Abstract
Slow Transit Constipation (STC) is characterized by impaired colonic motility, but its relationship with gut microbiota remains unclear. This study investigated the correlation between specific gut microbial populations and STC, focusing on the Lactobacillus acidophilus to Lactobacillus johnsonii (A/J) ratio. We used four rat groups: Control (CON), Loperamide-induced STC (LOP), antibiotic-treated (ABX), and antibiotic plus Loperamide (ABX + LOP). Fecal samples were analyzed using 16S rRNA gene sequencing, and serum metabolites were examined through LC–MS. The LOP group showed an increased A/J ratio, while ABX and ABX + LOP groups had decreased ratios. Notably, the ABX + LOP group did not develop STC symptoms. Metabolomic analysis revealed alterations in key metabolites across groups, including changes in levels of guanidinoacetate, glycine, l-glutamine, nicotine, and nicotinate d-ribonucleotide in the LOP group, and variations in l-glutamine, l-phenylalanine, l-tyrosine, histamine, d-ornithine, and lecithin in the ABX and ABX + LOP groups. Our findings suggest a correlation between the A/J ratio and STC development, offering insights into STC pathophysiology and potential microbiome-targeted therapies.
Similar content being viewed by others
Introduction
Slow Transit Constipation (STC), a specific type of functional constipation, results from reduced colonic motility, leading to prolonged transit times. Common symptoms include infrequent bowel movements, difficulty passing stools, hard and dry feces, and a feeling of incomplete evacuation1. A 2021 meta-analysis reported that STC affects 10.9% of adults in China2. Recently, there has been growing interest in the relationship between gut microbiota and STC3. Many studies have shown that gut microbiota plays a role in the development of functional gastrointestinal disorders4. The types of microbes present, their relative proportions, and the metabolites they produce all contribute to the development and characteristics of STC3. Initial theories suggest that an increase in beneficial bacteria might improve colonic motility, potentially helping to alleviate STC4. At the same time, research also indicates that metabolites produced by potentially harmful bacteria may slow down colonic motility, suggesting that adjusting the balance of these bacterial populations could significantly impact the gut environment5. The exact mechanisms by which gut microbiota affects STC are still unclear and require further investigation. Undoubtedly, there is a complex relationship between gut microbiota and STC.
To study the large and complex gut microbiota, researchers often use 16S rRNA gene sequencing6. This method allows for better identification of unusual or rarely isolated strains and can be used to identify new pathogens and uncultured bacteria. Its ability to provide a comprehensive overview of microbial communities has made it an essential tool in understanding the complex interactions between gut microbes and various gastrointestinal diseases. For example, the imbalance of microbial homeostasis can lead to the colonization and invasion of opportunistic pathogens in the gut, which promotes the development of IBD7. This technology has been used in many STC studies, for example, studies have shown that the gut microbiota could be a potential diagnostic and predictive biomarker for STC8. The research suggests that metabolomics is now also an important tool for the identification and diagnosis of gastrointestinal diseases.
Metabolomics, introduced by Professor Nicholson in the 1990s, is a high-throughput and sensitive method that follows genomics, transcriptomics, and proteomics in the field of biological research9,10. This advanced analytical technique enables the comprehensive profiling of metabolites, thereby facilitating the identification of potential biomarkers and elucidating the metabolic pathways involved in STC11. Consequently, metabolomics offers significant insights into the pathophysiology of STC, which could lead to improved diagnostic and therapeutic approaches.
To understand the interaction between gut microbiota and STC, our main goal was to compare the gut microbial composition and metabolic properties of four groups of rats: control, STC-model, antibiotic-treated, and antibiotic-treated then STC-model. We aimed to investigate whether changes in metabolites due to alterations in gut microbiota led to STC, identify STC-specific microbial characteristics, and understand the potential mechanisms involved. We hypothesized that the development and progression of STC might not only be related to intestinal motility but also to gut microbiota, possibly even linked to specific microbial groups.
