary rates to be estimated for each; (3) we inferred the best-fitting partitioning strategy with PartitionFinder231 for the alignment. The best fitting nucleotide substitution models were inferred with jModelTest232. Phylogenetic trees were visualized and edited with FigTree 1.4.433. Support for the ML tree branches was calculated using the nonparametric bootstrap method with 1000 replicates.
Data availability
The complete mitogenome sequence of Hygrobates turcicus Pešić, Esen & Dabert, 2017 has been submitted to GenBank with the accession number OM336267. Data are available on Zenodo as the full sequence of the mitogenome in fasta format and annotations in tbl format with the following link: https://doi.org/10.5281/zenodo.6940457.
References
Gerecke, R., Martin, P. & Gledhill, T. Water mites (Acari: Parasitengona: Hydrachnidia) as inhabitants of groundwater-influenced habitats - considerations following an update of Limnofauna Europaea. Limnologica 69, 81–93 (2018).
Martin, P., Dabert, M. & Dabert, J. Molecular evidence for species separation in the water mite Hygrobates nigromaculatus Lebert, 1879 (Acari, Hydrachnidia): evolutionary consequences of the loss of larval parasitism. Aquat Sci. 72, 347–360 (2010).
Pešić, V. et al. Six species in one: evidence of cryptic speciation in the Hygrobates fluviatilis complex (Acariformes, Hydrachnidia, Hygrobatidae). Syst Appl Acarol. 22, 9 (2017).
Pešić, V. et al. Re-established after hundred years: definition of Hygrobates prosiliens Koenike, 1915, based on molecular and morphological evidence, and redescription of H. longipalpis (Hermann, 1804) (Acariformes, Hydrachnidia, Hygrobatidae). Syst Appl Acarol. 24, 8 (2019).
Pešić, V., Saboori, A., Zawal, A. & Dabert, M. Hidden but not enough: DNA barcodes reveal two new species in Hygrobates fluviatilis complex from Iran (Acariformes, Hydrachnidia, Hygrobatidae). Syst Appl Acarol. 24, 12. https://doi.org/10.11158/saa.24.12.11 (2019).
Pešić, V. et al. Two new species from the Hygrobates nigromaculatus-complex (Acariformes, Hydrachnidia, Hygrobatidae), based on morphological and molecular evidence. Acarologia 60(4), 753–768 (2020).
Pešić, V. et al. Torrenticola dowlingi sp. nov. a new water mite from Iran based on morphometrical and molecular data (Acariformes, Hydrachnidia, Torrenticolidae). Int J Acarology 46, 5 (2020).
Pešić, V., Zawal, A., Bańkowska, A., Jovanović, M. & Dabert, M. A new crenobiontic water mite species of the genus Atractides Koch, 1837 from Montenegro and Bulgaria, based on morphological and molecular data (Acariformes, Hydrachnidia, Hygrobatidae). Syst Appl Acarol. 25, 10 (2020).
Pešić, V., Zawal, A., Manović, A., Bańkowska, A. & Jovanović, M. A DNA barcode library for the water mites of Montenegro. Biodivers Data J. 9, e78311. https://doi.org/10.3897/BDJ.9.e78311 (2021).
Pešić, V., Zawal, A., Saboori, A. & Smit, H. New records of water mites (Acari, Hydrachnidia) from Iran with the description of one new species based on morphology and DNA barcodes. Zootaxa 5082(5), 425–440. https://doi.org/10.11646/zootaxa.5082.5.2 (2021).
Gerecke, R., Gledhill, T., Pešić, V. & Smit, H. Chelicerata: Acari III in Süßwasserfauna von Mitteleuropa (ed. Gerecke, R.) 1–429 (Elsevier GmbH, Spektrum Akademischer Verlag München, 2016).
Davids, C., Di Sabatino, A., Gerecke, R., Gledhill, T., Smit, H. & Van der Hammen, H. Acari: Hydrachnidia I in Süßwasserfauna von Mitteleuropa (ed. Gerecke, S.) 241–376 (Elsevier GmbN, Spektrum Akademischer Verlag München, 2007).
Di Sabatino, A., Gerecke, R., Gledhil, T., Smit, H., Chelicerata: Acari II in Süßwasserfauna von Mitteleuropa (ed. Gerecke, S.) 1–134 (Elsevier GmbN, Spektrum Akademischer Verlag München, 2010).
