Abstract
SASH1 (SAM and SH3 domain containing 1) has been increasingly reported as a tumor suppressor gene. However, there is limited research on the role of SASH1 in breast cancer. This manuscript aims to investigate the mechanism of SASH1 in the occurrence, development, and prognosis of breast cancer. Firstly, we obtained RNA-sequencing data of the tumors from the Genomic Data Commons data portal website, along with the corresponding clinical information of patients. Pan-cancer analysis was performed to analyze the expression of SASH1 across all tumors. Univariate Cox regression analysis was used to assess the correlation between SASH1 expression and the prognosis of breast cancer patients. Then, immunohistochemistry was utilized to evaluate the expression levels of SASH1, p-Akt, p-PI3K, and p-mTOR in breast cancer tissue. Finally, a cell assay was employed to analyze the impact of SASH1 on the proliferation and invasion of breast cancer cells (MDA-MB-231). The results revealed that SASH1 expression is decreased in BRCA, LUSC, LUAD, CESC, ESCA, and COAD. Meta-analysis also found that SASH1 is downregulated in most tumor tissues, and the expression level of SASH1 in breast cancer was significantly lower than that in the control group (OR = 0.14, 95% CI = 0.08–0.25; P < 0.001). Further experimental validation showed that SASH1 expression is significantly downregulated in breast cancer tissue (38.33%, 23/60), and the overexpression of SASH1 can inhibit the proliferation and invasion of breast cancer cells accompanied by the suppression of PI3K-Akt-mTOR signaling pathway. Additionally, SASH1 overexpression can improve OS and RFS of breast cancer patients.
Similar content being viewed by others
Introduction
The development of tumors is a complex, multistage process influenced by various factors1. Key among these are the activation of proto-oncogenes and the inactivation of tumor suppressor genes1. Breast cancer, which originates from the epithelial tissue of the breast, is the most common malignancy affecting women, particularly those between the ages of 40 and 60 or around the peri-menopausal period2. Statistics show that the incidence of breast cancer has been steadily increasing in recent years3. While numerous factors can contribute to its development, genetic factors account for less than 10% of cases, leaving the mechanisms behind the remaining 90% still largely unknown4.
SAM and SH3 domain containing 1 (SASH1) is located on chromosome 6q24.3 and has a total length of 7709 bp5. It encodes 1247 amino acids and contains one SH3 domain and two SAM domains6. SASH1 belongs to the SLY family, which includes other protein members such as SAMSN1 and SASH36. SAMSN1 primarily functions in B-cell activation, while SASH3 is involved in the specific immune response of organisms7. Notably, SASH1 expression is reduced in various malignant tumors, such as breast, lung, gastric, and thyroid cancers, and its levels are closely linked to tumor proliferation, migration, and prognosis8. Current studies highlight a significant connection between SASH1 and tumor progression9. Yang et al.10 confirmed that low expression of SASH1 not only promotes the occurrence of gliomas but also is closely related to the grading of gliomas. In liver cancer and thyroid tumors, the mRNA expression level of SASH1 is notably decreased11. Studies have shown that SASH1 protein expression is significantly downregulated in 74% of breast cancer patients12. Gong et al.13 demonstrated that, compared to the control group, the positivity rate of SASH1 protein in breast cancer patients is significantly reduced (63/186, 33.9%). Our previous studies have also confirmed that low expression of SASH1 is significantly associated with the occurrence of breast cancer4. These findings suggest that SASH1 may play a crucial role in the initiation and progression of breast cancer.
The phosphoinositide 3-kinase (PI3K)-protein kinase B (Akt)-mammalian target of rapamycin (mTOR) signaling pathway relays signals from cell membrane receptors to the cytoplasm14. In-depth research on cancer molecular biology has revealed that the PI3K-Akt-mTOR signaling pathway is intimately linked with the initiation, progression, and metastasis of tumors14. Abnormal expression of this pathway has been detected in various tumors, including squamous cell carcinoma, prostate cancer, and liver cancer15. Activation of PI3K triggers the activation of Akt, which becomes phosphorylated to form p-Akt, subsequently regulating downstream gene expression, and mediating tumor cell proliferation and apoptosis16. Miao et al.17 discovered that PI3K-Akt-mTOR expression levels are significantly elevated in breast cancer, facilitating tumor proliferation and drug resistance. Pan et al.18 found that SASH1 can inhibit the proliferation and migration of pancreatic cancer cells by blocking the expression of the PI3K-Akt-mTOR signaling pathway. Nonetheless, there is currently insufficient research to confirm whether an interaction exists between SASH1 and the PI3K-Akt-mTOR signaling pathway in breast cancer. Examining the function and molecular mechanisms of the SASH1 gene could not only shed light on its role in tumor initiation and progression but also lay the groundwork for targeting it in breast cancer treatment9,19. Consequently, this study aims to investigate the role of SASH1 in the onset, development, and prognosis of breast cancer through multiple approaches, including clinical samples, cell experiments, and meta-analysis.
