Abstract
The pharmaceutical industry faces a significant challenge from the low water solubility of nearly 90% of newly developed Active Pharmaceutical Ingredients (APIs). Despite extensive efforts to improve solubility, approximately 40% of these APIs encounter commercialization hurdles, impacting drug efficacy. In this context, a promising strategy will be introduced in which nanosuspensions, particularly polyvinyl alcohol (PVA) as a stabilizer, are applied to increase drug solubility. In this work using molecular dynamics simulations, the nanosuspension of four poorly water-soluble drugs (flurbiprofen, bezafibrate, miconazole, and phenytoin) stabilized with PVA is investigated. The simulation data showed van der Waals energies between polyvinyl alcohol with flurbiprofen and bezafibrate are − 101.12 and − 58.42 kJ/mol, respectively. The results indicate that PVA is an effective stabilizer for these drugs, and superior interactions are obtained with flurbiprofen and bezafibrate. The study also explores the impact of PVA on water molecule diffusion, providing insights into the stability of nanosuspensions. Obtained results also provide valuable insights into hydrogen bond formation, diffusion coefficients, and nanosuspension stability, contributing to the rational design and optimization of pharmaceutical formulations.
Similar content being viewed by others
Introduction
The pharmaceutical arena grapples with a widespread challenge, as nearly 90% of newly developed Active Pharmaceutical Ingredients (APIs) exhibit low water solubility. Despite the existence of a multitude of pre-formulation and formulation strategies aimed at improving water solubility, a substantial 40% of these APIs encounter commercialization hurdles, primarily stemming from this inherent limitation1. The profound influence of solubility on drug effectiveness is indisputable. The solubility of a drug in water stands as a foundational characteristic that demands meticulous consideration2. In oral formulations, the amount of drug adsorption from the gastrointestinal tract into the systemic circulation, known as oral bioavailability, is crucial for ensuring sufficient efficacy3.
The bioavailability of a pharmaceutical active is often characterized by two primary properties: permeability and solubility. The Food and Drug Administration, along with the pharmaceutical industry, commonly categorizes drugs based on these characteristics within a four-quadrant Biopharmaceutical Classification System (BCS). Presently, drugs exhibiting both high solubility and high permeability (Class I) constitute approximately 35% of the currently marketed drugs. However, only 10% of future drug candidates in the drug production line fall into Class I. In contrast, a substantial majority (around 80%) of upcoming drug candidates are anticipated to possess low aqueous solubility (Classes II and IV), and this has led to extensive efforts towards formulation strategies to improve water solubility4.
The consequence of low solubility is more emphasized in oral drugs, where drugs with low solubility lead to a decrease in the presence of active pharmaceutical compounds in the systemic circulation.
A significant proportion, approximately 30–40%, of drugs developed to date fall into the challenging category of being very difficult to dissolve in water, defined by the United States Pharmacopeia (USP) as less than 0.1 mg/ml. Examples of such drugs include drugs categorized as BCS class II such as danazol, nifedipine, and phenytoin, and drugs categorized as BCS class IV drugs like furosemide, taxol, and hydrochlorothiazide2.
Various techniques are employed to address the issue of poor aqueous solubility of drugs. These techniques include, nanosuspension, particle size reduction, salt formation, hydrotropy, pH adjustment, solid dispersion, amorphous, and co-crystal compound formation5,6.
Among these techniques, nanonization stands out as a widely employed method for enhancing solubility and bioavailability, as indicated by a substantial body of literature and the existence of authorized marketed products such as Rapamune®, Emend®, Triglide®, Tricor®, and Megace ES®7.
Nanosuspension, defined as nanosized drug molecules prepared in an aqueous medium and stabilized by suitable stabilizers like surfactants and polymers, has emerged as a prominent approach8. The reduction in particle size is a well-established method for increasing dissolution rates and oral bioavailability. Beyond the advantages mentioned, nanocrystals offer easy transformation into solid forms such as tablets, capsules, and powders for redispersion. Researchers are expanding into nutraceuticals, utilizing nanocrystals to increase Maximum solubility and speed of dissolution while retaining the benefits of nanoformulation with the addition of a slight surfactant9. The advent of nanotechnology has revolutionized various fields, including chemical, physical, and life sciences, providing a new avenue for drug delivery in pharmaceuticals1,10.