Results
Rats with disturbed gut microbiota impeded STC development
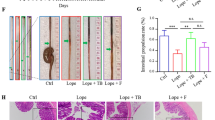
After the experiment, feces were collected from each group of rats as shown in Fig. 1A. The fecal water content in the LOP group (Fig. 1B) was the lowest among the four groups. Interestingly, the ABX and ABX + LOP groups did not show any significant difference compared to the CON group (P > 0.05). As shown in Fig. 1C and D, the intestinal propulsive ratio was significantly lower in the LOP group compared to the CON group (P < 0.05), while the ABX and ABX + LOP groups did not exhibit any decrease compared to the CON group (P > 0.05). H&E staining revealed a clear infiltration of inflammatory cells, decreased amounts, and irregularly arranged microvilli in the distal colon of the LOP group. Additionally, there was a disruption in the structure of tight junctions and a notable reduction in electron-dense materials in the LOP group (Fig. 1E). These results demonstrate the successful establishment of STC in the LOP group, while the ABX and ABX + LOP groups did not show any tendency towards STC.
Effect of STC and antibiotic cocktail on gut microbiota
Overview of the gut microbiota of each group
Usearch software was used to cluster Reads at a similarity level of 97.0% to obtain OTUs, the feature number column revealed that the CON and the LOP groups notably exceeded that observed in the ABX and the ABX + LOP groups in Fig. 2A. The Venn diagram also illustrates that the ABX and the ABX + LOP groups exhibited reduced microbial richness in comparison to the CON and the LOP groups, which indicated that most of the gut microbiota has been eliminated by using antibiotic cocktail (Fig. 2B).
Specifically, at the Phylum level, Firmicutes/Bacteroidota (F/B) ratio in the LOP group has shown an increase compared to the CON group12, this change observed in the LOP group aligns with previous researchers’ findings13,14, which demonstrated that the STC model was constructed. However, the F/B ratio in the ABX + LOP group also increased, so we focused on the Species level, both the ABX and the ABX + LOP groups were prominently characterized by the prevalence of M. morganii. Besides, we could note-worthy find that adjustments were evident in L. acidophilus and L. johnsonii, within the Firmicutes between the CON and the LOP group (Fig. 3). In the LOP group, L. acidophilus and L. johnsonii (A/J) ratio increased significantly, and the ABX and the ABX + LOP groups showed a decrease in the proportion of A/J ratio (Table 1). It may show that the STC was impacted by the A/J ratio.
Alterations in the biodiversity of the gut microbiota
The assessment using the Chao, Shannon, and Simpson index (Fig. 4A–C) notably highlighted decreased microbial species diversity within the ABX and the ABX + LOP groups compared to CON groups (P < 0.01), and the LOP group showed no significant difference to the CON group (P > 0.05). Based on the Specaccum analysis (Fig. 4D), the curve does not exhibit a steep incline, indicating that sampling was adequate, thus allowing for subsequent data analysis.
Results of Alpha diversity analysis, (A) Chao index, (B) Shannon index, and (C) Simpson index showed ABX and ABX + LOP groups’ diversity were significantly decreased; (D) Specaccum analysis curve declines steadily indicated sampling was adequate, thus allowing for subsequent data analysis. *P < 0.05, **P < 0.01, ***P < 0.001 compared with CON.
PCoA outcomes through visualization in Fig. 5, noteworthy the ABX and the ABX + LOP groups displayed a high degree of concordant clustering, which aligns harmoniously with the outcomes derived from taxonomic profiling. There were notable variations between the CON to the ABX and the ABX + LOP group. Besides, the LOP group showed changes in the composition of the colony in comparison with the CON group, which revealed exemplary separation.