Materials and methods
Reagents
Immunohistochemical detection reagents were purchased from OriGene Biotechnology Co., Ltd. (Wuxi, China). PCR primers synthesis and sequencing were performed by GenScript Biotechnology Co., Ltd. (Nanjing, China). Rabbit anti-human SASH1 polyclonal antibodies (cat. no. NBP-26650) were purchased from Novus (USA). Mouse monoclonal GAPDH antibody (cat. no. 97166), Akt total protein mouse monoclonal (cat. no. 2920) and Akt phosphoprotein rabbit monoclonal (cat. no. 4060) antibodies, PI3K total protein mouse monoclonal (cat. no. 13666) and PI3K phosphoprotein rabbit monoclonal (cat. no. 4228) antibodies, mTOR total protein mouse monoclonal (cat. no. 4517) and mTOR phosphoprotein rabbit monoclonal (cat. no. 5536) antibodies were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). TRIzol® reagent was purchased from Takara Biotechnology Co., Ltd. (Dalian, China); Cell protein extraction and western blot detection reagents RIPA lysis and extraction buffer were purchased from Beyotime Biotechnology Co., Ltd. (Shanghai, China).
Patients and Cell lines
Breast cancer tissue specimens were obtained from 60 patients who underwent surgical resection at Yongchuan People's Hospital of Chongqing from April 2023 to April 2024. All specimens were confirmed by pathological examination (HE staining) for diagnosis. The patients' ages ranged from 37 to 75 years old. This study received approval from the Ethics Committee of the Yongchuan People's Hospital of Chongqing. MDA-MB-231 breast cancer cells were purchased from the Cell Resource Center, Shanghai Institute for Biological Sciences (Chinese Academy of Sciences).
Cell culture and infection
MDA-MB-231 cells were cultured in L15 culture medium containing 10% fetal bovine serum (FBS) (including 1% penicillin and streptomycin) at 37 °C, in the absence of CO2. MDA-MB-231 cells in the exponential phase of growth were seeded at a density of 2 × 106/culture flask. After 24 h, the cells were subsequently infected with adenovirus vehicles that express pcDNA3.1 and pcDNA3.1-SASH1.
Immunohistochemistry
The breast cancer tumor tissues and adjacent normal tissues were embedded in paraffin and cut into 5 μm thick sections. Drip 3% H2O2 deionized water to cover the tissue and inactivate endogenous enzymes. Normal goat serum blocking solution was then added dropwise (at room temperature, 30 min), followed by the addition of rabbit anti-human SASH1, p-PI3K, p-Akt, and p-mTOR polyclonal antibodies (at 37 °C, 1 h), and incubated overnight at 4 °C. Biotin-labeled ready-to-use goat anti-rabbit IgG was added dropwise (at 37 °C, 30 min). Horseradish peroxidase-labeled streptavidin working solution was added dropwise (at 37 °C, 15 min), followed by DAB chromogenic staining. Finally, neutral resin was used for sealing, and positive results were observed and photographed under an optical microscope20.
Cell proliferation assay, wound-closure assay and transwell invasion assay
Cell proliferation was analyzed by Cell Counting Kit-8 (CCK-8) assay as described previously. The absorbance was measured daily for 5 days at 450 nm using a microplate reader. Each experiment was done in quintuplicate, and repeated three times. Then, a growth curve was delineated21.
The wound closure assay was performed as previously described. Cell migration was monitored by microscopy 24 h later and reported as wound healing rate. Wound healing rate = (initial scratch area − 24 h initial scratch area)/ initial scratch area. The experiment was repeated three times22.
The transwell invasion assay was performed as previously described. After 24 h, the cells migrating through the filter were dried for 10 min, stained with hematoxylin and eosin. Cells on the lower chamber were counted under a microscope. The experiment was repeated three times22.
RT‑qPCR
When the cells cover approximately 80% of the culture dish area, total RNA will be extracted from the breast cancer cells in each group using the TRIzol® method. Subsequently, the mRNA expression levels of SASH1 in each group will be detected using qPCR. Specifically, 2 μg of total RNA will be used to synthesize cDNA using a reverse transcription kit. Then, 1 μL of cDNA will be used as a template for PCR amplification. The following primer sequences were used: SASH1, sense, 5′-TCCCGTCACAGGAAGAAACG′-3 and antisense, 5′- GATACCCATCACGTCGGTCC-3′.
Western blotting
Different recombinant cells were collected, and the western blotting assay was performed as usual. The ECL chromogenic solution was used for imaging. GEL EQ (Bio-Rad Laboratories, Hercules, CA, USA) was used in the gel imager, and Quantity One® software (Bio-Rad Laboratories, Inc.) was used to analyze the relative optical densities. The relative expression level of the target gene protein was calculated according to the gray value of the target gene band/the gray value of the internal reference band20.
