Abstract
Interleukin-6 (IL-6) plays a crucial role in the pathogenesis of cardiovascular disease (CVD), and IL-6 receptor (IL-6R) blockade has emerged as a promising therapeutic option. However, their specific therapeutic effects in different types of CVDs remain unclear. This study aimed to assess the efficacy of IL-6R blockade in the management of various CVDs, including hypertension (HTN), coronary heart disease (CHD), myocardial infarction (MI), atrial fibrillation (AF), and heart failure (HF). The Mendelian randomization (MR) approach was utilized to investigate the therapeutic impact of IL-6R blockade on HTN, CHD, MI, AF, and HF based on the genome-wide association study (GWAS) summary statistics. MR-Egger intercept test, Cochran's Q test, and leave-one-out analysis were used for sensitivity analysis to verify the reliability of the MR results. The Bonferroni method was used to correct for bias caused by multiple comparisons. Inverse variance weighted (IVW) results demonstrated that IL-6R blockade significantly influenced CHD (odds ratio (OR) = 0.757, 95% confidence interval (CI): 0.690 - 0.832, P = 5.804 × 10–9) and MI (OR = 0.840, 95% CI: 0.744 - 0.949, P = 0.005). However, IL-6R blockade had no significant effect on HTN (OR = 1.015, 95% CI: 0.950 - 1.084, P = 0.663), AF (OR = 0.905, 95% CI: 0.800 - 1.025, P = 0.116) and HF (OR = 1.012, 95% CI: 0.921 - 1.113, P = 0.805). Genetically predicted IL-6R blockade was associated with a protective effect on CHD and MI, but not HTN, AF and HF. This study's findings offer valuable insights for tailoring IL-6R blockade treatment for different types of CVD, and serve as a reference for future research.
Similar content being viewed by others
Introduction
Cardiovascular disease (CVD) is a leading cause of global mortality1 and poses a significant challenge to public health worldwide. It is projected that by 2030, the prevalence of CVD in the global population aged over 65 will continue to rise, making it the primary cause of non-infectious disease-related deaths globally2. Therefore, there is great significance in identifying new treatment strategies and targets for CVD.
The pathogenesis of CVD is multifaceted, involving genetic, environmental, lifestyle, and other factors3. In recent years, inflammation has been recognized as playing a pivotal role in the development of CVDs and has garnered considerable attention4,5,6,7. Interleukin-6 (IL-6) is a multifunctional cytokine involved in regulating various biological processes such as immune response, inflammatory processes, and hematopoiesis8. Recent studies have established close associations between IL-6 and the development of various CVDs including atherosclerosis, inflammatory responses following myocardial infarction (MI), and heart failure (HF)9,10, suggesting that IL-6 may represent a potential target for treating CVD.
IL-6 receptor (IL-6R) blockade is a biological agent that inhibits the IL-6 signaling pathway by blocking the binding of IL-6 to its receptor, thereby reducing inflammation and immune response11. Currently, IL-6R blockade has demonstrated favorable efficacy and safety in treating conditions such as rheumatoid arthritis (RA)12 and COVID-1913 . However, studies on their application and effectiveness in treating CVDs remain limited.
Mendelian randomization (MR) studies, as a method to assess the causal relationship between exposure and outcome using genetic variants as instrumental variables (IV)14, are able to partially overcome the limitations of traditional observational studies. By selecting genetic variants associated with IL-6R expression as IV, MR studies can more effectively evaluate the role of IL-6R blockade in the treatment of CVD and their potential causal relationships. This study aims to reveal the feasibility and effectiveness of IL-6R blockade as potential drug targets for hypertension (HTN), coronary heart disease (CHD), MI, atrial fibrillation (AF), and HF, and to provide more precise treatment strategies for the prevention and treatment of CVD.
