Abstract
Dynamic changes in maternal‒zygotic transition (MZT) require complex regulation of zygote formation, maternal transcript decay, embryonic genome activation (EGA), and cell cycle progression. Although these changes are well described, some key regulatory factors are still elusive. Sirtuin-1 (SIRT1), an NAD+-dependent histone deacetylase, is a versatile driver of MZT via its epigenetic and nonepigenetic substrates. This study focused on the dynamics of SIRT1 in early embryos and its contribution to MZT. A conditional SIRT1-deficient knockout mouse model was used, accompanied by porcine and human embryos. Embryos across mammalian species showed the prominent localization of SIRT1 in the nucleus throughout early embryonic development. Accordingly, SIRT1 interacts with histone H4 on lysine K16 (H4K16) in both mouse and human blastocysts. While maternal SIRT1 is dispensable for MZT, at least one allele of embryonic Sirt1 is required for early embryonic development around the time of EGA. This role of SIRT1 is surprisingly mediated via a transcription-independent mode of action.
Similar content being viewed by others
Introduction
The mature oocyte clearly represents a key cell in reproduction. Progressive oocyte growth results in the accumulation of essential transcripts and proteins for the early stages of embryonic development and maternal-to-zygotic transition (MZT), including chromatin decondensation, pronucleus development, and embryonic genome activation (EGA)1. This seemingly precipitous beginning of life is perfectly tuned via the molecular machinery the mediates epigenetic changes, such as the decay of maternal transcripts2, the regulation of minor EGA3, and the transcriptional awakening of the embryonic genome4. However, the timing of these changes differs across animal species, and both post-transcriptional and post-translational modes of gene regulation vary even among mammalian models5. Despite differences in MZT, it seems likely that key regulators are conserved across species.
We consider SIRT1 (sirtuin-1, an NAD+-dependent deacetylase), the ortholog of Yeast SIR2 (Silent Information Regulator 2)6, to be a unique candidate for the regulation of the epigenetic transition from maternal to zygotic programs for the following reasons: i) the maternal transcript of SIRT1 is eliminated via sperm siRNAs7; ii) SIRT1 deacetylates zygotic chromatin and prevents further transcription of minor EGA-expressed genes8; and iii) SIRT1 is highly expressed after major EGA, suggesting that SIRT1 is required for further embryonic development3. Indeed, SIRT1 targets transcription factors9,10 and histone codes11,12,13, both of which are important for gene expression and transcriptional regulation.
With respect to the histone code, the deacetylation of histone 4 at lysine 16 (H4K16) is known to result from the epigenetic mode of action of SIRT111. Therefore, the acetylation level of H4K16 can be a valuable marker for assessing SIRT1 activity during the dynamic period of MZT and early embryonic development, particularly, pronucleus formation, transcriptional reinitiation, cell cycle progression, and cell lineage differentiation, consistent with the transcription of H4K16ac-marked genes14 and cell differentiation15. The origin and dynamics of expression of upregulated factors, such as SIRT1, are crucial for understanding the transformation of terminally differentiated oocytes into pluripotent embryonic cells through EGA and cell differentiation.
On the basis of the evolution of SIRT1 knockout mouse models16,17,18, we generated conditional knockout (cKO) models in our study; these models lack three exons (exons 4, 5, and 6)19, which constitute a major part of the SIRT1 catalytic domain (aa 236–490)20. The cKO model with SIRT1-deficient (Sirt1null) oocytes allowed us to evaluate the activity and necessity of maternal SIRT1 in MZT and embryonic development. In our study, in which the results obtained from the mouse genetic model are supported by investigations in porcine and human embryos, we reveal dynamic changes in SIRT1 and demonstrate its necessity for MZT. Our findings reveal that SIRT1 contributes to this process through a nontranscriptional mode of action and highlight its epigenetic impact on cell differentiation in embryos post-EGA.
Results
Female donors of Sirt1 null oocytes do not suffer from infertility or subfertility
We used a cKO mouse model to investigate maternal SIRT1 activity in mammalian oocytes and early embryos. Thus, we crossbred females in which exons 5–7 are flanked by loxP sites (Sirt1loxP/loxP) with Zp3-Cre males, following the Cre‒Lox principle21. Since Cre recombinase expression is driven by the Zp3 promoter expressed in growing oocytes, exons 5–7 were excised during oocyte growth in Cre-positive female carriers of Sirt1loxP/loxP (i.e., cKO). We genotyped female donors of SIRT1-deficient (Sirt1null) oocytes via PCR of genomic DNA, and Cre-negative;Sirt1loxP/loxP females were considered wild-type (wt) controls (Fig. 1A). The lack of Sirt1 mRNA in oocytes was confirmed by RT‒PCR (Fig. 1B). As expected, the ovarian follicles of ovaries in the cKO mice were not affected (Fig. 1C). Accordingly, neither the ovarian reserve nor hormone responsiveness was reduced in cKO females (Fig. 1D). Either mitochondrial abundance in Sirt1null oocytes was not affected (Fig. 1E). Indeed, the results of the reproductive trial did not differ between wt and cKO mice, and no subfertility or infertility was observed (Fig. 1F).