Data acquisition and pre-processing
To download and organize the RNA-sequencing data of the TCGA-ALL project with the STAR pipeline from the TCGA database (https://portal.gdc.cancer.gov) and extract the data in FPKM format for corresponding paired tumor and normal samples. The following tumor types were included in this study: bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical and endocervical cancer (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC)23.
Expression level and prognostic evaluation
R software was used for statistical analysis. We downloaded two breast cancer datasets (GSE22820 and GSE42568) from the GEO database. The ‘normalizeBetweenArrays’ function from the limma package was used to normalize the data. Patients were divided into a breast cancer group and a healthy control group. A linear model combined with the Empirical Bayes approach in the limma package was used to perform differential analysis between the two groups. To assess the relationship between SASH1 gene expression and various survival outcomes in breast cancer, univariate Cox regression analysis was employed. Specifically, this analysis evaluated the correlation between SASH1 gene expression and four key survival metrics: overall survival (OS), distant metastasis-free survival (DMFS), recurrence-free survival (RFS), and post-progression survival (PPS). According to the median value of SASH1 gene expression, the patients were divided into SASH1 high expression group and SASH1 low expression group. The Kaplan–Meier method was employed to construct survival curves, providing a visual representation of survival outcomes. Subsequently, the log-rank test was utilized to analyze the differences in survival status between groups. To assess the expression of the SASH1 gene between tumor and normal tissue, a t-test was employed. P < 0.05 was considered statistically significant.
Statistical methods
SPSS version 25.0 (IBM Corp.) was used for processing clinical and experimental data. For the analysis of immunohistochemical results, all data were divided into the breast cancer tissue group and the adjacent normal tissue group. The chi-square test was used to analyze whether there is differential expression of SASH1 in breast cancer tissues. Spearman's correlation analysis was used to evaluate the correlation between SASH1 and PI3K, Akt, and mTOR. For cell experiments, we categorized the MDA-MB-231 breast cancer cells into three groups: Blank, pcDNA3.1, and pcDNA3.1-SASH1, unpaired student's t‑test was employed perform the statistical analysis. For meta-analysis, the STATA 12.0 software (StataCorp LLC) was used to perform statistic analysis. When two or more articles examined the association between SASH1 expression and the risk of cancers, statistical analyses were performed. In cases where there were fewer than two relevant articles, descriptive analyses were conducted instead. The Chi-square test and I2 test were used to assess heterogeneity among studies. To pool the data, when I2 test ≥ 50 or q ≤ 0.1, significant heterogeneity between studies is considered, and random effects model (REM) is adopted. When I2 < 50 or q > 0.1, there was no significant heterogeneity between the studies and fixed effects model (FEM) is adopted. P > 0.05 was considered to have no publication bias.
Results
SASH1 expression is decreased in cancer
We observed that SASH1 is commonly downregulated across various tumor types. Among the 23 types of tumors related to the expression level of SASH1 gene in the TCGA database, 16 types of tumors have statistically significant differences in SASH1 gene expression compared with normal tissues (all P < 0.05). SASH1 showed low expression levels in BLCA, BRCA, COAD, ESCA, HNSC, KICH, KIRC, KIRP, LUAD, LUSC, READ, STAD, THCA, and UCEC, whereas it exhibited higher expression levels in CHOL and PRAD (Fig. 1). In addition, we conducted a meta-analysis to explore the role of SASH1 in tumorigenesis. A total of 253 articles were identified. Following the elimination of duplicate studies and the application of the specified criteria, 16 articles were dedicated to exploring the correlation between the expression levels of SASH1 and eight distinct types of cancer11,13,24,25,26,27,28,29,30,31,32,33,34,35,36,37 (Fig. 2; Table 1). Specifically, among these 16 articles, 5 studies examined the correlation between SASH1 expression levels and the risk of breast cancer 13,24,25,26,27 (Fig. 3; Table 1). After statistical analysis, it was found that the expression level of SASH1 in tumor tissue was significantly lower than that in the control group (OR = 0.16, 95% CI = 0.09–0.28; P < 0.001), with the difference being statistically significant (Fig. 3). We conducted a stratified analysis according to the type of tumor. The results showed that only breast cancer related SASH1 studies were relatively more, and other tumor related SASH1 studies were few (less than or equal to 2 studies) (Fig. 3). So, the association between the SASH1 expression level and the risk of breast cancer was analyzed in five datasets. It was found that the expression level of SASH1 in breast cancer was significantly lower than that in control group (OR = 0.14, 95% CI = 0.08–0.25; P < 0.001), with the difference being statistically significant (Fig. 3; Table 2).
The expression level of SASH1 gene in tumors and their corresponding normal tissues. bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical and endocervical cancer (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC).