Identification of particular bacterial species
The LEfSe outcomes demonstrated that the identification of biomarkers in Fig. 6A,B did not reveal statistically significant results for the ABX + LOP group, suggesting a lack of distinctive biomarkers within this particular group, then the CON, LOP, and ABX group had their biomarkers. Analysis of variance findings was presented in Fig. 6C. At the Species level, considerable disparities in microbial taxa were observed between the CON group the CON group held L. johnsonii statistically significant differences compared to the LOP group, and the LOP group showed more L. acidophilus (P < 0.01), which mean STC may be associated with A/J ratio.
(A) LEfSe analysis with an LDA value of > 4 displayed the dominant microbiomes in each group; (B) The diameter of the circles was proportional to the relative abundance of the species. Species with no significant differences were uniformly colored in yellow, while different species were colored according to the subgroup with the highest abundance in which the species is located; we draw by the Huttenhower Lab Galaxy Server 2.0, http://199.94.60.28/galaxy/. (C) ANOVA showed the Species level significant differences; variance findings column demonstrated that both CON and LOP group compared to both ABX and ABX + LOP group, the CON group hold more L. johnsonii and the LOP group showed L. acidophilus. *P < 0.05, **P < 0.01, ***P < 0.001.
The changes in gut microbiota composition were reflected in altered metabolites in STC and antibiotic cocktails by KEGG enrichment pathways analysis
To unravel the intricate mechanisms governing the complex interplay between gut microbiota and metabolites, we conducted a pathway enrichment analysis (Fig. 7). The outcomes of this analysis underscored the importance of pathways closely linked to metabolites. Notably, when comparing the CON group to the LOP group, we observed an upregulation of metabolic pathways (Fig. 7A); when compared to the CON group to the ABX group, alterations shifted towards ABC transporters (Fig. 7B). Histidine metabolism was the top pathway compared the ABX + LOP group to the LOP group in Fig. 7C. There were only two pathways comparing the ABX group to the ABX + LOP group, the metabolic pathway was a top one. (Fig. 7D).
Simultaneously, we conducted further by using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to get the pathway analysis bubble plot in Fig. 8, results showed when comparing the CON group with the LOP group, the predominant pathway of importance is the Glycine, serine, and threonine metabolism. The LOP group compared to the ABX + LOP group was identified as the d-glutamine and d-glutamate metabolism, the Phenylalanine, tyrosine, and tryptophan biosynthesis, and the Phenylalanine metabolism15,16,17. Moreover, the Histidine metabolism was demonstrated in the pathway between the ABX group and the ABX + LOP group.
Glycine, l-glutamine, and nicotine are decreased in rats suffering from STC
In searching for modified metabolites from enriched pathways, multivariate statistical analyses, including principal component analysis (PCA), volcano plots, and metabolic heatmaps, were employed to depict the clustering and separation among the CON group and other groups. The PCA multivariate statistical analysis demonstrated that there was a well-defined separation among the groups, providing evidence of significant differences between them (Fig. 9). Heatmap showed the specifical metabolite changes in serum between the CON group to the LOP and ABX group certainly in Fig. 10. Then when focused on the volcano plots in Fig. 11, accompanied by reduced levels of glycine, l-glutamine, and nicotine in the LOP group. In contrast, the metabolic profiles of the ABX groups exhibited the changes when compared to the CON group which changes included increased levels of l-glutamine, l-phenylalanine, l-tyrosine, and histamine, along with decreased levels of d-ornithine and lecithin.
The univariate statistical analysis of the Volcano plot showed the metabolite changes in serum between each different group, the red area indicated the existence of heightened levels of metabolites, the blue indicated a reduction, and the gray area indicated no changes. (A) CON VS LOP; (B) ABX VS CON.