Generation of the mouse Sirt1loxPloxP;Zp3-Cre (cKO) model and reproductive trial of cKO females. (A) The PCR products indicate the recombined Sirt1 allele (the Sirt1 allele containing loxP-flanked exons 5–7) and Cre recombinase coding locus. (B) Quantitative RT‒PCR of Sirt1 cDNA in wt and Sirt1null oocytes. The data are shown as the means ± SEMs of three independent replicates, each including at least 20 oocytes; significant differences were identified via unpaired t tests. (C) Histology of the ovaries of wt and cKO females; H&E staining. Scale bar: 500 µm. (D) Ovarian capacity is indicated by the ovarian reserve (i.e., the yield of GV oocytes per PMSG-treated female) and hormone responsiveness (i.e., IVO oocytes per PMSG-hCG-treated female). The results are shown as the means ± SEMs; the numbers of females are noted in brackets. (E) Oocyte mitochondrial abudance expressed by mtDNA copy number. Columns show mean ± SEMs of three independent experiments. Significant difference was tested using the unpaired t-test. (F) In the assessment of the reproductive phenotype, the conception rate was calculated as the number of matings per pregnancy; lines represent the median. The number of pups per litter (1st, 2nd, and 3rd) is shown as the mean ± SEM of three females. Wt: wild-type, cKO: conditional knockout, GV: germinal vesicle, IVO: in vivo ovulated.
Translation-independent appearance of maternal SIRT1 and escape from sperm-driven mRNA decay
SIRT1 activity is beneficial for oocyte quality and fertilization22,23, but SIRT1 dynamics in the zygote remain unknown. However, it is known that sperm contain siRNA targeting maternal Sirt1 mRNA7; therefore, we also investigated parthenogenetic zygotes. While Sirt1 mRNA was detected in wt oocytes (Fig. 1), we did not observe any SIRT1 signal in IVO oocytes (i.e., eggs; Fig. 2A; Supplemental Figure S1). Surprisingly, SIRT1 appeared once pronuclei formed in parthenogenetic zygotes (Fig. 2B). The injection of Sirt1-Eyfp into Sirt1null parthenotes and time-lapse images of zygotes revealed a strong SIRT1 affinity for the nucleus, which disappeared once the nuclear envelope was disrupted (i.e., NEBD, nuclear envelope breakdown). SIRT1 reappeared as soon as the interphase nucleus formed (Fig. 2C). The density of SIRT1 peaked in middle two-cell (2C) embryos, decreased again in late 2C embryos and completely disappeared with subsequent nuclear envelope breakdown (see Supplemental Video S1). Owing to the decay of maternal Sirt1 transcripts by sperm siRNA7, we expected to observe the absence of SIRT1 in the pronuclei of fertilized eggs, in contrast to the results obtained for parthenotic zygotes. However, we detected abundant signals of SIRT1 in both pronuclei of wt zygotes. This pronuclear SIRT1 was produced entirely by oocytes, whereas Sirt1null fertilized oocytes did not present any signal (Fig. 2D). H4K16, a main target of SIRT111, was hyperacetylated in the male pronucleus of Sirt1null zygotes, but morphometry was not affected; these findings indicate that SIRT1 deacetylates H4K16 in the pronucleus rather than deacetylating soluble histones before the genesis of pronuclei (Fig. 2D). In further experiments, we proposed that SIRT1 is recruited to the nucleus from already available cytoplasmic maternal SIRT1 (below the detection limit owing to dilution in the large volume of the zygote cytoplasm). Therefore, we used cycloheximide (CHX), an inhibitor of proteosynthesis, and verified that the reappearance of SIRT1 in the nucleolema-bound structure was translation independent (Fig. 2E). Similarly, the observation of equal H4K16 acetylation levels confirmed that SIRT1 activity was unchanged (Fig. 2E). Taken together, these data reveal that SIRT1 dynamics depend strictly on the envelope-bound nucleus; SIRT1 is not resynthesized from maternal Sirt1 mRNA and, therefore, functions regardless of the degree of sperm-induced maternal Sirt1 mRNA decay (Fig. 2F).
The reappearance of SIRT1 in one-cell zygotes. (A) Immunocytochemical staining of SIRT1 in wt and Sirt1null IVO oocytes. (B) SIRT1 is present in one-cell zygotes generated via parthenogenetic activation of wt and Sirt1null oocytes. (C) Distribution and dynamics of SIRT1 during nuclear envelope breakdown (NEBD) in parthenogenetic zygotes; embryos expressing H2B‐mCHERRY (red) and SIRT1-EYFP (green) are shown. (D) Analysis of SIRT1 and H4K16ac in the male pronuclei of in vitro fertilized WT and Sirt1null oocytes. The rectangle delimits the pronuclei (asterisk indicates the male pronucleus). Quantification of H4K16ac integrated density (IntDen) in pronuclei of wt and Sirt1null zygotes and morphometry of IVF zygotes. Correlation analysis of H4K16ac with SIRT1 in wt zygotes. The Spearman coefficient is shown and was considered significant if P was ≤ 0.05 (bold). (E) Distribution and quantity of SIRT1 integrated density (IntDen) in wt parthenote zygotes after CHX treatment. (F) Overview of the relationship between the maternal SIRT1 protein and the Sirt1 transcript in the zygote (created in BioRender.com). The dots represent individual zygotes, and the lines represent the medians; significance was tested via the Mann‒Whitney U test (**, P < 0.01). IVO: in vivo ovulated; PA: parthenogenetic activation; NEBD: nuclear envelope breakdown; IVF: in vitro fertilization; VC: vehicle control; CHX: cycloheximide. Scale bar: 25 µm.