Materials and Methods
Selection of IVs
The IL6R gene encodes the IL-6 receptor protein, which plays an important role in inflammation and immune responses. The serum level of C-reactive protein (CRP) is a reliable downstream biomarker of IL-6 signal transduction. Therefore, the association between single nucleotide polymorphisms (SNPs) within the IL6R gene region and CRP levels can reflect the functional status of the IL-6 signaling pathway. IL-6R blockade inhibits downstream signal transduction by blocking the binding of IL-6 to its receptor, thereby achieving the purpose of anti-inflammation. Therefore, to simulate the effect of IL-6R blockade, we obtained SNPs within a 100 kb range on both sides of the IL-6R locus gene (chromosome 1: 154377819 - 154441926) that are associated with CRP levels, to estimate the downstream effect of IL-6R blockades on CVD risk.
In this study, the summary data of CRP were from a study by Sakaue S et al.15, which included 353,466 Europeans and 19,057,467 SNPs, mainly from the UK Biobank. First, we screened SNPs that were strongly correlated with CRP levels with the condition of P < 5E-08. Secondly, to ensure the independence of IVs, we excluded the interference of linkage disequilibrium (LD) (KB = 100, R2 < 0.3). Given the limited number of SNPs available for analysis under more stringent thresholds, we selected a threshold of R2 < 0.3 based on previous MR analyses of drug targets16,17,18,19. This threshold not only ensures an adequate number of SNPs but also controls the potential influence of LD. Finally, this study only considered common SNPs with an effect allele frequency (EAF) > 0.01. Through this process, we can more accurately simulate the effect of IL-6R blockades and thereby accurately estimate the downstream effect of IL-6R blockades on the risk of CVDs.
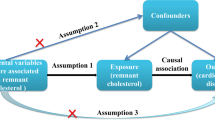
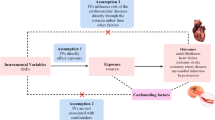
Besides, the selection of SNPs in MR analysis was based on three core assumptions: (1) SNPs must be strongly associated with the exposure variables; (2) SNPs should be independent of any potential confounders; and (3) SNPs influence the outcome solely through the exposure variables20.
Additionally, in order to ensure the stability of the results, we acquired additional GWAS data for CRP from the IEU Open GWAS project (204,402 Europeans) and conducted a repeated analysis using the aforementioned method to obtain IVs for IL-6R21. The data are mainly sourced from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Inflammation Working Group (CIWG)21, thereby avoiding the risk of sample overlap. The specific process of this study is illustrated in Fig. 1.
Source of results
This study analyzed the causal relationship between IL-6R blockade and five types of CVD, including HTN, CHD, MI, AF and HF. IL-6R blockade has been approved for the treatment of RA12, therefore RA was selected as a positive control in this study. All datasets used in this study were sourced from European populations to ensure demographic consistency. Furthermore, to avoid bias caused by sample overlap, the GWAS data for RA in this study were from an independent dataset provided by Okada Y et al.12. This dataset contains 14,361 RA cases and 43,923 controls, mainly from 18 European studies such as BRASS2, CANADA2, EIRA2, NARAC12, NARAC22, WTCCC2, Rheumatoid Arthritis Consortium International for Immunochip (RACI). The GWAS data for the five CVDs including CHD (51,098 cases and 402,635 controls), HF (33,250 cases and 420,483 controls), HTN (137,312 cases and 316,345 controls), MI (28,546 cases and 378,019 controls), and AF (55,853 cases and 231,952 controls) were obtained from the latest FinnGen consortium R11 version22. The data sources for this study are presented in Supplementary Table 1.
In this study, inverse variance weighted (IVW) was used as the main analysis method, and weighted median (WME) method, MR Egger method, weighted mode (WM) method and simple mode (SM) method were used for supplementary analyses. Given that the IV representing IL-6R blockade was weighted by CRP, the odds ratios (ORs) were proportional to the natural logarithm of the decrease in CRP.