Nanosuspensions can be prepared employing two main methods: bottom-up and top-down. Stabilizing agents may consist of polymers like hydroxypropyl methylcellulose (HPMC), polyvinylpyrrolidone (PVP K30), and surfactants such as ionic sodium lauryl sulfate (SLS) and nonionic polysorbate (Tween 80)11.
PVA is biocompatible and non-toxic, making it safe for use in drug formulations. Its hydrophilic nature improves the wettability of poorly soluble drugs, enhancing their dispersion in aqueous environments. PVA is chemically stable and resistant to degradation, providing long-term stability to formulations. PVA can form hydrogen bonds with APIs, which can improve the stability and dispersion of nanosuspensions. These properties make PVA an effective stabilizer, ensuring the consistency, efficacy, and safety of pharmaceutical products12,13,14.
The choice of stabilizers, a critical aspect of the bottom-up method, can be a tedious task, requiring considerable effort. The successful application of nanosuspensions has been demonstrated in improving the activity of various anti-infectives, including triclosan, ciprofloxacin, itraconazole, and miconazole10,15,16. Nanocrystals, in addition to improving drug dissolution, have shown promise in realizing sustained release and targeting specific tissues or organs. An important advantage of nanosuspensions is their administration routes, including oral, parenteral, ocular, and pulmonary delivery, which surpass traditional formulation products17.
In contemporary pharmaceutical research, the utilization of Computer Aided Formulation Design (CAFD) methods has become widespread. These methodologies play a pivotal role in the selection of an optimal combination of stabilizers. Through the application of rational predictive CAFD methods, researchers can achieve desirable outcomes with a reduced number of experiments, expediting the attainment of results even before the initiation of actual laboratory experiments. The computational simulations contribute to comprehending the intermolecular interactions occurring at the molecular level among the formulation components. This understanding provides valuable insights into the observed physical and morphological changes within the formulation8.
Molecular Dynamics (MD) is a computational method based on classical mechanics, which simulates the physical movements of atoms and furnishes atomic-scale information not easily attainable through experimental investigations. MD offers insights that can assist in the development of drug formulations in a cost-effective and time-efficient manner18.
Styliari et al. and colleagues conducted an experimental and computational study on a method for coating drug nanoparticles with an amphiphilic polymer (mPEG-b-PCL) to enhance drug delivery. Their research found that after the evaporation of acetone, the polymer surrounds the drug molecules, resulting in a drug nanoparticle coated with the polymer with an efficiency of 99%19.
The article by Tian et al. presents computational research on screening stabilizers and exploring stabilization mechanisms for irbesartan nanosuspensions, which improve the dissolution of poorly soluble irbesartan polymorphs. Soluplus-P407 and TPGS-HPMCE5 were identified as suitable stabilizers through spatial conformation and thermodynamic energy analyses. The prepared nanosuspensions enhanced dissolution at different pH values. The stabilization mechanism was analyzed using molecular docking, molecular dynamics simulations, FTIR, and Raman spectroscopy, suggesting decreased enthalpy and Gibbs free energy due to synergistic external and internal interactions contributing to stabilization20.
Singh et al. utilized the emulsion solvent evaporation technique to create drug-loaded polymer nanoparticles for the oral formulation of tolbutamide using a biodegradable polymer (ε-caprolactone). The goal was to improve the bioavailability and therapeutic efficacy of tolbutamide by reducing its particle size to the nanoscale and achieving sustained drug release from the polymeric nanoparticles. Characterization techniques like FTIR, DSC, particle size analysis, zeta potential, and drug release studies were performed. The absence of new peaks or shifting of existing peaks in the FTIR spectra of the drug-excipient mixture compared to the pure drug indicates no chemical interaction and compatibility between the drug and polymers. DSC can detect any physical interactions or incompatibilities between the drug and excipients by analyzing changes in the melting endotherms, glass transition temperatures, or the appearance of new thermal events. The absence of any new thermal events or significant shifts in the melting/glass transition temperatures of the drug-excipient mixture, when compared to the pure drug, indicates that there are no physical interactions and that the components are compatible. The prepared drug-loaded poly(ε-caprolactone) nanoparticles were found to be in the nanometer size range with spherical morphology. The particle size was measured by dynamic light scattering using a Nicomp particle sizer. The zeta potential was measured to determine the stability of the nanoparticles. The prepared nanoparticles were found to be stable, with the zeta potential being influenced by the stabilizer type and concentration21.