Maternal SIRT1 is progressively replaced by embryonic SIRT1 during EGA
To track the dynamics of the maternal SIRT1 protein, we studied two-cell (2C) embryos since embryonic genome activation (EGA) occurs at this time in these mice. Therefore, we suppressed mRNA synthesis after EGA in parthenotes and IVF embryos via treatment of pronuclear zygotes with α-amanitin (α-AM), an inhibitor of RNA polymerase II and III (Supplemental Figure S2), by late 2C embryos. A decrease in SIRT1 levels indicated the persistence of approximately 50% of maternal SIRT1, even in post-EGA 2C embryos (Fig. 3A). In accordance with previous findings (Fig. 2), there was no difference between parthenotes and IVF embryos after α-AM treatment, suggesting that transcription and translation from the maternal Sirt1 mRNA Were not needed. In accordance with the fact that the Sirt1 gene reaches peak activity during EGA3, we observed a significant amount of SIRT1 in Sirt1null-derived IVF embryos, demonstrating new expression (transcription and translation) of the paternal (functional) Sirt1 allele (Fig. 3B). Moreover, as Sirt1null-derived IVF embryos express roughly ¼ of SIRT1 in wt embryos, Sirt1 transcription and translation seemingly occur in a gene dosage-dependent manner in post-EGA embryos, suggesting the coexistence of maternal and embryonic SIRT1 in two-cell embryos (Fig. 3C). On the basis of these observations, we presumed that SIRT1 would be sequentially diluted through the cell cycle; the gradual elimination of maternal SIRT1 was further investigated in a pig model because of the later timepoint of EGA (during the four-to-eight-cell embryo transition). Pig zygotes were treated with α-AM by the stage of pre- and post-EGA embryos (4C and 8C, respectively) and, indeed, in α-AM-treated pig embryos, the level of SIRT1 in 4C embryos did not differ, whereas the level of SIRT1 in 8C embryos was dramatically lower than that in vehicle control embryos (Fig. 3D, Supplemental Figure S3). These results support the ‘dilution’ hypothesis and suggest that maternal SIRT1 is progressively eliminated and that replacement by embryonic SIRT1 is related to the timing of EGA (Fig. 3E).
Assessment of SIRT1 origin and maternal-to-embryonic SIRT1 exchange in two-cell (2C) embryos. (A) SIRT1 presence and quantification in two-cell parthenotes and IVF-produced embryos treated with α-amanitin (α-AM). (B) SIRT1 integrated density (IntDen) in two-cell (2C) IVF embryos; Sirt1+/+ (wt) and Sirt1+/- embryos were generated via fertilization of wt and Sirt1null oocytes with wt sperm. (C) Summary of different sources of SIRT1 in two-cell wt embryos (created in BioRender.com). (D) Distribution and quantification of SIRT1 in pig four-cell (4C) and eight-cell (8C) parthenogenetic embryos treated with α-AM. (E) Interpretation of the findings obtained in mouse and porcine embryos after α-amanitin treatment (created from BioRender.com). Lines represent the median, and differences were evaluated via an unpaired Mann‒Whitney test (****, P < 0.0001). PN: pronuclear zygote; 2C/4C/8C: two-/four-/eight-cell embryo; PA: parthenogenetic activation; IVF: in vitro fertilization; VC: vehicle control; α-AM: α-amanitin; EGA: embryonic genome activation; ICC: immunocytochemistry. Scale bar: 50 µm.
Embryonic SIRT1 appears to be dispensable in post-EGA embryonic development
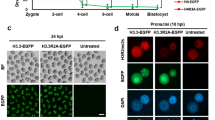
Next, we analysed preimplantation embryonic development and quantified SIRT1 in blastocysts. Consistent with the results of the reproductive trial, we did not observe any failure of early embryonic development (Fig. 4A) or any negative impact on the blastomere count and transcriptional activity (Fig. 4B; Supplemental Figure S4). However, SIRT1 levels in blastocysts dramatically decreased (to below 50%), i.e., out of a gene dosage-dependent manner (Fig. 4B), and we considered the necessity of SIRT1 in the post-EGA embryonic development. Therefore, we tested the effect of sirtinol, a selective SIRT1 inhibitor22,23, on the developmental capacity of post-EGA embryos (Fig. 4C). This experiment confirmed that embryonic SIRT1 expression post-EGA is generally not necessary to reach the blastocyst stage (Fig. 4D). However, we observed hyperacetylation of H4K16 in sirtinol-treated embryos (Fig. 4E), clearly demonstrating that SIRT1 is present and active even though it is dispensable for post-EGA embryonic development.
Developmental competence of Sirt1+/- embryos and inhibition of SIRT1 in post-EGA embryos. (A) IVF output and blastocyst quality. In vitro fertilization of wt and Sirt1null oocytes with wt spermatozoa generated the Sirt1+/+ and Sirt1+/- genotypes, respectively. The columns represent the means ± SEMs of the fertilization rate, cleavage rate, blastocyst rate, and hatching rate in the five IVF assays; differences were evaluated via paired t tests. (B) SIRT1 localization in IVF blastocysts. The dots represent the SIRT1 integrated density (IntDen) of individual blastocysts, and the lines represent the median; differences were evaluated via the Mann‒Whitney U test (****, P < 0.0001). Scale bar: 50 µm. (C) Scheme of SIRT1 inhibition in post-EGA (wt) embryos subjected to in vitro culture of in- vivo vivo-produced post-EGA (2C) embryos. Sirtinol was used as a selective SIRT1 inhibitor during embryo culture. (D) Embryonic development of embryos treated with vehicle (0.1% DMSO) or sirtinol (10 µM). The columns represent the means ± SEMs of the blastocyst rates and hatching rates from three independent assays; differences were evaluated via paired t tests. Scale bar: 500 µm. (E) SIRT1 and H4K16ac colocalization in sirtinol-treated blastocysts. The dots represent the integrated density (IntDen) of H4K16ac and SIRT1 in individual blastocysts, and the lines represent the median; differences were evaluated via the Mann‒Whitney U test (****, P < 0.0001). Scale bar: 50 µm.