Van Haaren, T. & Tempelman, D. The Dutch species of Limnesia, with ecological and biological notes (Acari: Hydrachnidia: Limnesiidae). Nederlandse Faunistische Mededelingen. 30, 53–73 (2009).
Tuzovskij, P. V. Water mites of the genus Tiphys Koch, 1836 (Acariformes: Pionidae) in Russia. Acarina 19(2), 113–212 (2011).
Tuzovskij, P. V. Water mites of the genus Ljania thor, 1898 (Acari: Hydrachnidia: Aturidae) in Russia. Zootaxa 3249, 1–17 (2012).
Stryjecki, R. et al. The use of molecular techniques in the taxonomy of water mites (Hydrachnidia, Acari). Acta Biol. 23, 117–126 (2016).
Dabert, M., Proctor, H. & Dabert, J. Higher-level molecular phylogeny of the water mites (Acariformes: Prostigmata: Parasitengonina: Hydrachnidiae). Mol Phylogenet Evol. 101, 75–90 (2016).
Shao, R., Mitani, H., Barker, S. C., Takahashi, M. & Fukunaga, M. Novel mitochondrial gene content and gene arrangement indicate illegitimate inter-mtDNA recombination in the chigger mite, Leptotrombidium pallidum. J. Mol. Evol. 60, 764–773 (2005).
Edwards, D. D., Jackson, L. E., Johnson, A. J. & Ernsting, B. R. Mitochondrial genome sequence of Unionicola parkeri (Acari: Trombidiformes: Unionicolidae): molecular synapomorphies between closely-related Unionicola gill mites. Exp Appl Acarol. 54, 105–117 (2011).
Ernsting, B. R., Edwards, D. D., Aldred, K. J., Fites, J. S. & Neff, C. R. Mitochondrial genome sequence of Unionicola foili (Acari: Unionicolidae): a unique gene order with implications for phylogenetic inference. Exp Appl Acarol. 49, 305 (2009).
Hiruta, S.F., Morimoto, S., Yoshinari, G., Goldschmidt, T, Nishikawa, K. & Shimano, S. Complete mitochondrial genomes of two water mite species: Hygrobates (H.) longiporus and Hygrobates (rivobates) taniguchii (Acari, Trombidiformes, Hygrobatoidea). Mitochondrial DNA PART B 5(3), 2969–2971 (2020).
Macher, J., Drakou, K., Papatheodoulou, A., van der Hoorn, B. & Vasquez, M. The mitochondrial genomes of 11 aquatic macroinvertebrate species from Cyprus. Metabarcoding Metagenom. 4, 58259. https://doi.org/10.3897/mbmg.4.58259 (2020).
Zawal, A. et al. New records of water mites (Acari: Hydrachnidia) from Sri Lanka with description of four new species and some remarks of relationships. Syst Appl Acarol. 25(9), 1589–1610 (2020).
Blattner, K., Ebner, J. N., Zopfi, J. & von Fumetti, S. Targeted non-invasive bioindicator species detection in eDNA water samples to assess and monitor the integrity of vulnerable alpine freshwater environments. Ecol Indic. 129, 107916. https://doi.org/10.1016/j.ecolind.2021.107916 (2021).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19, 5 (2012).
Gordon, D., Abajian, C. & Green, P. Consed: a graphical tool for sequence finishing. Genome Res. 8, 195–202 (1998).
Bernt, M. et al. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2), 313–319 (2013).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4), 772–780 (2013).
Kozlov, A. M. et al. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 21 (2019).
Lanfear, R. et al. ParitionFinder2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34, 772–773 (2017).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9, 772 (2012).
Rambaut, A., FigTree v. 1.4.4. http://tree.bio.ed.ac.uk/software/figtree/ (2014).
Funding
This study is partly supported by Polish National Science Centre, Poland, grant no. 2017/27/N/NZ8/01568 and also by the 2017–2022 research funds granted for implementation of a co-financed international research project from the Polish Ministry of Science and Higher Education.
Author information
Authors and Affiliations
Contributions
L.S. initiated the project. Data collection were performed by A.Z., A.B., G.M.; A.Z. L.S., R.G. and I.S.-K. conducted experiments. Bioinformatic analyses were performed by R.G. The first draft of the manuscript was written by A.Z. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zawal, A., Skuza, L., Michoński, G. et al. Complete mitochondrial genome of Hygrobates turcicus Pešić, Esen & Dabert, 2017 (Acari, Hydrachnidia, Hygrobatoidea). Sci Rep 12, 22063 (2022). https://doi.org/10.1038/s41598-022-26188-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-022-26188-w