In this work, MD was employed to simulate the nanosuspension of four active pharmaceutical compounds: flurbiprofen, bezafibrate, miconazole, and phenytoin. These compounds, known using their low solubility in water, were simulated with a polyvinyl alcohol polymer stabilizer. The MD and subsequent analysis provided valuable insights at the molecular level. The examination of hydrogen bonding interactions, radial distribution functions, diffusion coefficients, and energy profiles permitted us to observe the virtual formation of particles.
Flurbiprofen (C15H13FO2) and phenytoin (C15H12N2O2) both have functional groups, which are crucial for their pharmacological properties, and they are classified as class II drugs in the biopharmaceutics classification system BCS, indicating high permeability and low solubility4,22. Flurbiprofen’s structure includes a biphenyl system with a fluorine atom and a propanoic acid moiety, while phenytoin features an imidazolidine ring with carbonyl and phenyl groups.
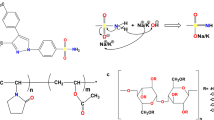
Miconazole (C18H14Cl4N2O) and bezafibrate (C19H20ClNO4) are also class II drug in the BCS23,24. Miconazole includes an imidazole ring with dichlorophenyl groups, a methoxy group, and an ethyl chain. Bezafibrate features two benzene rings, an amide group, a carboxylic acid group, and a chlorine substituent. The drug’s structure is shown in Fig. 1.
The molecular structures of flurbiprofen, phenytoin, miconazole, and bezafibrate differ owing to the presence of unique chemical groups and ring structures. These structural differences play a significant role in determining the pharmacological activities and therapeutic uses of the compounds.
In this study, we used these drugs because of their low solubility and their placement in classification under BCS class II.
Method
MD simulations are a wide technique in the field of drug delivery, serving tasks such as understanding drug interactions with proteins25,26, membranes27,28, and polymers29,30.
Using a simulation approach allows for predicting the physicochemical properties like particle size distribution, surface charge, stability, and solubility of nanosuspensions. Biopharmaceutical, it aids in forecasting aspects such as bioavailability, drug release profiles, permeability, and interactions with biological components. These insights help in tailoring nanosuspension formulations for enhanced therapeutic efficacy and targeted drug delivery.
In this study, MD was utilized to model the nanosuspension of four drugs bezafibrate, flurbiprofen, miconazole, and phenytoin stabilized with polyvinyl alcohol polymer. The GROMACS 2021.1 package (see http://www.gromacs.org) package was utilized, and the force field parameters, as well as the structural details of the drugs and polymer, were sourced from the SwissParam website. The structures of drugs and polyvinyl alcohol are depicted in Figs. 1 and 2, respectively. Four simulation boxes, each with dimensions of 10 × 10 × 10 \({\text{nm}}^{3}\), were examined. These boxes contained an equivalent number of polyvinyl alcohol molecules but varied in the quantity of drug molecules. The number of active drug compounds and polymer molecules was determined based on the molecular mass ratio of miconazole to other APIs, ensuring compatibility, given that miconazole exhibited the highest molecular mass.
Taking into account the extended chain structure of the polyvinyl alcohol polymer and, consequently, its high molecular weight, five vinyl alcohol monomers were employed. Detailed information is presented in Table 1.
The non-bonded electrostatic and Lennard–Jones interactions were managed using the particle mesh Ewald method. Van der Waals (vdW) interactions were explored with a cutoff distance set at 1.4 nm. The Nose–Hoover algorithm and the Parrinello-Rahman thermostat coupling were utilized to maintain pressure at 1 bar and temperature at 300 K, respectively. Equilibrium was maintained by applying the LINk algorithm to restrict all simulation connections. The simulation system underwent relaxation through energy minimization employing the steepest descent algorithm. The simulation spanned over a total duration of 105 ns, with a time step of 2 fs under periodic boundary conditions. Water molecules in the simulation boxes were explicitly treated using the TIP3P model, and \({Na}^{+}\) , \({Cl}^{-}\) particles were introduced to neutralize the system. Visualization of the simulation results was performed using the VMD software.