SIRT1 decline is accompanied by H4K16ac abundance in the blastocyst stage
In accordance with the finding that SIRT1 modulates the acetylation of H4K16, we further studied the SIRT1-H4K16ac status of the Sirt1 cKO model and human blastocysts. First, we flushed investigated in vivo-produced blastocysts of the Sirt1+/- genotype and confirmed the ability of SIRT1 to deacetylate H4K16 (Fig. 5A). Surprisingly, SIRT1 insufficiency seems to be beneficial, as indicated by the positive correlation of H4K16ac with blastomere counts (Fig. 5B). To understand the relationship between SIRT1 and H4K16ac in human embryos, we performed comprehensive immunocytochemical analysis of human blastocysts. The definition of three-dimensional (3D) regions of interest (ROIs) in individual blastomeres allows the precise analysis of the sum intensity of SIRT1 and H4K16ac. The analysis did not reveal any association of SIRT1-H4K16ac with the number of cells in the blastocysts (Fig. 5C). Although the broad analysis did not reveal any correlation between SIRT1 and H4K16ac through the entire blastocyst (Fig. 5C), we detected a substantial subpopulation of blastomeres with an obvious negative correlation between SIRT1 levels and H4K16ac levels (Fig. 5D). Indeed, this subpopulation of blastomeres demonstrated a negative correlation in almost all investigated patients (Fig. 5E). Complex analysis of human blastomeres revealed that H4K16ac is a marker of SIRT1 activity in embryonic cells and occurs in some blastomeres in blastocysts.
Analysis of the SIRT1-H4K16ac interaction in mouse and human blastocysts. (A) SIRT1 and H4K16ac localization in in vivo-produced blastocysts produced via mating wt and cKO females with wt males. The dots represent the H4K16ac integrated density (IntDen) of individual blastocysts, and the lines represent the median; differences were evaluated via the Mann‒Whitney U test (**, P < 0.01; ***, P < 0.001). (B) Correlation of blastocyst parameters and regression analysis of H4K16ac-blastomere counts. (C) Maximum intensity projection of Z-stacks of SIRT1 and H4K16ac in human blastocysts. Three-dimensional (3D) regions of interest (ROIs) of individual blastomeres defined by the DAPI signal. Correlation analysis of the sum intensity and number of cells in the blastocyst. (D) Co-localization analysis of SIRT1 and H4K16ac in blastomeres. The image represents the analysed blastocysts, and the rectangle highlights the cells used for H4K16ac and SIRT1 colocalization analysis. The number (N) of analysed cells of 13 blastocysts is indicated. (E) Linear regression and correlation analysis of the mean intensity of SIRT1 and H4K16ac in the subpopulation of human blastomeres. Maximum intensity projection was used, and the rectangle shows an example of an ROI for the analysed blastomeres (10 blastomeres per blastocyst). The Spearman coefficient was calculated, and P values are noted.
Embryonic SIRT1 is required for MZT and acts in a transcription-independent manner
Following Sirt1+/- embryo analyses, we generated embryos with the Sirt1-/- genotype. To exclude both Sirt1 alleles, we mated cKO females with Sirt1+/- hemizygote males that were generated and characterized previously24. The reproductive trial revealed a significantly reduced litter size after the elimination of the Sirt1-/- genotype in the offspring population, as the incidence of the Sirt1-/- genotype was significantly lower (P˂0.0001) than the theoretical incidence (i.e., 50%) (Fig. 6A). Therefore, parthenogenetic activation of Sirt1null oocytes was used as a unique way to study the Sirt1-/- genotype and to determine the period during which the development of Sirt1-/- embryos fails. Moreover, the parthenogenetic activation allowed to avoid paternal contribution and the necessity of maternal Sirt1 for the embryonic development could be assessed exclusively. Despite the nonessential role of SIRT1 post-EGA embryos, we observed developmental arrest of Sirt1null embryos at the 2C stage (Fig. 6B; Supplemental Figure S5; Supplemental Videos S2 and S3). Live-cell imaging of parthenotes revealed that zygote formation is impaired when SIRT1 is lacking, mostly because of DNA disintegration and first-cell cycle incompetence (Fig. 6C). SIRT1 is a well-known regulator of gene transcription and acts via several transcription factors; this led us to investigate transcriptional activity in Sirt1null 2C embryos via labelling of 5-ethynyl uridine (5-EU) incorporated into nascent mRNA during embryo culture. We applied two schemes of 5-EU treatment, starting either before or after minor EGA (Fig. 6D). Surprisingly, we did not observe any difference in the amount of mRNA translated in late two-cell embryos (Fig. 6E). This finding is supported by (1) the lack of observed difference in the di-methylation of histone H3 at lysine K4 (H3K4me2), a transcription-positive epigenetic marker of EGA25, and (2) the lack of re-localization of the phosphorylated form of Ser/Thr-protein kinase (pCHK2), another marker of transcriptional activity26 (Supplemental Figure S6). Notably, there was no hyperacetylation of H4K16 in Sirt1null embryos (Fig. 6F), in contrast to the relationship between SIRT1 and H4K16ac observed in other embryonic stages. Unexpectedly, qualitative analysis of the transcriptome of post-EGA embryos revealed no significant changes in the Sirt1null embryos (Fig. 6G; Supplemental Figure S7), and only a few transcripts were differentially expressed in the Sirt1-/- embryos: Stag3, Fbxl3, Tex13b, and SIRT1 mRNA itself (Fig. 6H). We conclude that the necessity of SIRT1 for EGA is independent of transcription. Additionally, this SIRT1 function does not seem to be mediated by H4K16 deacetylation, contrary to what we observed in zygotes and post-EGA embryos. The unique mode of action of SIRT1 during EGA warrants further investigation.