Furthermore, the analysis also revealed that elevated expression levels of SASH1 in lung cancer, ovarian cancer, gastric cancer, esophageal cancer, cervical cancer, and osteosarcoma were associated with a suppression of tumor occurrence compared to normal tissues. Conversely, high expression of SASH1 in colon cancer was associated with the promotion of tumor occurrence compared to normal tissues (Fig. 3; Table 2). It is exciting to note that this finding is consistent with the conclusion of our pan-cancer. However, it's noteworthy that the available clinical information on the role of SASH1 in these malignancies is currently insufficient. Consequently, additional research is warranted to delve deeper into the role of SASH1 in these types of cancer.
SASH1 is differentially expressed in breast cancer
The cBioPortal was employed to assess the frequency of the genetic alteration of SASH1 in breast cancer patients. The findings indicated that genetic alterations of SASH1 were observed in 6% of breast cancer patients. Interestingly, the genetic alterations of SASH1 is primarily characterized by decreased mRNA levels. This further indicates that the expression of SASH1 is down in breast cancer (Fig. 4A).
The rate of SASH1 mutations in breast cancer. (A), the mutation rate of SASH1 is relatively high in breast cancer. (B), the differential expression of SASH1 in breast cancer was observed in both breast cancer datasets. Patients were divided into a breast cancer group and a healthy control group. A linear model combined with the Empirical Bayes approach in the limma package was used to perform differential analysis between the two groups. (C), SASH1 has a high predictive value for the risk of breast cancer occurrence. SASH1, SAM—and SH3 domain containing 1.
We downloaded two breast cancer datasets (GSE22820 and GSE42568) from the GEO database. After obtaining the differentially expressed genes from both datasets and taking the intersection, we found 985 common differentially expressed genes. Additionally, SASH1 showed differential expression in both datasets (Fig. 4B). ROC analysis of SASH1 expression data yielded an AUC (Area Under the Curve) value of 0.915, demonstrating that SASH1 is highly accurate in predicting the risk of breast cancer (Fig. 4C).
The expression levels of SASH1 are negatively correlated with p-Akt, p-PI3K, and p-mTOR in breast cancer tissues
In the 60 cases of breast cancer tissues and their paracancerous tissues, immunohistochemical results revealed that the expression level of SASH1 in tumor tissues was 38.33% (23/60), while in paracancerous tissues, it was 70% (42/60). The expression level of SASH1 in tumor tissues was significantly lower than that in paracancerous tissues (P < 0.05). Furthermore, compared to paracancerous tissues, the expression levels of p-Akt (39/60, 65.00%), p-PI3K (44/60, 73.33%), and p-mTOR (46/60, 76.67%) were significantly increased in tumor tissues (Fig. 5A–C). We performed a correlation analysis on the expression levels of SASH1, p-Akt, p-PI3K, and p-mTOR in tumor tissues. The results showed a significant negative correlation between SASH1 and p-Akt (r = − 0.447, P < 0.01), p-PI3K (r = − 0.385, P < 0.01), and p-mTOR (r = − 0.390, P < 0.01) (Fig. 5B).
The expression levels of SASH1 are negatively correlated with p-Akt, p-PI3K, and p-mTOR in breast cancer tissues. (A, C), The expression levels of SASH1, p-Akt, p-PI3K, and p-mTOR in breast cancer tissues. (B), The correlation analysis between SASH1 and p-Akt, p-PI3K, and p-mTOR. SASH1, SAM—and SH3 domain containing 1; PI3K, phosphoinositide 3-kinase; Akt, protein kinase B; mTOR, mammalian target of rapamycin.
SASH1 can inhibit the proliferation and invasion of MDA-MB-231 cells
The wound closure assay and transwell invasion assay were utilized to explore the impact of SASH1 on the migration and invasion of MDA-MB-231 cells. The results indicated, that compared to MDA-MB-231/Blank cells (80.15%) and MDA-MB-231/pcDNA3.1 cells (80.34%), the wound healing rate of MDA-MB-231/pcDNA3.1-SASH1 cells (17.31%) was significantly reduced (P < 0.05) (Fig. 6A,B). Additionally, the invasion assay revealed that the invasive ability of MDA-MB-231/pcDNA3.1-SASH1 cells was significantly lower compared to that of MDA-MB-231/pcDNA3.1 cells (P < 0.05) (Fig. 6C,D). These findings suggest that SASH1 can inhibit the migration and invasion of breast cancer cells.
The overexpression of SASH1 can inhibit the proliferation, migration, and invasion abilities of MDA-MB-231 cells. (A–D), The wound-closure assay and transwell invasion assay shown that SASH1 can inhibit the migration and invasion of MDA-MB-231 cells. (E), CCK8 assay shown that SASH1 can inhibit the proliferation of MDA-MB-231 cells. SASH1, SAM—and SH3 domain containing 1.