Results and discussion
Stabilizing nanosuspensions
Four distinct systems were established for MD simulations. The initial and final snapshots captured from these systems are presented in Fig. 3.
Upon examination of the final snapshots, it is evident that all the drug molecules have aggregated with polyvinyl alcohol stabilizer. It can be inferred that polyvinyl alcohol is a suitable stabilizer for the preparation of nanosuspensions of flurbiprofen, bezafibrate, miconazole, and phenytoin.
A closer inspection of the snapshots reveals that the accumulation of flurbiprofen and bezafibrate molecules with the polymer is more pronounced than that of miconazole and phenytoin molecules with polyvinyl alcohol. Consequently, PVA proves to be a superior stabilizer for the latter two drugs.
Interaction energy
Throughout the simulation process, it is observed that API molecules initially interact with PVA molecules, forming clusters, and subsequently come into contact with water molecules. Screening test results demonstrate that the interaction between the drug and stabilizers may be a crucial factor influencing the properties of the drug. To further explore this relationship, van der Waals and electrostatic energies were calculated. The results are depicted in Fig. 4, with average values presented in Table 2. The energy values between drug and polymer molecules suggest that API molecules interact with PVA. Particle size is a critical feature of any nanosuspension system as it significantly affects physical stability and biological performance. It is important to note that smaller particle sizes result in greater interaction between the drug and stabilizer, preventing the accumulation of drug particles and forming a stable nanosuspension12,14. A stronger interaction between API and PVA leads to better stabilization of the nanosuspension. Our results show that the interaction of polyvinyl alcohol with flurbiprofen, with a value of − 101.12 kJ/mol, surpasses that of other drugs. Therefore, it can be concluded that the combination of flurbiprofen with polyvinyl alcohol produces a more stable nanosuspension.
Radial distribution function
In molecular dynamics simulations, the radial distribution function (RDF) is a powerful analysis used to investigate the spatial distribution of particles in a system. It is a type of pairwise distribution function that expresses the probability of finding a particle at a given distance from a reference particle. RDF analysis is widely used in molecular dynamics simulations in various scientific disciplines and provides insights into phenomena such as solvation shells, phase transitions, and structural changes in a system. The RDF (g(r)) is calculated using the following equation (Eq. 1);
where V represents the system’s volume, N denotes the particle count, and dr indicates the width of the shell employed for the averaging process.
In this study, computed the radial distribution function between polyvinyl alcohol and API molecules, and the corresponding results are illustrated in Fig. 5. The outcomes reveal that the most prominent peak corresponds to flurbiprofen, positioned at a distance of 0.6 nm from the polymer molecules. As discussed in former sections, this proximity suggests the potential formation of van der Waals interactions between the drug and polymer molecules. Subsequently, the flurbiprofen molecule exhibits the highest peak.
This observation leads to the conclusion that the polyvinyl alcohol molecule serves as a more effective stabilizer for bezafibrate and flurbiprofen molecules. Notably, these findings align with the energy results obtained in our study.
Mean square displacement of water molecules
The Mean Squared Displacement (MSD) serves as the statistical average of particle trajectories and shows changes caused by the diffusion behavior of water molecules. The dynamic translation involves the examination of the Diffusion coefficient (Di) evaluated from the mean squared displacement (Eq. 2) by the Einstein relation (Eq. 3)31:
where \({R}_{i}\left(t\right)\) represents the instantaneous position of atom i at time step t, \({R}_{i}\left({t}_{0}\right)\) denotes the initial position of atom i, and N represents the total number of particles.
where \({r}_{i}\) (t) represents the position of the studied molecule, while D and t denote the diffusion coefficient and simulation time, respectively.
The viscosity of the dispersion medium affects the stability of nanosuspensions. Higher viscosity reduces the diffusion of particles, which slows down particle growth and promotes stability. The movement of water molecules, which is related to viscosity and diffusion, also indirectly affects stability.