Discussion
Due to STC rat yielded findings that closely corresponded to clinical symptoms of constipation18, the LOP-induced STC model was adopted to evaluate the potential interrelation between progression of STC and gut microbiota19,20,21. In this study, the intestinal transit rate of rats in the ABX and the ABX + LOP groups was not significantly different from that of the CON group, and fecal water content in the ABX and ABX + LOP groups was significantly higher compared to the CON group. Loperamide can inhibit the contraction of intestinal smooth muscle and reduce intestinal peristalsis, however, in this research, we found the ABX + LOP group has not appeared STC. The above results indicated that eliminating major gut microbiota could hinder the establishment of the STC model. This prompts speculation that the emergence and advancement of STC might be intricately linked to some gut microbiota.
At the species level, the findings indicated a significant increase in the A/J ratio in the LOP group compared to the CON group. Conversely, the ABX and ABX + LOP groups exhibited a decreased A/J ratio relative to the CON group, accompanied by symptoms of diarrhea. This suggests a significant association between the A/J ratio and intestinal homeostasis. Additionally, M. morganii, a pathogen known to cause diarrhea, was predominantly found in both the ABX and ABX + LOP groups, potentially explaining the mild diarrhea observed in these groups. It is hypothesized that higher A/J ratios may lead to STC, while lower ratios result may in diarrhea.
Gut microbiota-derived metabolites play a crucial role in modulating host physiology. To understand these influences, a comprehensive metabolomics analysis was conducted on serum samples from the CON, LOP, and ABX groups. Compared to the CON group, the LOP group showed increased guanidinoacetate levels and decreased levels of glycine, l-glutamine, and nicotine. l-glutamine, an important excitatory neurotransmitter in the enteric nervous system, induces depolarization in gastrointestinal neurons, enhancing intestinal motility22,23. This supports the consistent establishment of the STC model as expected. The metabolic changes in the ABX and ABX + LOP groups were similar to those in the LOP group, with elevated levels of l-glutamine, l-phenylalanine, l-tyrosine, and histamine, and decreased levels of d-ornithine and lecithin. Glutamine, beyond its neurotransmitter role, is a vital fuel for intestinal tissues and is converted into glutamate23,24, which plays a key role in amino acid production and the tricarboxylic acid (TCA) cycle25. Macrophages and monocytes use glutamine to synthesize d-ornithine. Notably, M. morganii produces ornithine decarboxylase, reducing d-ornithine levels in the ABX and ABX + LOP groups, leading to decreased synthesis of immune cells and contributing to inflammatory responses24.
In this study, firstly, carried out on 16S rRNA gene sequencing, we found that augmentation in the A/J ratio within the probiotic Lactobacillales order may potentially contribute to shaping the development of STC. The results of metabolomics suggested that apart from the effects on common excitatory neurotransmitters such as glycine, l-glutamine, and nicotine, the proportions changes of the A/J ratio may be associated with the enrichment of the Glycine, serine, and threonine metabolism pathway. However, certain limitations still remain in our study, and future work focuses on further validating are still needed.
Above all, this study used 16S rRNA gene sequencing to reveal that an increased A/J ratio within the probiotic Lactobacillales order may contribute to the development of STC. Metabolomics results indicated that changes in the A/J ratio are linked to the glycine, serine, and threonine metabolism pathway, beyond just affecting common excitatory neurotransmitters like glycine, l-glutamine, and nicotine. Despite these findings, there are limitations in our study, and further validation is needed in future research.
Methods
Animal and reagent
Male Sprague–Dawley (SD) rats (180 ± 20 g) were procured from Shanghai SLAC Laboratory Animal Co., Ltd. The experimental animal production license number was SCXK (Hu) 2017-0005. Rats were housed in the barrier environment of the New Drug Safety Evaluation Center at Fujian Medical University, accommodated in plastic rodent cages, provided with standard animal feed, and offered ad libitum access to water. The housing conditions maintained a temperature of 24 ± 2 °C, relative humidity of 55 ± 10%, and a 12-h light–dark cycle.