The CCK-88 assay was utilized to explore the impact of SASH1 on the proliferation of MDA-MB-231 cells. The results revealed that, starting from the 3rd day, the proliferative capacity of MDA-MB-231/pcDNA3.1-SASH1 cells was significantly lower compared to that of MDA-MB-231/Blank cells and MDA-MB-231/pcDNA3.1 cells. Overexpression of SASH1 significantly inhibited the proliferation of breast cancer MDA-MB-231 cells (P < 0.05). The proliferative capacity did not show a significant difference between MDA-MB-231/Blank cells and MDA-MB-231/pcDNA3.1 cells (Fig. 6E).
The overexpression of SASH1 can inhibit the PI3K-Akt-mTOR signaling pathway
In breast cancer tissues, we found that the expression levels of SASH1 were negatively correlated with the levels of p-Akt, p-PI3K, and p-mTOR, indicating that the overexpression of SASH1 can inhibit the PI3K-Akt-mTOR signaling pathway. To further validate this conclusion, we conducted cell experiments. Q-PCR results showed the successful construction of SASH1 overexpression cells (MDA-MB-231/pcDNA3.1-SASH1) (Fig. 7A). Western blotting results revealed that, compared to MDA-MB-231/Blank cells and MDA-MB-231/pcDNA3.1 cells, the expression levels of p-Akt, p-PI3K, and p-mTOR were significantly decreased in breast cancer cells with SASH1 overexpression (MDA-MB-231/pcDNA3.1-SASH1). This further supports the notion that SASH1 may regulate the progression of breast cancer via the PI3K-Akt-mTOR signaling pathway (Fig. 7B).
The overexpression of SASH1 can inhibit the PI3K-Akt-mTOR signaling pathway. (A), Q-PCR showed successful construction of SASH1 overexpression cells (MDA-MB-231/pcDNA3.1-SASH1). (B), The expression levels of p-Akt, p-PI3K, and p-mTOR were significantly decreased in breast cancer cells of SASH1 overexpression (MDA-MB-231/pcDNA3.1-SASH1). SASH1, SAM—and SH3 domain containing 1; PI3K, phosphoinositide 3-kinase (This study explores PIK3 p110); Akt, protein kinase B (This study explores AKt-Thr308 phosphorylation sites); mTOR, mammalian target of rapamycin (This study explores mTORC1).
High expression of SASH1 can improve OS, RFS, and reduce PPS of breast cancer patients
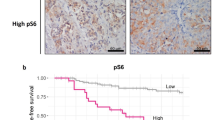
Through TCGA database, the data on the relationship between the expression level of SASH1 and the prognosis of breast cancer were downloaded, including three datasets (213236_at, 226022_at, and 41644_at) (Fig. 8). According to the median value of SASH1 gene expression, breast cancer patients were divided into a SASH1 high expression group and a SASH1 low expression group. Survival analysis found that high expression of SASH1 could increase OS (213236_at: HR = 0.75, 226022_at: HR = 0.75, and 41644_at: HR = 0.81) and RFS (213236_at: HR = 0.82, 226022_at: HR = 0.78, and 41644_at: HR = 0.83) of patients, and the difference is statistically significant (all P < 0.05) (Fig. 8). However, the high expression of SASH1 could reduce PPS (213236_at: HR = 1.45 and 41644_at: HR = 1.4) of patients, and the difference is statistically significant (all P < 0.05) (Fig. 8). High expression of SASH1 has no effect on the DMFS of patients (Fig. 8).
The relationship between the expression level of SASH1 and clinicopathological parameters in breast cancer patients
In the Breast Cancer Gene Expression Miner v4.9 database, clinicopathological data of breast cancer patients are available, including information from three datasets (213236_at, 226022_at, and 41644_at)38. We found that the expression level of SASH1 was related to age and SBR grading. The expression level of SASH1 increased in patients older than 51 years. The finding shows that SASH1 might improve the prognosis of elderly patients (> 51 years old). With the increase of SBR grading, the expression level of SASH1 gradually decreased, indicating that the higher the level of SASH1, the better the prognosis of breast cancer patients (Supplementary figs. 1, 2, 3).
Discussion
Despite being a member of the SLY family, SASH1's expression is limited in mature lymphocytes, indicating that it may have functions beyond the immune system39. Recent studies provide growing evidence that SASH1 acts as a tumor suppressor gene30,33. It is expressed in a variety of human tissues, but its expression is notably reduced or lost in solid tumors33,40. Reintroducing SASH1 has been shown to significantly suppress tumor cell growth, proliferation, and migration, suggesting that SASH1 could be a valuable therapeutic target with potential clinical applications in cancer therapy40,41.