Stabilizers like surfactants and polymers are used to increase viscosity and create steric/electrostatic barriers around particles, preventing agglomeration32.
The MSD graphs of water molecules are presented in Fig. 6. Average MSD values for 105 ns were used for comparing the diffusion of water molecules in the simulated system. The resulting diffusion coefficients are listed in Table 3.
Comparing our results to Kennt’s experimental study33, where the diffusion coefficient of pure water at 298.15 K and 0.1 MPa pressure is established as \(2.3\times {10}^{-9}\) \({m}^{2}/s\), our research yielded an average diffusion coefficient for the water of \(0.4\times {10}^{-9}{m}^{2}/s\)
This suggests that the presence of PVA slows down the diffusion of water molecules, indicating PVA’s ability to adsorb water molecules around API molecules, thereby impeding their movement and increasing contact between API molecules and water. Consequently, lower diffusion coefficient values in water suggest increased stability in nanosuspensions prepared with polyvinyl alcohol.
Hydrogen bond determination
Hydrogen bond analysis in molecular dynamics simulations is a crucial tool for understanding the behavior and interactions of molecules in a system. Hydrogen bonds are attractive interactions between a hydrogen atom attached to an electronegative atom and another electronegative atom. In MD simulations, hydrogen bond analysis helps researchers study the dynamics of these bonds over time, providing insights into the structure and stability of molecular systems.
Hydrogen bond formation can play a role in the preparation of nanosuspensions, which are colloidal dispersions of submicron-sized drug particles in a liquid medium. While the primary focus in nanosuspension preparation is often on particle size reduction and stabilization, hydrogen bonding can influence the process in several ways. The formation of interfacial hydrogen bonds between the nanoparticles and stabilizers (polymers, surfactants) is crucial for creating a stable interface and preventing particle agglomeration.
Increasing the number of hydrogen bond donors/acceptors on the nanoparticle and stabilizer surfaces can enhance the formation probability of interfacial hydrogen bonds, leading to improved nanosuspension stability7,34. Stabilizers with functional groups capable of hydrogen bonding can adsorb onto the nanoparticle surface, providing a protective layer that prevents particle aggregation or precipitation. Hydrogen bonding between the drug molecules and the dispersion medium (usually water) can enhance the solubility of the drug in the liquid phase. This improved solubility can aid in the formation and stabilization of nano-sized drug particles during the preparation process.
Researchers and pharmaceutical scientists carefully consider the intermolecular interactions, including hydrogen bonding, to design effective nanosuspensions that meet the desired criteria for drug delivery and therapeutic applications.
In this study, we computed the hydrogen bonds formed between water molecules and drug molecules, as well as those between drug and polymer molecules. The results are illustrated in Fig. 7. According to the findings presented, it is evident that the quantity of hydrogen bonds between flurbiprofen with polyvinyl alcohol surpasses that of other drugs. Consequently, it can be inferred that polyvinyl alcohol serves as a superior stabilizer for the aforementioned drugs. This conclusion aligns with the results mentioned earlier.
As evident from the molecular structures of the four drugs, miconazole contains the fewest hydrogen bond-donating groups. Consequently, it was expected to have the lowest number of hydrogen bonds with water. The next drug is flurbiprofen, which exhibits the least hydrogen bonding with water. This can be attributed to the strong interaction between polyvinyl alcohol and flurbiprofen molecules, resulting in the formation of hydrogen bonds between them. As a result, the ability of flurbiprofen to form hydrogen bonds with water decreases.
Essentially, the polyvinyl alcohol polymer forms a protective coating around the drug core, preventing interaction and the formation of hydrogen bonds with water molecules.
Solubility
The SASA (solvent-accessible surface area) represents the surface area of compounds available for contact with solvent molecules35. According to the initial algorithm introduced by Lee and Richards36, the solvent-accessible surface area of an atom is characterized as the surface area on a sphere with a radius R. At every location on this surface, the solvent molecule’s center can be positioned in contact with the atom, ensuring no penetration of other atoms within the molecule. The radius R is established by combining the van der Waals radius of the atom and the selected radius of the solvent molecule. Equations 4 and 5 states that in a certain atom, the length of an arc on a specific section (denoted as Li) is considered.