The strength of IVs was evaluated using the F-statistic, with a value greater than 10 indicating no weak instrument bias23. Heterogeneity was assessed using Cochran’s Q statistics based on IVW and MR Egger methods24. In cases of heterogeneity, the inverse variance weighted multiplicative random effects (IVW-MRE) model was employed for MR Analysis25. Otherwise, the inverse variance weighted fixed effects (IVW-FE) model is applied. MR-Egger intercept test was used to detect pleiotropy, MR-PRESSO was used to identify and remove outliers, and leave-one-out sensitivity test was used to assess whether individual SNPs had a significant effect on the overall results.
All statistical analyses were performed using R 4.3.2 and the R package (TwoSampleMR and MR-PRESSO). To correct for bias due to multiple comparisons, P values were adjusted through Bonferroni correction with a significance threshold set at P < 0.01 (0.05/5 outcomes)26.
Results
Selection of IVs
After screening IVs as described above, a total of 44 SNPs were included for the MR Analysis between IL-6R blockade and CVDs and 10 SNPs were included for the repeated MR Analysis (Supplementary Table 2 and Supplementary Table 3). The F-statistics of the SNPs related to IL-6R blockade screened in this study ranged from 31.614 to 1002.100, and the F-statistics of the SNPs in the repeated analysis ranged from 38.554 to 744.476, indicating that the selected IVs were all highly associated with exposure.
Positive control analysis
As expected, the results of IVW-MRE showed that IL-6R blockade significantly reduced the risk of RA (OR = 0.407, 95% CI: 0.299 - 0.554, P = 1.084 × 10−8).
MR Analysis between IL-6R blockade and CVD
The IVW results showed that, after applying the Bonferroni correction (Bonferroni-corrected significance level: P-value < 0.01), IL-6R blockade had a significant effect on CHD (OR = 0.757, 95% CI: 0.690 - 0.832, P = 5.804 × 10–9) and MI (OR = 0.840, 95% CI: 0.744 - 0.949, P = 0.005) (Fig. 2 and Supplementary Table 4). However, IL-6R blockade had no significant effect on HTN (OR = 1.015, 95% CI: 0.950 - 1.084, P = 0.663), AF (OR = 0.905, 95% CI: 0.800 - 1.025, P = 0.116) and HF (OR = 1.012, 95% CI: 0.921 - 1.113, P = 0.805) (Supplementary Table 4). Furthermore, similar conclusions were drawn when repeating the analysis with the five CVDs using another GWAS dataset (Fig. 3 and Supplementary Table 5).
Results of MR analysis between IL-6R blockade and CVDs. Nsnp, number of single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; WME, weighted median; IVW-FE, inverse variance weighted fixed effects; IVW-MRE, inverse variance weighted multiplicative random effects; SM, simple mode; WM, weighted mode. This figure shows the results of the MR analysis, namely that the IL-6R blockade has a protective effect on CHD and MI, but has no significant effect on HTN, AF and HF. RA is used as a positive control. The IVW is the primary analysis method. When heterogeneity is detected, the IVW-MRE method is used to reduce bias, otherwise, the IVW-FE method is applied. MR Egger, WM, SM, and WM serve as supplementary methods. Significance is determined by a Bonferroni-corrected IVW p-values < 0.01 and consistent OR directions across methods. In this picture, the dots represent the OR values and the line segments represent the 95% confidence intervals. An OR less than 1 represents a protective effect of IL-6R blockade, and an OR greater than 1 represents a risk effect of IL-6R blockade.
Results of MR analysis between IL-6R blockade and CVDs for repeated analysis. Nsnp, number of single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; WME, weighted median; IVW-FE, inverse variance weighted fixed effects; IVW-MRE, inverse variance weighted multiplicative random effects; SM, simple mode; WM, weighted mode. This figure shows the results of the repeated MR analysis after changing the data source of CRP, namely that the IL-6R blockade has a protective effect on CHD and MI, but has no significant effect on HTN, AF and HF. RA is used as a positive control. The IVW method is the primary analysis method. When heterogeneity is detected, the IVW-MRE method is used to reduce bias, otherwise, the IVW-FE method is applied. The MR Egger, WME, SM, and WM serve as supplementary methods. Significance is determined by a Bonferroni-corrected IVW p-values < 0.01 and consistent OR directions across methods. The dots represent the OR values and the line segments represent the 95% confidence intervals, where an OR less than 1 suggests a protective effect of IL-6R blockade, and an OR greater than 1 indicates an increased risk.