Generation of the Sirt1-/- genotype, embryonic development and transcriptomic analysis of Sirt1null embryos. (A) Number of pups per litter after mating with a Sirt1+/- male. The dots represent individual litters, and the lines represent the medians; significance was tested via the Mann‒Whitney U test (**, P < 0.01). Ratio of offspring genotypes following mating with Sirt1+/- males. Stacked columns show the cumulative proportion of recorded genotypes. A one sample t test was used to compare the recorded genotype ratio with a hypothetical value (i.e., 0.5). (B) Live-cell imaging and time analysis of early embryonic development of the Sirt1+/+ and Sirt1-/- genotypes following the activation of wt and Sirt1null oocytes, respectively (red: H2B‐tdTomato). Assessment of cleavage (24 h) and blastocyst rates (96 h). The columns represent the means ± SEMs of three independent IVF assays, and significance was evaluated via paired t tests (**, P < 0.01). (C) Zygote development and viability. Stacked columns show the cumulative proportions of recorded phenotypes of zygotes; significance was evaluated via the chi-square test (***, P < 0.001). (D) Design of two schemes of nascent mRNA analysis using 5-ethynyl uridine (5-EU) treatment. (E) Representative images and quantification of 5-EU incorporated into nascent mRNA in parthenogenetically activated wt and Sirt1null oocytes. (F) Immunocytochemical staining and analysis of H4K16ac in two-cell parthenote embryos. Dot plots show individual values (i.e., embryos) of integrated density (IntDen), and the lines represent the median; significance was evaluated via the Mann‒Whitney U test. (G) Heatmap of transcripts expressed in bulk samples of wt and Sirt1null parthenote embryos. (H) Qualitative RNA analysis and volcano plot of the RNA-seq data. FDR = false discovery rate; |LFC|= absolute value of log2 fold change. Scale bar: 50 µm.
Discussion
In this study, we demonstrated the activity and necessity of SIRT1 during the dynamic period of MZT. More specifically, SIRT1 activity is essential for EGA but not for early embryonic development. We tracked H4K16ac as a marker of SIRT1 activity11, and although we confirmed the inverse relationship between SIRT1 and H4K16ac levels in embryos across species, this pattern was not observed during EGA; we thus propose an alternative mechanism of SIRT1 action.
Therefore, we speculate that other histone targets are modulated, leading to successful MZT. Although the deacetylation of H3K27ac by SIRT1 is obviously needed for exit from minor zygotic gene activation8, our experiments highlight other mechanisms of action of SIRT1 and its necessity for MZT; indeed, Sirt1null-fertilized oocytes (x wt) showed competence in development to the blastocyst stage, although we suspected persistent transcription of minor zygotic genes. Moreover, flushed Sirt1+/- blastocysts presented increased blastomere counts and SIRT1 insufficiency, which seems to be beneficial for blastocyst expansion.
Alternatively, transcription factors such as p53 and FOXO3A9,27,28 were proposed to be candidate SIRT1 targets that regulate transcription activity during EGA; therefore, we tested the transcriptional activity of post-EGA embryos. Surprisingly, neither the mRNA amount nor the transcriptome composition of Sirt1null (Sirt1-/-) embryos differed from those of wt embryos. Even other assumed checkpoints essential for MZT, such as the DNA damage response and cell cycle regulation, were not affected by SIRT1 deficiency in post-EGA embryos (Supplemental Figure S6), although the cross-talk of both with SIRT1 has been reported29,30.
It is noteworthy that although EGA is highly conserved across mammalian species, seemingly due to the similarity of their egg, developed and fertilized in the safety of the female reproductive tract, the timing of EGA differs in models used in this study. Therefore, by the nature of EGA, we should consider mechanisms of EGA conserved across mammals regardless of timing; unlike nucleocytoplasmic ratio and introduction of gap (G2) phase into the cell cycle4, we suggest nucleolus as a candidate element for EGA driving. Inter-species nucleolar transfer results show nucleolar material's control role in initiating embryonic transcription31,32. Accordingly, we aim to elucidate SIRT1 involvement in forming maternal nucleolar material and its contribution to EGA in oncoming experiments.