Statement
We confirm that all methods were performed in accordance with the relevant guidelines and regulations, all experimental protocols were approved by the Institutional Ethics Committee of FUJIAN MEDICAL UNIVERSITY (Protocol No.: FJMU IACUC 2021-0039), and we confirmed the study was reported in accordance with ARRIVE guidelines, the rats were euthanized by CO2.
Loperamide hydrochloride (LOP), Ampicillin Sodium Salt and Metronidazole was purchased from Aladdin Reagent Co., Ltd., (Batch No.: L129465; No.: A105483; No.: M109874); Dimethyl Sulfoxide (DMSO) was purchased from China National Pharmaceutical Group Chemical Reagent Co., Ltd., (Batch No.: 20150228); Neomycin Sulfate Hydrate was purchased from Adamas Reagent Co., Ltd., (Batch No.: 22966B).
1 g of Ampicillin, 1 g of Neomycin, and 0.5 g of Metronidazole were weighed, all of which were introduced into a 1000 mL volumetric flask, serving as an antibiotic cocktail (ABX).
Experimental design
A total of 32 rats were subjected to random allocation, forming 4 distinct cohorts with eight rats in each group:
-
CON group: i.g. of 0.5% DMSO solution;
-
LOP group: i.g. of LOP solution;
-
ABX group: i.g. of 0.5% DMSO solution then fed with antibiotic cocktail;
-
ABX + LOP group: i.g. of LOP solution then fed with antibiotic cocktail.
The rats in the LOP group and ABX + LOP group were orally administered with 1 mL of 5 mg/kg LOP suspension once daily for a continuous period of 7 d to establish STC in rats, while CON group i.g. received an equivalent volume of 0.5% DMSO for 7 consecutive days (Fig. 12).
Throughout the experimental duration both the ABX and the ABX + LOP groups were induced with an antibiotic cocktail consisting of the drinking water, which was refreshed every 3 d. The antibiotic cocktail facilitated the depletion of gut microbiota within the rats, thereby engendering a pseudonym-free state (Fig. 12).
Freshly extruded feces were collected in time after the STC induction or microbiota depletion in the 7th d for fecal parameters measurement and 16S rRNA gene sequencing analysis.
Evaluation of fecal water content and intestinal propulsive ratio
Feces were obtained from each experimental group within 12 h, the initial feces weight (mwet) was reordered, followed by drying the feces in the oven at a constant temperature of 60 °C for 8 h, and the dried weight (mdry) of the feces was determined. Fecal water content be described:
Evaluation of intestinal propulsive ratio
After a 24 h period of food deprivation while maintaining access to water, the rats received a 2 mL oral administration of ink via gavage. 15 min later sacrificed rats, and took total intestine, measured the length of intestine and ink propulsion distance, the rate of intestinal ink propulsion was precisely quantified. Intestinal propulsive ratio be described:
16s RNA gene sequencing
200 mg of glass beads and 50–100 mg of feces sample were weighed into a 2 mL centrifuge tube and placed the tube on ice, then added 300 µL of SP1 and 10 µL of Proteinase K, and vortex at maximum speed for 5 min until thoroughly mixed. The samples were incubated at 70 °C for 10 min (13 min for frozen samples), during which time they were vortexed and mixed twice, 2 min on ice. After that added 100 µL SP2, vortexed for 30 s and 5 min on ice. 13,000–20,000×g centrifuged for 5 min. Carefully transferred the supernatant into a centrifuge tube supplied with a volume of 1.5 mL without touching the sediment at the bottom of the tube, and added 200 µL of HTR solution and vortexed for 10 s. Allowed to stand at room temperature for 2 min then centrifuged at > 13,000×g for 2 min. Transferred 250 µL of supernatant to a new 2 mL tube, then added 250 µl of anhydrous ethanol and 250 µL of BL buffer. Vortexed and mixed for 10 s, and transferred the entire solution to a HiBind column. Placed in a 2 mL collection tube, centrifuged for 1 min at > 13,000×g, discarded the filtrate and collection tube. Replaced the collection tube with a new 2 mL tube. Added 500 µl of HB buffer and centrifuged for 1 min at > 13,000×g, discarded the filtrate and the collection tube. Added 500 µL of HB buffer and centrifuged for 1 min at > 13,000×g, discarded the filtrate and the collection tube. Added 500 µL of HB buffer and centrifuged for 1 min at > 13,000×g, discarded the filtrate and the collection tube. Transferred to a new 2 mL collection tube and added 750 µL RNA Wash buffer. Centrifuged at > 13,000×g for 1 min, discarded the filtrate, and put the column back into the new collection tube, discarded the filtrate again, and returned the column to the collection tube. The collection tube was subjected to > 13,000×g in vacuo for 2 min before being transferred to a new 1.5 mL centrifuge tube. Next, it was eluted with 200 µL of preheated Elution Buffer at temperatures between 60 and 70 °C. Following this, the column was left to stand at room temperature for 2 min and subsequently centrifuged at > 13,000×g for 1 min. We got the rRNA for the gene sequencing.