Zi represents the perpendicular distance from the center of the sphere to section i, while ΔZ is the spacing between sections. Δ′Z is defined as the minimum value between ΔZ/2 and the difference between the sphere’s radius (R) and Zi. The summation is performed over all the arcs drawn for the specified atom37.
The SASA analysis is relevant in understanding the bioavailability of a molecule. A higher SASA can suggest greater exposure to the solvent, potentially influencing how a molecule is adsorbed or interacts in biological systems38.
To assess the solubility of polyvinyl alcohol-stabilized nanosuspension, SASA analysis was employed. The results are presented in Fig. 8. During the simulation period, it is evident that the drug’s contact surface with the water molecules decreased, leading to the accumulation of the drug with the polymer.
Flurbiprofen exhibited the most significant reduction in SASA, indicating superior stabilization with polyvinyl alcohol. Conversely, miconazole demonstrated less accumulation with polyvinyl alcohol and the lowest interaction energy with the polymer, resulting in an almost constant trend in its SASA value.
Conclusion
Nanosuspensions, employing PVA as a stabilizer, prove to be a viable approach for enhancing the solubility and bioavailability of poorly water-soluble drugs. Molecular dynamics simulations have provided crucial insights into the nanosuspension behavior of four poorly water-soluble drugs flurbiprofen, bezafibrate, miconazole, and phenytoin.
The outcomes affirm PVA as an effective stabilizer, with notable interactions observed, particularly in the cases of flurbiprofen and benzafibrate. These findings not only contribute to the growing body of knowledge on nanosuspensions but also shed light on the intricate molecular-level dynamics governing the stabilization process. The correlation between hydrogen bond formation, diffusion coefficients, and nanosuspension stability emphasizes the multifaceted nature of this formulation strategy.
The utilization of molecular dynamics simulations and computer-aided formulation design methods showcases the potential for a more streamlined and informed approach to drug development. the insights gained from this study not only advance our understanding of nanosuspensions but also underscore the potential of computational tools in optimizing drug formulations. The prospect of tailoring stabilizers through rational design, informed by molecular dynamics simulations, opens avenues for future research and innovation in drug delivery. Ultimately, our findings contribute to the ongoing efforts to enhance the efficacy and commercial viability of pharmaceutical products facing solubility challenges.
Data availability
The authors confirm that the simulation data are presented in the form of graphs and tables and are analyzed within the text of the article. If you require the original data in specialized files for molecular dynamics simulation due to their large volume, you can send an email to sedigheh.abdollahi@birjand.ac.ir, and the data will be made available to you.
Code availability
The codes that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Ghosh, P., Rasmuson, A. & Hudson, S. P. Impact of additives on drug particles during liquid antisolvent crystallization and subsequent freeze-drying. Org. Process Res. Dev. 27, 2020–2034 (2023).
Ainurofiq, A., Putro, D. S., Ramadhani, D. A., Putra, G. M. & Santo, L. D. C. D. E. A review on solubility enhancement methods for poorly water-soluble drugs. J. Rep. Pharm. Sci. 10, 137–147 (2021).
Yoshida, T. & Kojima, H. Oral drug delivery systems applied to launched products: Value for the patients and industrial considerations. Mol. Pharm. 20, 5312–5331 (2023).
Johnson, L. M. & Hillmyer, M. A. Critical excipient properties for the dissolution enhancement of phenytoin. ACS Omega 4, 19116–19127 (2019).
Kalepu, S. & Nekkanti, V. Insoluble drug delivery strategies: Review of recent advances and business prospects. Acta Pharm. Sin. B 5, 442–453 (2015).
Kawabata, Y., Wada, K., Nakatani, M., Yamada, S. & Onoue, S. Formulation design for poorly water-soluble drugs based on biopharmaceutics classification system: Basic approaches and practical applications. Int. J. Pharm. 420, 1–10 (2011).
Ahire, E., Thakkar, S., Darshanwad, M. & Misra, M. Parenteral nanosuspensions: A brief review from solubility enhancement to more novel and specific applications. Acta Pharm. Sin. B 8, 733–755 (2018).