Sensitivity analysis
The MR-Egger intercept test indicated no horizontal pleiotropy in all the results of this study (P > 0.05). The Cochran’s Q test revealed heterogeneity in the MR analyses between IL-6R blockade and RA (IVW: Q = 25.713, P = 0.028), CHD (IVW: Q = 66.963, P = 0.011), HTN (IVW: Q = 66.109, P = 0.013), AF (IVW: Q = 76.512, P = 3.691 × 10−7) and MI (IVW: Q = 66.613, P = 0.012) . In the repeated analysis, heterogeneity was detected between IL-6R blockade and RA (IVW: Q = 18.266, P = 0.019), so we performed the MR analyses using the IVW-MRE method to eliminate bias caused by heterogeneity27. However, in the remaining MR analyses, we did not detect the existence of heterogeneity. Therefore, we used the IVW-FE method. The MR-PRESSO test results showed that outliers existed in the MR analyses between IL-6R blockade and RA (rs12083537 and rs2274988), and AF (rs116710125). In the repeated analysis, we detected outliers in the MR analyses of the IL-6R blockade and RA (rs10752641), CHD (rs11265608), AF (rs4129267), and MI (rs11265608). Therefore, we repeated the MR analysis after removing these outliers to ensure the robustness of the results. The results of sensitivity analysis are presented in Supplementary Table 6 and Supplementary Table 7. The leave-one-out analysis demonstrated that no individual SNP had a significant effect on the overall results across MR analyses between IL-6R blockade and RA, CHD, and MI (Fig. 4 and Fig. 5). In conclusion, it can be stated that the MR results of this study are robust.
The leave-one-out analysis of IL-6R on RA and CVDs. (A)RA, (B) HTN, (C) CHD, (D) MI, (E) AF, (F) HF. This figure presents the leave-one-out sensitivity analysis of IL-6R on RA and CVDs, namely that in the MR analyses between IL-6R blockade and RA, CHD, and MI, no individual SNP has a significant influence on the overall results. Each plot displays the results of excluding one SNP at a time to test the robustness of the findings. The x-axis represents the MR leave-one-out sensitivity analysis estimates, while the y-axis lists the SNPs. The black dots indicate the effect estimates for each leave-one-out analysis, with horizontal lines showing the 95% confidence intervals. The red line represents the overall estimate without excluding any SNPs.
The leave-one-out analysis of IL-6R on RA and CVDs for repeated analyses. (A) RA, (B) HTN, (C) CHD, (D) MI, (E) AF, (F) HF. This figure displays the results of a repeated leave-one-out sensitivity analysis with alternative datasets, that is, in the repeated MR analyses between IL-6R blockade and RA, CHD and MI, no individual SNP had a significant influence on the overall results. In each plot, individual SNPs are excluded one at a time to assess the robustness of the results. The x-axis represents the MR leave-one-out sensitivity analysis estimates, while the y-axis lists the SNPs. The black dots indicate the effect estimates for each leave-one-out analysis, with horizontal lines representing the 95% confidence intervals. The red line represents the overall estimate without excluding any SNPs.
Discussion
The present study investigated the potential role of IL-6R blockade in the prevention and treatment of five types of CVD, including HTN, CHD, MI, AF, and HF, using MR methods. The results showed that the drug had a significant effect on the treatment of CHD and MI but had no obvious effect on the treatment of HTN, AF and HF. These findings provide an important basis for the clinical application of IL-6R blockade in the treatment of CVD and offer theoretical support and practical evidence for utilizing genetic information to guide precise CVD treatment.