Biotree Co. (Shanghai, China) measured fecal parameters and 16S rRNA. This entailed subjecting the high variability regions of the PCR products to high-throughput sequencing, thereby facilitating an in-depth investigation into the alterations within the rat gut microbiota. 341F and 806R were used to amplify 16S V3-V4 region, 341F was 5′-CCTACGGGNGGCWGCAG-3′ and 806R was 5′-GACTACHVGGGTATCTAATCC-3′. PCR production was purified by AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA), then quantified by Qubit (Invitrogen, USA). Agilent 2100 Bioanalyzer and Illumina library quantification kit (Kapa Biosciences, Woburn, MA, USA) were selected to evaluate the production. The qualified libraries (Index sequences are not repeatable) were diluted in a gradient, mixed according to the required sequencing volume in the appropriate ratio, and denatured by NaOH to single-stranded for online sequencing; 2 × 250 bp double-end sequencing was performed using NovaSeq 6000 sequencer, and the corresponding reagent was NovaSeq 6000 SP Reagent Kit (500 cycles).
Building upon this foundation, an array of diversity analyses, including Alpha and Beta diversity assessments, were executed. These encompassed evaluations such as Chaos, Shannon, and Simpson index curves, which depicted microbial diversity trends, and Principal Coordinates Analysis (PCoA) outcomes, providing insights into the principal components of the microbial community.
Metabolites extraction
100 μL of serum sample was transferred to an EP tube. After the addition of 400 μL of extract solution (acetonitrile:methanol = 1:1, containing isotopically-labeled internal standard mixture), the samples were vortexed for 30 s, sonicated for 10 min in ice-water bath, and incubated for 1 h at – 40 °C to precipitate proteins. Then the sample was centrifuged at 12,000 rpm (RCF = 13,800(×g), R = 8.6 cm) for 15 min at 4 °C. The resulting supernatant was transferred to a fresh glass vial for analysis. The quality control (QC) sample was prepared by mixing an equal aliquot of the supernatants from all of the samples.
LC–MS/MS analysis
In parallel, comprehensive compositional analyses were performed on serum specimens using the liquid chromatography-mass spectrometry (LC–MS) methodology. LC–MS/MS analyses were performed using a UHPLC system (Vanquish, Thermo Fisher Scientific) with a UPLC BEH Amide column (2.1 mm × 100 mm, 1.7 μm) coupled to Orbitrap Exploris 120 mass spectrometer (Orbitrap MS, Thermo). The mobile phase consisted of 25 mmol/L ammonium acetate and 25 ammonia hydroxide in water (pH = 9.75) (A) and acetonitrile (B). The auto-sampler temperature was 4 °C, and the injection volume was 2 μL. The Orbitrap Exploris 120 mass spectrometer was used for its ability to acquire MS/MS spectra on information-dependent acquisition (IDA) mode in the control of the acquisition software (Xcalibur, Thermo). In this mode, the acquisition software continuously evaluates the full scan MS spectrum. The ESI source conditions were set as follows: sheath gas flow rate as 50 Arb, Aux gas flow rate as 15 Arb, capillary temperature 320 °C, full MS resolution as 60,000, MS/MS resolution as 15,000 collision energy as 10/30/60 in NCE mode, spray Voltage as 3.8 kV (positive) or − 3.4 kV (negative), respectively.