Studies have demonstrated that SASH1 can inhibit cell proliferation41. An increase in SASH1 expression leads to a rise in cell adhesion proteins, which in turn inhibits cell migration5. On the other hand, reduced SASH1 expression diminishes cell adhesion capabilities and facilitates tumor cell migration19,42. Gong et al.13 discovered that the expression level of SASH1 in tumor tissue (33.9%, 63/186) was significantly lower than that in patients with breast ductal hyperplasia (62.0%, 31/50), and the expression of SASH1 was associated with edge burr sign and lymph node metastasis. In our study, we conducted a comprehensive analysis of SASH1 expression in various tumors. We found that SASH1 expression levels were decreased in tumors (OR = 0.16, 95% CI = 0.09–0.28; P < 0.001), indicating its role as a tumor suppressor gene. These findings were also supported by the results of the pan-cancer analysis and immunohistochemistry assay. Moreover, we further analyzed the impact of SASH1 on the progression of breast cancer and found that the expression level of SASH1 in breast cancer tissue was significantly reduced. Cell experiments also revealed that overexpression of SASH1 can significantly reduce the proliferation, migration, and invasion of breast cancer cells. We examined the correlation between SASH1 and clinical pathological parameters in breast cancer patients, uncovering a relationship between SASH1 expression and both age and SBR grade. Low SASH1 expression was correlated with a higher malignancy level in breast cancer. These studies collectively suggest that SASH1 is downregulated in tumor tissues and is associated with tumor cell proliferation and metastasis.
As a scaffold protein, SASH1 is located in both the cytoplasm and nucleus of cells, recruiting numerous signaling molecule proteins4,8. SASH1 can enhance LPS-induced cell migration by binding to TRAF6/TAK1/IKK/IKK, and it can also bind to MAP2K2/MAP4K4, thereby inhibiting ERK1/2 phosphorylation and suppressing tumor cell proliferation and migration8,27. The overexpression of SASH1 may involve multiple cellular signaling pathways, necessitating further research to explore its spatiotemporal expression characteristics27. In our prior studies, we discovered that SASH1 is downregulated in breast cancer4. SASH1 interacts with IQGAP1, and its decreased expression leads to reduced E-cadherin levels, which promotes epithelial-mesenchymal transition (EMT), lowers cell adhesion between tumor cells, and facilitates tumor cell migration4. SASH1 can influence tumor progression through various signaling pathways, with the PI3K-Akt-mTOR pathway being particularly crucial18. The abnormal activation of the PI3K-Akt-mTOR pathway in various tumors results in the dysregulation of downstream proteins related to the cell cycle, proliferation, and apoptosis, thereby contributing to tumor initiation and progression43. The overexpression of the PI3K-Akt-mTOR pathway has been observed in cervical cancer tissues, potentially driving the occurrence, progression, and metastasis of cervical cancer44. SASH1 can directly or indirectly interact with PI3K, inhibiting its kinase activity and reducing the production of PIP36. This limits the membrane localization and activation of AKT6. By inhibiting PI3K, SASH1 reduces the phosphorylation of AKT40. Additionally, SASH1 may directly interact with AKT or indirectly affect its activation through upstream negative regulatory proteins (such as PTEN)40. Since the activities of mTORC1 and mTORC2 depend on the phosphorylation state of AKT, SASH1 further influences downstream mTOR signaling by regulating AKT activity, thereby affecting cell growth, survival, and metabolism6. However, whether SASH1 can exert its anticancer effects through the PI3K-Akt-mTOR signaling pathway in breast cancer remains understudied.
In this manuscript, we found through immunohistochemistry that the expression levels of SASH1 in breast cancer tissues are negatively correlated with the levels of p-Akt, p-PI3K, and p-mTOR, suggesting that overexpression of SASH1 can regulate the proliferation of breast cancer cells through the PI3K-Akt-mTOR signaling pathway. Our western blotting results further confirmed this conclusion. However, due to the lack of follow-up data from breast cancer patients, we downloaded data on the relationship between SASH1 and the prognosis of breast cancer patients from the TCGA database. The Kaplan–Meier method was used to plot the survival curve. The results revealed that SASH1 overexpression can improve the OS, RFS, and reduce the PPS of breast cancer patients.
In conclusion, our manuscript found that the expression level of SASH1 is downregulated in multiple types of tumors, suggesting a potential tumor-suppressive role for SASH1. The downregulation of SASH1 can promote the occurrence, proliferation, and metastasis of breast cancer. Overexpressing SASH1 can suppress the proliferation and invasion of breast cancer accompanied by the inhibition of the PI3K-Akt-mTOR signaling pathway. Moreover, SASH1 overexpression may improve OS and RFS in breast cancer patients. Therefore, SASH1 may serve as a novel therapeutic target in the treatment of tumors. However, our study also has certain limitations. For instance, we only validated that SASH1 regulates breast cancer proliferation and metastasis through the PI3K-Akt-mTOR signaling pathway in a single cell line. Additionally, we did not follow up on the survival time of breast cancer patients to investigate the impact of SASH1 on patient prognosis. These issues will need to be addressed in future research.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Zheng, R. S. et al. Cancer statistics in China, 2016. Zhonghua zhong liu za zhi [Chinese journal of oncology]. 45(3), 212–220 (2023).