First, the potential therapeutic effects of IL-6R blockade on CHD and MI may be related to their anti-inflammatory properties. Inflammation plays a crucial role in the occurrence and development of atherosclerosis as well as its related CVDs. The study by Held C et al.28 demonstrated that IL-6 was independently associated with the risk of major coronary events (hazard ratio (HR) 1.60, 95% CI: 1.30 - 1.97, P < 0.0001) and MI (HR: 1.53, 95% CI: 1.14 - 2.04, P < 0.05), and suggested that anti-inflammatory drugs targeting IL-6 might be a potential future target for the treatment of stable coronary heart disease. This result is consistent with our observation in this study that IL-6R blockade has a protective effect on the treatment of CHD and MI. Besides, previous studies have indicated that IL-6 is closely associated with the formation and progression of atherosclerotic plaques29,30. IL-6 can not only promote macrophage uptake of low-density lipoprotein leading to accelerated lipid particle deposition and foam cell formation but also enhance CD44 expression in macrophages while promoting positive feedback secretion of IL-6 by macrophages. This further exacerbates inflammatory responses by increasing vascular endothelial growth factor (VEGF) production resulting in intravascular atherosclerotic plaque formation. Therefore, we hypothesized that inhibition of IL-6 signaling by targeting IL-6R may reduce inflammatory responses, slow the progression of atherosclerosis, and have therapeutic effects on CHD and MI.
The results of this study indicated that there was no significant causal relationship between IL-6R blockade and AF. However, the study by Jing et al.31 showed that inhibiting IL-6 gene expression could significantly improve myocardial infarction-induced atrial fibrosis and myocardial remodeling. Gupta S et al.32 reported a reduced risk of AF in COVID-19 patients treated with tocilizumab compared to those not treated (9.0% vs 14.5%). Although some previous studies suggested that IL-6R blockade was associated with a reduced risk of AF, most of these studies were observational studies, which might be affected by reverse causality and potential confounding factors and could not provide definite causal inferences. Our MR analysis showed that there was no significant causal relationship between IL-6R blockade and AF, suggesting that the association observed in previous studies might only be a correlation rather than a causal relationship. However, since MR analysis also has its limitations, we believe that future studies require larger sample sizes and higher statistical power to explore the causal relationship between IL-6R and AF in a more powerful way.
Besides, this study failed to demonstrate a significant therapeutic effect of IL-6R blockade for HTN and HF. This may be due to the fact that the role of IL-6 in HTN and HF is not as direct or critical as it is in CHD and MI. For example, the development of HTN involves various mechanisms such as the renin-angiotensin system, sympathetic nerve activity, and vascular smooth muscle cell function33, suggesting that IL-6 may not play a major regulatory role in this process. This finding was supported by Gonzalez34, who observed that loss of IL-6 reduced myocardial inflammation and fibrosis, thereby improving cardiac function but did not prevent the development of HTN. Similarly, HF also involves complex mechanisms including abnormal calcium circulation, activation of neurohumoral system, myocardial cell injury, inflammatory response and ventricular remodeling35. These processes may be more regulated by other cytokines and signaling pathways. Additionally, studies on the correlation between IL-6R blockade and HF are limited and require further exploration in future research.
At the same time, this study has certain limitations. First of all, all the data in the study were from populations of European ancestry. Although this reduced the bias due to ethnic and regional differences, it also limited the generalizability of the study results to other ethnic groups. Therefore, our results are mainly applicable to populations of European ancestry, and the applicability to other ethnic groups is unknown. Secondly, the MR results rely on three assumptions, which may not always be fully achievable. Although we adopted various sensitivity analysis methods in the study, potential horizontal pleiotropy and potential confounding factors may still exist. Moreover, due to the limited number of SNPs available for analysis under more stringent thresholds, we chose a relatively relaxed R2 threshold of < 0.3. Although a few previous studies have employed a similar threshold, this is not a standard approach and may risk inflating results. Future studies should consider employing larger datasets that allow for the application of more stringent thresholds without compromising on the number of SNPs available for analysis. In addition, although we used multiple data sources to improve the robustness of the analysis and repeated MR analyses showed consistent results, future studies still require larger-scale clinical trials and in-depth mechanistic studies to verify and expand our findings. Finally, our study results are also limited by the coverage and quality of the existing GWAS datasets. With the increase in the availability of more high-quality genetic data from diverse populations, we will be able to estimate the impact of genetic variations on the risk of CVDs more accurately.