Data preprocessing and annotation
The raw data were converted to the mzXML format using ProteoWizard and processed with an in-house program, which was developed using R and based on XCMS, for peak detection, extraction, alignment, and integration. Then an in-house MS2 database at BiotreeDB was applied to metabolite annotation. The cutoff for annotation was set at 0.3.
Histopathology of colon tissues
Rats from different groups’ distal colon and the spinal cord were obtained and embedded in paraffin. Sections of the distal colon were stained with hematoxylin and eosin (H&E) to reveal adrenal hyperplasia.
Statistical analysis
Data were expressed as mean ± standard deviation (SD) by GraphPad Prism 8. One-way ANOVA or Student’s t-test was performed for the rest comparison between groups. When ANOVA showed significant differences, pair-wise comparisons between means were tested by Bonferroni post-hoc test or Dunnett’s tests. Values of P < 0.05 were considered statistically significant.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files). The datasets generated and analysed during the current study are available in the Science Data Bank repository, https://doi.org/10.57760/sciencedb.07579.
References
Sailer, M. Slow transit constipation. Zentralbl. Chir. 144, 179–189. https://doi.org/10.1055/a-0860-5960 (2019).
Yang, Z. et al. Prevalence of chronic constipation in Chinese adults: A meta-analysis. Chin. Gen. Pract. 24, 2092–2097. https://doi.org/10.12114/j.issn.1007-9572.2021.00.092 (2021).
Dimidi, E., Christodoulides, S., Scott, S. M. & Whelan, K. Mechanisms of action of probiotics and the gastrointestinal microbiota on gut motility and constipation. Adv. Nutr. 8, 484–494. https://doi.org/10.3945/an.116.014407 (2017).
Sun, D. et al. Lacidophilin tablets alleviate constipation through the regulation of intestinal microflora by promoting the colonization of Akkermansia sps. Sci. Rep. 14, 7152. https://doi.org/10.1038/s41598-024-57732-5 (2024).
Lin, X. et al. Constipation induced gut microbiota dysbiosis exacerbates experimental autoimmune encephalomyelitis in C57BL/6 mice. J. Transl. Med. 19, 317. https://doi.org/10.1186/s12967-021-02995-z (2021).
Usi, M. et al. 16S rRNA nanopore sequencing for rapid diagnosis of causative bacteria in bovine mastitis. Res. Vet. Sci. 161, 45–49. https://doi.org/10.1016/j.rvsc.2023.06.006 (2023).
Qiu, P. et al. The gut microbiota in inflammatory bowel disease. Front. Cell. Infect. Microbiol. 12, 733992. https://doi.org/10.3389/fcimb.2022.733992 (2022).
Tian, H. L. et al. Gut metagenome as a potential diagnostic and predictive biomarker in slow transit constipation. Front. Med. 8, 77961. https://doi.org/10.3389/fmed.2021.777961 (2021).
Lindon, J. C., Holmes, E. & Nicholson, J. K. Metabonomics and its role in drug development and disease diagnosis. Expert Rev. Mol. Diagn. 4, 189–199. https://doi.org/10.1586/14737159.4.2.189 (2004).
Lodes, M. J. et al. Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Investig. 113, 1296–1306. https://doi.org/10.1172/jci20295 (2004).