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clin. 71(3), 209–249 (2021).
Xia, C. et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 135(5), 584–590 (2022).
Li, S. et al. SASH1-IQGAP1-E-cadherin signal cascade may regulate breast cancer metastasis. Tumor. 37(06), 633–641 (2017).
Burgess, J. et al. The overexpression and cleavage of SASH1 by caspase-3 stimulates cell death in lung cancer cells. J. Thorac. Oncol. 12(1), S1158–S1159 (2017).
Sun, C. Y., Zhang, Z. H., He, P., Zhou, Y. & Xie, X. H. Involvement of PI3K/Akt pathway in the inhibition of hepatocarcinoma cell invasion and metastasis induced by SASH1 through downregulating Shh-Glil signaling. Int. J. Biochem. Cell Biol. 89, 95–100 (2017).
Zhou, N. SASH1, a potential therapeutic target for cancer-reply. Human Pathol. 80, 247 (2018).
Wang, Q. Q. & Dai, H. F. SASH1, a potential therapeutic target for cancer. Human Pathol. 80, 246 (2018).
Nitsche, U. et al. Analysis of the metastasis/tumor-suppressing effects of SASH1. Eur. J. Cancer. 50, S120–S121 (2014).
Yang, L. et al. miR-17 enhances proliferation and migration and inhibits apoptosis in glioma cells by regulating SASH1 expression. Transl. Cancer Res. 5(2), 140–151 (2016).
Rimkus, C. et al. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br. J. Cancer. 95(10), 1419–1423 (2006).
Zeller, C. et al. SASH1: A candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene 22(19), 2972–2983 (2003).
Gong, X. C. et al. Correlation of SASH1 expression and ultrasonographic features in breast cancer. OncoTargets Ther. 10, 271–276 (2017).
Sathe, A. & Nawroth, R. Targeting the PI3K/AKT/mTOR pathway in bladder cancer. Methods Mol. Biol. 1655, 335–350 (2018).
Tewari, D., Patni, P., Bishayee, A., Sah, A. N. & Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Seminars Cancer Biol. (2019).
Zhang, W., Jiang, H. & Kong, Y. Exosomes derived from platelet-rich plasma activate YAP and promote the fibrogenic activity of Muller cells via the PI3K/Akt pathway. Exp. Eye Res. 193, 107973 (2020).
. (!!! INVALID CITATION !!! [17]).
Pan, J. & Liu, Y. R. SASH1 inhibits hypoxia-induced epithelial-to-mesenchymal transition via suppression of the PI3K/Akt pathway in human pancreatic cancer cells. Int. J. Clin. Exp. Pathol. 9(2), 750–757 (2016).
He, P., Zhang, H. X., Sun, C. Y., Chen, C. Y. & Jiang, H. Q. Overexpression of SASH1 inhibits the proliferation, invasion, and EMT in hepatocarcinoma cells. Oncol. Res. 24(1), 25–32 (2016).
Li, S. et al. BMP9 inhibits the growth of breast cancer cells by downregulation of the PI3K/Akt signaling pathway. Oncol. Rep. 40(3), 1743–1751 (2018).
Xia, M. et al. miR-10b-5p promotes tumor growth by regulating cell metabolism in liver cancer via targeting SLC38A2. Cancer Biol. Ther. 25(1), 2315651 (2024).
Ren, W. et al. BMP9 inhibits proliferation and metastasis of HER2-positive SK-BR-3 breast cancer cells through ERK1/2 and PI3K/AKT pathways. PloS one. 9(5), e96816 (2014).
S. X. Role of CAD gene in the prognosis evaluation of pan‑cancer. J. Basic Clin. Oncol. 33: 485–490 (2020).
Qinwen, Z., Li, S., Wang, K. & Li, B. SASH1 is a new biomarker for the prognosis of breast cancer patients. J. Shanxi Med. Univ. 50(03), 313–320 (2019).
Lin, Z., Dong, Q., Zhang, Y. & Zhou, X. The expression and clinical significance of SASH1 and p-ERK in infiltrating ductal carcinoma of breast. Progress Anat. Sci. 25(05), 540–543 (2019).
Binbin, W. Expression and clinical significance of SASH1 and MMP-9 in breast cancer. J. Bengbu Med. College 2, 44–46 (2015).
Jing, W. et al. SASH1 may interact with ERK signaling pathways through MAP2K2 and MAP4K4. Basic Clin. Med. 34(11), 1530–1536 (2014).