Ultimately, proteosynthesis can be affected at a point upstream of transcription when SIRT1 activity is insufficient or deficient33,34. Regardless of transcriptome quantity and composition, disrupted deacetylation interplay might lead to a decline of the NAT10-p53-MDM2 pathway35, followed by decreased mRNAs' acetylation and translation into a protein36. Alternatively, SIRT1 can directly deacetylase some target proteins37, presumably also during EGA, and can thus regulate their activity without the need for new translation.
Embryonic SIRT1 is essential for MZT around the time of EGA, although not via transcription. Indeed, the inhibition of embryonic SIRT1 in post-EGA embryos did not affect developmental potential. Similarly, maternal SIRT1 is dispensable for fertilization and MZT. The hypothesis of ‘maternal SIRT1 dispensability’ is supported by tracking of SIRT1 in oocytes: there was no SIRT1 signal in mature oocytes from the C57Bl6/J strain (the same genetic background of the Sirt1 cKO), although we described the spindle-like pattern observed in the CD-1 outbred strain previously22. Indeed, we compared CD-1 and C57Bl6/J mice and revealed the promiscuous behaviour of SIRT1 across strains (Supplemental Figure S1). Nevertheless, we observed a prominent SIRT1 signal in the zygotic pronucleus in both mouse strains. Further investigation demonstrated the recruitment of soluble SIRT1 protein into the pronucleus even in apparently SIRT1-free C57BL6/J oocytes. This translation-independent SIRT1 recruitment enables escape from sperm-driven decay by siRNAs7; therefore, we can detect maternal SIRT1 even in late two-cell (2C) IVF embryos. Nucleus-bound localization of SIRT1 was thus observed, while maternal SIRT1 was progressively replaced by embryonic SIRT1 during post-EGA embryonic development through the blastocyst stage. To confirm this ‘dilution’ of the maternal transcript, we inhibited transcription in porcine embryos that is occurring physiologically later than in mouse embryos. We accordingly observed the ultimate disappearance of SIRT1 in 8C embryos, supporting the idea of a competition of paternal and maternal factors during MZT38.
In the nucleus, SIRT1 deacetylates H4K16 in zygotes and blastocysts, as described previously in somatic cells11. Despite the SIRT1-H4K16 interaction observed in both one-cell zygotes and blastocysts, we presume the existence of different impacts during early embryonic development: while the silencing of minor EGA genes can be assumed in zygotes, the contribution of SIRT1 to the differentiation of blastomeres into sequential lineages through H4K16 deacetylation is very likely in late blastocysts15. On the other hand, the acetylation of H4K16 is not changed in Sirt1null-derived 2C embryos, and we assume that one-cell zygote inherits hyperacetylated H4K16 from the cytoplasmic pool of the Sirt1null oocyte, whereas embryonic histone residuum of H4K16 is not a target of SIRT1 in the post-EGA embryo.
Although this late SIRT1 contribution seems to be dispensable for the development of blastocysts, we consider SIRT1 to contribute to postimplantation embryo development for two reasons. First, we can observe few Sirt1-/- blastocysts with developed inner cell mass and even foetuses of this genotype (Fig. 6; Supplemental Figure S8). Second, we did not observe SIRT1-deficient (Sirt1-/-) pups. This second wave of SIRT1 necessity is apparently enforced during organogenesis, where severe defects have been described in the absence of SIRT116,39. In addition to embryogenesis failure, we can assume the inter-/transgenerational transmission of inappropriate epigenetic H4K16ac-derived imprinting40 in SIRT1-insufficient individuals (Sirt1+/-), although no fatal failure of preimplantation or postimplantation development was observed.
Taken together, these findings indicate that embryonic Sirt1 expression is required for MZT within the limited time window around EGA, whereas maternal SIRT1 is dispensable. Concurrently, post-EGA embryos can lack the activity of embryonic SIRT1 for blastocyst development. While H4K16 is a target of SIRT1 in noncritical stages of embryogenesis (i.e., one-cell zygotes and blastocysts) and has different impacts, the mode of action of SIRT1 during the essential period of embryonic development around EGA needs to be elucidated.
Materials and methods
Mice
Mouse strains harbouring loxP sites flanking exons 5–7 of the Sirt1 gene were generated at the School of Life Sciences19 and donated by Prof. J. Auwerx. To generate conditional knockout Sirt1loxPloxP;Zp3-Cre mice (cKO), female mice carrying the Sirt1 floxed alleles were crossed with Zp3-Cre males (Jackson Laboratories; C57BL/6-Tg(Zp3-cre)93Knw/J, #003,651). Cre-positive females were used as donors of SIRT1-deficient oocytes, along with oocytes provided by Cre-negative females as wild-type (wt) controls. The mice were housed under a 12 h–12 h light–dark cycle with constant temperature and food and water provided ad libitum. Genotyping for LoxP and Cre was carried out via PCR amplification of tail snip genomic DNA. The primers for Sirt1LoxP (Forward: 5’—ATCATTGGAGGTTATAACATGAATTG- 3’, Reverse: 5’–GTTACCTATTGAACGCCCTACGAGAG—3’) and Zp3-Cre (Forward: 5’—GCGGTCTGGCAGTAAAAACTATC- 3’, Reverse: 5’—GTGAAACAGCATTGCTGTCACTT-3’, Internal control Forward: 5’—CTAGGCCACAGAATTGAAAGATCT- 3’, Internal control Reverse: 5’—GTAGGTGGA AATTCTAGCATCATC C- 3’) were used at a concentration of 20 pMol, using FastMix French PCR beads (Bulldog Bio, #25,401) following the manufacturer’s protocol. DNA was stained using SYBR Safe DNA gel stain (S33102; Thermo Fisher Scientific, Waltham, MA, USA) and scanned on a gel station (Universal Hood II; Bio-Rad, France).