Conclusions
In summary, our study provides evidence from genetic proxies for the potential benefit of IL-6R blockade in the treatment of CHD and MI. At the same time, it also suggests that other therapeutic targets may need to be explored in the treatment of HTN, AF and HF. Future studies should further investigate the therapeutic mechanism of IL-6R blockade in different CVDs and evaluate their safety and efficacy in clinical application.
Data availability
The datasets supporting this study are available from GWAS Catalog (https://www.ebi.ac.uk/gwas/), FinnGen consortium (https://www.finngen.fi/en) and the IEU Open GWAS project (https://gwas.mrcieu.ac.uk/). Corresponding author Xinyu Chen will provide the data upon reasonable request.
References
Roth, G. A. et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. Coll. Cardiol. 70(1), 1–25 (2017).
Ferdinandy, P. et al. Interaction of cardiovascular nonmodifiable risk factors, comorbidities and comedications with ischemia/reperfusion injury and cardioprotection by pharmacological treatments and ischemic conditioning. Pharmacol. Rev. 75(1), 159–216 (2023).
Frąk, W. et al. pathophysiology of cardiovascular diseases: new insights into molecular mechanisms of atherosclerosis, arterial hypertension, and coronary artery disease. Biomedicines 10(8), 1938 (2022).
Suzuki, K. Chronic inflammation as an immunological abnormality and effectiveness of exercise. Biomolecules 9(6), 223 (2019).
Prabhu, S. D. & Frangogiannis, N. G. the biological basis for cardiac repair after myocardial infarction: From inflammation to fibrosis. Circ. Res. 119(1), 91–112 (2016).
Del Pinto, R. & Ferri, C. Inflammation-accelerated senescence and the cardiovascular system: Mechanisms and perspectives. Int. J. Mo.l Sci. 19(12), 3701 (2018).
Ruparelia, N. & Choudhury, R. Inflammation and atherosclerosis: What is on the horizon. Heart 106(1), 80–85 (2020).
Hunter, C. A. & Jones, S. A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 16(5), 448–457 (2015).
Ridker, P. M. & Rane, M. Interleukin-6 signaling and anti-interleukin-6 therapeutics in cardiovascular disease. Circ. Res. 128(11), 1728–1746 (2021).
Kaptoge, S. et al. C-reactive protein, fibrinogen, and cardiovascular disease prediction. N. Engl. J. Med. 367(14), 1310–1320 (2012).
Rose-John, S. Interleukin-6 Family Cytokines. Cold Spring Harb. Perspect Biol. 10(2), a028415 (2018).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506(7488), 376–381 (2014).
Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2021. 397(10285) 1637–1645.
Davies, N. M., Holmes, M. V. & Davey, S. G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. 362, k601 (2018).
Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53(10), 1415–1424 (2021).
Hu, X. et al. Identification of lipid-modifying drug targets for autoimmune diseases: Insights from drug target mendelian randomization. Lipids Health Dis. 23(1), 193 (2024).
Su, Y., Zhang, Y. & Xu, J. Genetic variations in anti-diabetic drug targets and COPD risk: Evidence from mendelian randomization. BMC Pulm. Med. 24(1), 240 (2024).
Duan, H. et al. Causal relationship between PCSK9 inhibitor and primary glomerular disease: A drug target Mendelian randomization study. Front. Endocrinol. (Lausanne). 15, 1335489 (2024).