Smirnov, K. S. et al. Challenges of metabolomics in human gut microbiota research. Int. J. Med. Microbiol. 306, 266–279. https://doi.org/10.1016/j.ijmm.2016.03.006 (2016).
Sobol, C. V. Stimulatory effect of Lactobacillus metabolites on colonic contractions in newborn rats. Int. J. Mol. Sci. https://doi.org/10.3390/ijms24010662 (2022).
Boudreau, M. D. et al. From the cover: Aloin, a component of the Aloe vera plant leaf, induces pathological changes and modulates the composition of microbiota in the large intestines of F344/N male rats. Toxicol. Sci. 158, 302–318. https://doi.org/10.1093/toxsci/kfx105 (2017).
Zhang, X. et al. Chitosan oligosaccharides attenuate loperamide-induced constipation through regulation of gut microbiota in mice. Carbohydr. Polym. 253, 117218. https://doi.org/10.1016/j.carbpol.2020.117218 (2021).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M. et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592. https://doi.org/10.1093/nar/gkac963 (2023).
Zhang, Y. et al. Therapeutic effect of protease-activated receptor 2 agonist SLIGRL-NH(2) on loperamide-induced Sprague–Dawley rat constipation model and the related mechanism. Drug Des. Dev. Ther. 12, 2403–2411. https://doi.org/10.2147/dddt.S160628 (2018).
Ge, X. et al. Potential role of fecal microbiota from patients with slow transit constipation in the regulation of gastrointestinal motility. Sci. Rep. 7, 441. https://doi.org/10.1038/s41598-017-00612-y (2017).
Mancabelli, L. et al. Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci. Rep. 7, 9879. https://doi.org/10.1038/s41598-017-10663-w (2017).
Inatomi, T. & Honma, M. Effects of probiotics on loperamide-induced constipation in rats. Sci. Rep. 11, 24098. https://doi.org/10.1038/s41598-021-02931-7 (2021).
Liu, M. T., Rothstein, J. D., Gershon, M. D. & Kirchgessner, A. L. Glutamatergic enteric neurons. J. Neurosci. 17, 4764–4784. https://doi.org/10.1523/jneurosci.17-12-04764.1997 (1997).
Newsholme, P., Procopio, J., Lima, M. M., Pithon-Curi, T. C. & Curi, R. Glutamine and glutamate—their central role in cell metabolism and function. Cell Biochem. Funct. 21, 1–9. https://doi.org/10.1002/cbf.1003 (2003).
Cao, H. et al. Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci. Rep. 7, 10322. https://doi.org/10.1038/s41598-017-10835-8 (2017).
Kimura, R. E., LaPine, T. R., Johnston, J. & Ilich, J. Z. The effect of fasting on rat portal venous and aortic blood glucose, lactate, alanine, and glutamine. Pediatr. Res. 23, 241–244. https://doi.org/10.1203/00006450-198802000-00022 (1988).
Acknowledgements
This work was kindly supported by grants from the Scientific Research Initiation Fund of Fujian Medical University (No. 60000166), the Fujian Provincial Department of Science and Technology Joint Innovation Project Fund (No. 2021Y4011), and the Key Laboratory of Pharmaceutical Analysis and Laboratory Medicine (Putian University), Fujian Province University (No. PALM 202208).
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.L. and Y.W.; methodology, Y.L.; software, Y.W.; validation, W.H., N.Q. and Z.L.; formal analysis, J.Z.; investigation, J.W.; resources, H.Z.; data curation, Y.Y.; writing—original draft preparation, Y.L.; writing—review and editing, Y.W.; visualization, M.G.; supervision, Y.Y.; project administration, X.Y.; funding acquisition, Y.Y. All authors have read and agreed to the published version of the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liao, Y., Wang, Y., Huang, W. et al. L. acidophilus/L. johnsonii ratio affects slow transit constipation in rats. Sci Rep 14, 21088 (2024). https://doi.org/10.1038/s41598-024-71945-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-71945-8