All methods were carried out in accordance with relevant guidelines and regulations. Animal procedures and experimental protocols were conducted in accordance with Act No. 246/1992 Coll. on the Protection of Animals against Cruelty, under the supervision of the Animal Welfare Advisory Committee of Charles University, Faculty of Medicine in Pilsen, and approved by the Animal Welfare Advisory Committee at the Ministry of Education, Youth and Sports of the Czech Republic (approval number: MSMT-249/2017–4). Reports concerning experimental animals followed the recommendations in the ARRIVE guidelines (https://arriveguidelines.org).
Fertility trials
Sexually mature female 8- to 12-week-old wt and cKO mice were mated overnight with wild-type C57Bl/6 males. The number of matings of hormonally stimulated females was recorded and is shown as the conception rate. The number of pups in the 1st, 2nd, and 3rd litters was recorded and is presented as the litter size. Moreover, female fertility was assessed via hormone responsiveness, and the number of ovulated oocytes was recorded.
Histology
The ovaries were fixed for 7 days in Bouin’s solution and then embedded in paraffin blocks (LEICA TP1020 tissue processor, Leica Biosystems Inc. USA). Five-micron-thick cross-sections were prepared (LEICA RM 2145 microtome, Leica Biosystems Inc., USA). The sections were stained with haematoxylin–eosin for general observation of follicles.
Collection of mouse oocytes
Eight- to twelve-week-old female mice were used as oocyte donors. For the collection of IVO mature oocytes, PMSG administration was followed by hCG 48 h later; 15–16 h after hCG injection, ovulated cumulus–oocyte complexes were flushed from the fallopian tube and treated with 0.1 mg/ml hyaluronidase (Sigma‒-Aldrich, #H3506) in M2 medium for 5 min to remove cumulus cells. These oocytes were subsequently used.
Parthenogenetic activation (PA) of mouse oocytes
Mature IVO oocytes were incubated in potassium simplex optimization medium (KSOM) supplemented with 0.1% bovine serum albumin (BSA; mKSOM). To prepare the activating medium, mKSOM was supplemented with 2 mM EGTA, 10 mM SrCl2, and 5 μg/mL cytochalasin B for 5.5 h at 37 °C and 5% CO2. Cycloheximide (CHX; 15 µg/ml) was used for the inhibition of proteosynthesis during oocyte activation. Afterward, the embryos were moved into pure mKSOM and cultured for 24 h and 96 h until the cleaved embryo and blastocyst stages, respectively. Transcription in pre-EGA parthenotes was inhibited via the use of α-amanitin (11 µg/ml), an RNA polymerase II and III inhibitor, for 26 h following activation.
In vitro fertilization (IVF) assay
Fourteen-week-old male mice were euthanized by cervical dislocation, and cauda epididymis were dissected. Spermatozoa were then isolated in human tubal fluid (HTF) supplemented with 0.4% BSA (mHTF). Next, in vitro capacitation was allowed for at least 1 h under 5% CO2 at 37 °C. Moreover, cumulus–oocyte complexes were isolated from the ampulla of PMSG-hCG-stimulated females. The complexes were then coincubated with the capacitated sperm for 5.5 h in mHTF medium under the same conditions applied for capacitation. Thereafter, the zygotes were denuded from the remaining cumulus cells and cultured in mKSOM for 4 days under the described conditions. The cleavage and blastocyst rates were recorded 24 h and 96 h later, respectively.
In vitro culture of mouse embryos
Females were subjected to hormonal stimulation as described above. The females were mated overnight, and the morning was considered embryonic day E0.5. Females were euthanized by cervical dislocation at E1.5, and late two-cell embryos were flushed from the fallopian tubes. The embryos were cultured for an additional four days in vitro, using mKSOM covered with mineral oil at 37 °C and 5% CO2. At the end of embryo culture, the blastocyst rate and hatching rate were recorded; blastocysts were fixed and used for immunocytochemistry.
Production of porcine parthenotes
Porcine oocytes were obtained from gilts at a local slaughterhouse. Immature porcine GV oocytes were isolated from antral follicles via aspiration. The oocytes were washed and cultured in FLI medium41 for 44 h at 38.5 °C in a humidified atmosphere of 5% CO2. The cumulus-enclosed porcine oocytes were treated with 0.3% hyaluronidase, and denuded oocytes were activated as described previously42. Briefly, mature oocytes were treated with 10 μM ionomycin in PBS supplemented with 0.4% BSA for 5 min and cultured in PZM-3 medium43 supplemented with 2 mM 6-DMAP44 for 5 h. After this, presumed zygotes were cultured in pure PZM-3 medium until the 4C and 8C stages. For the inhibition of transcription in parthenotes, PZM-3 was supplemented with α-amanitin (25 µg/ml) for zygote culture.