Huang, A., Wu, X., Lin, J., Wei, C. & Xu, W. Genetic insights into repurposing statins for hyperthyroidism prevention: A drug-target Mendelian randomization study. Front. Endocrinol. (Lausanne). 15, 1331031 (2024).
Birney, E. Mendelian Randomization. Cold Spring Harb. Perspect Med. 12(4), a041302 (2022).
Ligthart, S. et al. Genome analyses of >200,000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am. J. Hum. Genet. 103(5), 691–706 (2018).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613(7944), 508–518 (2023).
Sanderson, E. et al. Mendelian randomization. Nat. Rev. Methods Prim. 2, 6 (2022).
Greco, M. F. D., Minelli, C., Sheehan, N. A. & Thompson, J. R. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat. Med. 34(21), 2926–2940 (2015).
Bowden, J. et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int. J. Epidemiol. 47(4), 1264–1278 (2018).
Sedgwick, P. Multiple hypothesis testing and Bonferroni’s correction. BMJ. 349, g6284 (2014).
Papadimitriou, N. et al. Physical activity and risks of breast and colorectal cancer: A Mendelian randomisation analysis. Nat. Commun. 11(1), 597 (2020).
Held, C. et al. Inflammatory biomarkers interleukin-6 and C-reactive protein and outcomes in stable coronary heart disease: Experiences from the STABILITY (Stabilization of atherosclerotic plaque by initiation of darapladib therapy) trial. J Am Heart Assoc. 6(10), e005077 (2017).
Tardif, J. C. et al. Efficacy and safety of low-dose colchicine after myocardial infarction. N. Engl. J. Med. 381(26), 2497–2505 (2019).
Okazaki, S. et al. Association of interleukin-6 with the progression of carotid atherosclerosis: A 9 year follow-up study. Stroke. 45(10), 2924–2929 (2014).
Jing, R., Long, T. Y., Pan, W., Li, F. & Xie, Q. Y. IL-6 knockout ameliorates myocardial remodeling after myocardial infarction by regulating activation of M2 macrophages and fibroblast cells. Eur. Rev. Med. Pharmacol. Sci. 23(14), 6283–6291 (2019).
Gupta, S. et al. Association between early treatment with tocilizumab and mortality among critically Ill patients with COVID-19. JAMA Intern. Med. 181(1), 41–51 (2021).
Guzik, T. J. & Touyz, R. M. Oxidative stress, inflammation, and vascular aging in hypertension. Hypertension 70(4), 660–667 (2017).
González, G. E. et al. Deletion of interleukin-6 prevents cardiac inflammation, fibrosis and dysfunction without affecting blood pressure in angiotensin II-high salt-induced hypertension. J. Hypertens. 33(1), 144–152 (2015).
Braunwald, E. Heart failure. JACC Heart Fail. 1(1), 1–20 (2013).
Acknowledgements
We sincerely thank the investigators who provided the summary data on GWAS.
Funding
This study was funded by National Natural Science Foundation of China (81704061, 81173213), the National Famous Elderly Chinese Medicine Expert Chen Xinyu Inheritance Workshop Construction Project [National Chinese Medicine Human Education Letter (2022) No. 75], the R & D Plan for Key Areas of Hunan Provincial Department of Science and Technology (2019SK2321) and the Hunan Science and Technology Talent Hosting Project (2020TJ-N01).
Author information
Authors and Affiliations
Contributions
G.O., H.C. and K.Y designed the study. Z.Q., Y.Y. and Y.C. analyzed and interpreted the data. G.O. and X.C. wrote the manuscript and revised the manuscript. All authors approve of the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ou, G., Cai, H., Yao, K. et al. Exploring the therapeutic potential of interleukin-6 receptor blockade in cardiovascular disease treatment through Mendelian randomization. Sci Rep 14, 21452 (2024). https://doi.org/10.1038/s41598-024-72195-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-72195-4