Mehta, C. H. et al. Molecular simulation driven experiment for formulation of fixed dose combination of Darunavir and Ritonavir as anti-HIV nanosuspension. J. Mol. Liq. 293, 111469 (2019).
Omolo, C. A. et al. Formulation and molecular dynamics simulations of a fusidic acid nanosuspension for simultaneously enhancing solubility and antibacterial activity. Mol. Pharm. 15, 3512–3526 (2018).
Tahara, K. et al. In vitro and in vivo characterization of drug nanoparticles prepared using PureNano™ continuous crystallizer to improve the bioavailability of poorly water soluble drugs. Pharm. Res. 33, 2259–2268 (2016).
Cheng, H., Mao, L., Zhang, S. & Lv, H. Impacts of polymeric additives on nucleation and crystal growth of indomethacin from supersaturated solutions. AAPS PharmSciTech 20, 1–10 (2019).
Wang, Y., Zheng, Y., Zhang, L., Wang, Q. & Zhang, D. Stability of nanosuspensions in drug delivery. J. Control. Release 172, 1126–1141 (2013).
Bartos, C. et al. Optimization of a combined wet milling process in order to produce poly (vinyl alcohol) stabilized nanosuspension. Drug Des. Dev. Ther. 12, 1567–1580 (2018).
Bi, Y. et al. Particle size control and the interactions between drug and stabilizers in an amorphous nanosuspension system. J. Drug Deliv. Sci. Technol. 29, 167–172 (2015).
Ansari, M. J. et al. Enhanced antibacterial effects of ciprofloxacin enclosed in cyclodextrin and nano-suspension carrier systems. Bull. Env. Pharmacol. Life Sci. 4, 14–18 (2015).
Parmentier, J., Tan, E. H., Low, A. & Möschwitzer, J. P. Downstream drug product processing of itraconazole nanosuspension: Factors influencing drug particle size and dissolution from nanosuspension-layered beads. Int. J. Pharm. 524, 443–453 (2017).
Jassim, Z. E. & Rajab, N. A. Review on preparation, characterization, and pharmaceutical application of nanosuspension as an approach of solubility and dissolution enhancement. J. Pharm. Res. 12, 771–774 (2018).
Guruge, A. G., Warren, D. B., Pouton, C. W. & Chalmers, D. K. Molecular dynamics simulation studies of bile, bile salts, lipid-based drug formulations, and mRNA–lipid nanoparticles: A review. Mol. Pharm. 20, 2781 (2023).
Styliari, I. D. et al. Nanoformulation-by-design: an experimental and molecular dynamics study for polymer coated drug nanoparticles. RSC Adv. 10, 19521–19533 (2020).
Tian, J. et al. Spatial-thermodynamic understanding of stabilization mechanism using computational approaches and molecular-level elucidation of the mechanism of crystal transformation in polymorphic irbesartan nanosuspensions. Int. J. Pharm. 612, 121350 (2022).
Singh, N., Verma, P. K. & Nanda, S. Nanotechnology based oral formulations of tolbutamide by using biodegradable polymer. Int. J. Pharm. Sci. 10, 5599–5605 (2019).
Surov, A. O. et al. New pharmaceutical cocrystal forms of flurbiprofen: Structural, physicochemical, and thermodynamic characterization. Cryst. Growth Des. 19, 5751–5761 (2019).
Sun, R. et al. Electrosprayed polymeric nanospheres for enhanced solubility, dissolution rate, oral bioavailability and antihyperlipidemic activity of bezafibrate. Int. J. Nanomed. 15, 705–715 (2020).
Hussain, A., Altamimi, M. A., Ramzan, M. & Khuroo, T. Hansen solubility parameters and QbD-oriented HPLC method development and validation for dermatokinetic study of a Miconazole-Loaded cationic nanoemulsion in rat model. ACS Omega 8, 34746–34759 (2023).
Agrawal, N. & Skelton, A. A. 12-crown-4 ether disrupts the patient brain-derived amyloid-$β$-fibril trimer: Insight from all-atom molecular dynamics simulations. ACS Chem. Neurosci. 7, 1433–1441 (2016).