Xiaoli, Z. et al. Expression and clinical significance of SASH1 in small cell lung cancer. China Med. Pharm. 11(10), 195–198 (2021).
Nan, Z., Zhang, X., Wang, Y., Jin, Q. & Wang, W. Clinical and prognostic significance of SASH1 expression in non-small cell lung cancer. Med. J. Commun. 31(05), 418–421 (2017).
Zhou, N. et al. Downregulated SASH1 expression indicates poor clinical prognosis in gastric cancer. Human Pathol. 74, 83–91 (2018).
Qiuhong, W., Du, G., Deng, G., Qiao, T. & Nie, Y. Expression and significance of SASH1 and p-ERK in gastric adenocarcinoma. Chin. J. Diagn. Pathol. 24(06), 440–444 (2017).
Yudi, W., zhang, J. & Liu, Y. The expression and significance of SASH1 in ovarian cancer. China Rural Health. 14(81) (2017).
Ren, X. Y. et al. Downregulation of SASH1 correlates with tumor progression and poor prognosis in ovarian carcinoma. Oncol. Lett. 11(5), 3123–3130 (2016).
Xie, J., Zhang, W., Zhang, J., Lv, Q. Y. & Luan, Y. F. Downregulation of SASH1 correlates with poor prognosis in cervical cancer. Eur. Rev. Med. Pharm. Sci. 21(17), 3781–3786 (2017).
Xiaoyin, D. & Guo, C. Expressions of SASH1 and FBXO32 protein in esophageal squamous cell carcinoma tissue. J. Zhengzhou Univ. 49(05), 611–614 (2014).
Qiuju, L., He, Y., Xu, L., Zhao, L. & Wang, Q. Research progress of SASH1 gene. Chin. J. Clin. 7(22), 10223–10226 (2013).
Meng, Q. B. et al. SASH1 regulates proliferation, apoptosis, and invasion of osteosarcoma cell. Mol. Cell. Biochem. 373(1–2), 201–210 (2013).
Jézéquel, P. et al. bc-GenExMiner 4.5: New mining module computes breast cancer differential gene expression analyses. Database: J. Biol. Databases Curation 2021(18), baab007 (2021).
Zhou, D. A. et al. p53 regulates ERK1/2/CREB cascade via a novel SASH1/MAP2K2 crosstalk to induce hyperpigmentation. J. Cell. Mol. Med. 21(10), 2465–2480 (2017).
Dawei, S. et al. SASH1 inhibits proliferation and invasion of thyroid cancer cells through PI3K/Akt signaling pathway. Int. J. Clin. Exp. Pathol. 8(10), 12276–12283 (2015).
Burgess, J. T. et al. SASH1 mediates sensitivity of breast cancer cells to chloropyramine and is associated with prognosis in breast cancer. Oncotarget. 7(45), 72807–72818 (2016).
O’Byrne, K. et al. The overexpression of SASH1 stimulates cell death in lung cancer cells. J. Thorac. Oncol. 11(4), S62–S63 (2016).
Liu, Y. et al. MiR-199a-5p targets ZEB1 to inhibit the epithelial-mesenchymal transition of ovarian ectopic endometrial stromal cells via PI3K/Akt/mTOR signal pathway in vitro and in vivo. Reprod. Sci. 27, 110 (2020).
Zhang, W. et al. The exosome-mediated PI3k/Akt/mTOR signaling pathway in cervical cancer. Int. J. Clin. Exp. Pathol. 12(7), 2474–2484 (2019).
Funding
This work was supported by the Natural Science Foundation of Chongqing (CSTB2022NSCQ-BHX0692), Foundation of Chongqing Science and Technology Bureau and Health Commission Joint medical research project (2022QNXM037), the National Natural Science Foundation of China (82201858), Senior Medical Talents Program of Chongqing for Young and Middle-aged, Yongchuan District Natural Science Foundation (2022yc-jckx20048).
Author information
Authors and Affiliations
Contributions
SL conceptualized the study, visualized the data, developed the methodology used, applied the software, performed the statistical analysis, wrote the original draft, and reviewed and edited the manuscript. YS applied the software and performed the statistical analysis. XW and LD developed the methodology used. JT and KW visualized the data. CL conceptualized the study, visualized the data, developed the methodology, and wrote, reviewed and edited the manuscript. SL and CL confirm the authenticity of all the raw data. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was performed in accordance with the Declaration of Helsinki, and all patients signed the informed consent form. This study received approval from the Ethics Committee of the Yongchuan People's Hospital of Chongqing (YCQRMY.Y2023008).
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, S., Su, Y., Tian, J. et al. The downregulation of SASH1 expression promotes breast cancer occurrence and invasion accompanied by the activation of PI3K-Akt-mTOR signaling pathway. Sci Rep 14, 21914 (2024). https://doi.org/10.1038/s41598-024-72562-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-72562-1