Agrawal, N. & Skelton, A. A. Binding of 12-crown-4 with Alzheimer’s A$β$40 and A$β$42 monomers and its effect on their conformation: Insight from molecular dynamics simulations. Mol. Pharm. 15, 289–299 (2018).
Bemporad, D., Luttmann, C. & Essex, J. W. Behaviour of small solutes and large drugs in a lipid bilayer from computer simulations. Biochim. Biophys. Acta (BBA) Biomembr. 1718, 1–21 (2005).
Bemporad, D., Luttmann, C. & Essex, J. W. Computer simulation of small molecule permeation across a lipid bilayer: Dependence on bilayer properties and solute volume, size, and cross-sectional area. Biophys. J. 87, 1–13 (2004).
Subashini, M., Devarajan, P. V., Sonavane, G. S. & Doble, M. Molecular dynamics simulation of drug uptake by polymer. J. Mol. Model. 17, 1141–1147 (2011).
Gao, Y. & Olsen, K. W. Drug–polymer interactions at water–crystal interfaces and implications for crystallization inhibition: Molecular dynamics simulations of amphiphilic block copolymer interactions with tolazamide crystals. J. Pharm. Sci. 104, 2132–2141 (2015).
Kialashaki, M., Sayyad Amin, J. & Zendehboudi, S. Molecular dynamics simulation to evaluate the stability of tetra-n-butyl ammonium/phosphonium bromide semiclathrate hydrates in the presence and absence of methane, carbon dioxide, methanol, and ethanol molecules. Ind. Eng. Chem. Res. 62, 7175–7196 (2023).
Ahmadi Tehrani, A., Omranpoor, M. M., Vatanara, A., Seyedabadi, M. & Ramezani, V. Formation of nanosuspensions in bottom-up approach: Theories and optimization. DARU J. Pharm. Sci. 27, 451–473 (2019).
Harris, K. R. & Woolf, L. A. Pressure and temperature dependence of the self diffusion coefficient of water and oxygen-18 water. J. Chem. Soc. Faraday Trans. 1 Phys. Chem. Condens. Phases 76, 377–385 (1980).
Tang, C., Li, X., Li, Z. & Hao, J. Interfacial hydrogen bonds and their influence mechanism on increasing the thermal stability of nano-SiO2-modified meta-aramid fibres. Polymers (Basel) 9, 504 (2017).
Faujan, N. H., Karjiban, R. A., Kashaban, I., Basri, M. & Basri, H. Computational simulation of palm kernel oil-based esters nano-emulsions aggregation as a potential parenteral drug delivery system. Arab. J. Chem. 12, 2372–2383 (2019).
Lee, B. & Richards, F. M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55, 379-IN4 (1971).
Ausaf Ali, S., Hassan, I., Islam, A. & Ahmad, F. A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr. Protein Pept. Sci. 15, 456–476 (2014).
Mishra, A., Ranganathan, S., Jayaram, B. & Sattar, A. Role of solvent accessibility for aggregation-prone patches in protein folding. Sci. Rep. 8, 12896 (2018).
Author information
Authors and Affiliations
Contributions
A: Sedigheh Abdollahi: Devised the computational protocol and prepared the model systems, performed all calculations, analyzed the data, and wrote and edited the original and the revised manuscript. B: Heidar Raissi: Supervision. Reviewing- Editing, edited the original and the revised version of the manuscript. C: Farzaneh Farzad: Reviewing-Editing, edited the original and the revised version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Abdollahi, S., Raissi, H. & Farzad, F. Examine stability polyvinyl alcohol-stabilized nanosuspensions to overcome the challenge of poor drug solubility utilizing molecular dynamic simulation. Sci Rep 14, 17386 (2024). https://doi.org/10.1038/s41598-024-68362-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-68362-2
This article is cited by
-
Functionalized MoS2 nanosheets as high-affinity nanocarriers for gemcitabine: a molecular simulation study
Scientific Reports (2025)
-
Voriconazole nanosuspension-loaded ocular bilayer dissolving microneedle patch for the management of fungal keratitis
Drug Delivery and Translational Research (2025)