Production of cRNA and microinjection
For cRNA preparation, the pYX-H2B-tdTomato plasmid was linearized with SfiI and purified with a NucleoSpin Gel and PCR Clean-up Kit (Macherey–Nagel, #740,609). The SIRT1 coding sequence was cloned by PCR from pAd-Track Flag-SIRT1 (Addgene, #8438) into pYX-EYFP plasmid to create pYX-SIRT1-EYFP plasmid as described earlier45. cRNA was produced via in vitro transcriptionusing the mMessage mMachine™ T3 Kit (Ambion, #1348) and was polyadenylated via the Poly(A) Tailing Kit (Ambion, #AM1350) according to the manufacturer’s protocols. cRNAs were purified using the RNeasy Mini Kit (Qiagen, #74,104) and stored at – 80 °C. The activated oocytes were microinjected with ̴10 pl of 35 ng/µl H2b-tdTomato and ̴10 pl of a 100 ng/µl Sirt1-Eyfp.
Light-sheet live-cell imaging of mouse parthenotes
Long-term time-lapse fluorescence imaging of activated oocytes was performed on a Viventis LS1 live light-sheet microscope (Viventis Microscopy, Sarl, Switzerland), as described previously46. The embryos were placed in culture medium covered with oil (OVOILTM, Vitrolife, 10,029) in a multiwell sample holder (Viventis Microscopy, SHM1_4W). Images were captured using a pixel resolution of 1,024 × 1,024 with a pixel size of 173 nm, 3 µm optical sections, and 10–30 min time intervals. Images are available in the BioStudies database (https://www.ebi.ac.uk/biostudies/studies)47 under accession number S-BSST1217.
Immunocytochemistry
Oocytes and embryos were fixed in two ways: either i) in 4% paraformaldehyde in PBS with 0.1% polyvinyl alcohol (PVA) for 30 min at room temperature or ii) for H4K16ac and H3K4me2 imaging, in PFA supplemented with 0.04% Triton X-100 and 0.3% Tween 20 for 15 min at 37 °C. Oocytes and embryos were permeabilized in PBS containing 0.1% Triton X-100 for 15 min and blocked in 1% BSA in PBS with 0.01% Tween 20 for 15 min afterwards. Incubation of the oocytes with specific antibodies (all diluted 1:200) was performed overnight at 4 °C with the following antibodies: anti-SIRT1 (Abcam; ab104833; 1:200), anti-H3K4me2 (Abcam; ab7766; 1:200), and anti-H4K16ac (Abcam; ab109463; 1:200). Afterwards, the samples were washed and incubated for 1 h with a cocktail of anti-mouse-AlexaFluor 488 and anti-rabbit AlexaFluor 647 (1:200) antibodies. After washing, β-actin was stained with Alexa 594-conjugated phalloidin (Thermo Fisher Scientific, USA; 1:200). Oocytes and embryos were mounted onto slides in Vectashield medium supplemented with 4′6´-diamidino-2-phenylindole (DAPI; Vector Laboratories Inc., Burlingame, CA, USA). Images were acquired with an Olympus IX83 spinning disk confocal microscope (Olympus, Germany) and VisiView® software (Visitron Systems GmbH, Germany). Immunostained oocytes and embryos were subjected to measurement of the “integrated density” (IntDen) in appropriate channels using ImageJ software (NIH, Bethesda, CA, USA).
Differential staining of blastocysts
Differential staining was carried out as described previously48 with slight modification. Blastocysts were fixed in PFA supplemented with 0.04% Triton X-100 and 0.3% Tween 20 for 15 min at 37 °C. The total blastomere count and the number of cells in the inner cell mass (ICM) and trophectoderm (TE) were recorded.
Imaging and analysis of human blastocysts
Human blastocysts were obtained at the Pronatal Sanatorium of the ART clinic (Prague, Czech Republic) under the supervision of the Ethical Committee of the University Hospital Pilsen and the Faculty of Medicine in Pilsen, Charles University (approval No. 117/2021). Informed consent was obtained from all subjects, and only blastocysts excluded from the cryopreservation program were used. Blastocysts were immunostained as described above and mounted onto Teflon-coated microchamber slides (Thermo Fisher Scientific, USA). The samples were imaged with a Nikon AX R scanning confocal microscope equipped with a high-speed resonant scanner (Nikon, Japan). Images were processed via 3D analysis using the GA3 module of NIS Elements Advanced Research software (Laboratory Imaging Inc., Czech Republic). The “sum intensity” was measured in the 3D ROI of the nuclei, defined on the basis of the DAPI signal. The values of SIRT1 and H4K16ac sum intensity were correlated with the number of blastocyst cells. In addition, the “maximum intensity projection” was applied, and the “ROI mean intensity” of SIRT1 and H4K16ac in 10 representative cells per blastocyst was correlated. Representative cells were imaged using a high-resolution galvano scanner; colocalization analysis was conducted, and Pearson’s coefficients and overlap coefficients were calculated for the ROI in the nucleus.
Quantification of mtDNA copy number
Matured IVO oocytes were placed in mtDNA lysis buffer, consisting of 50 mM Tris–HCl, pH 8.5, with 0.5% Tween 20 and 200 ng ml–1 proteinase K. Samples were incubated at 55 °C for 30 min, followed by incubation at 95 °C in for 15 min, and stored at – 20 °C until use. Oocyte mtDNA copy number was calculated by absolute quantification, using primers specific for mouse mtDNA49, and a standard curve using PCR-generated template. All samples and standards were measured in technical triplicates. Quantitative real-time PCR (qPCR) was performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad), using the following protocol: pre-incubation at 95 °C for 5 min (1 cycle); denaturation at 95 °C for 10 s, annealing and extension at 60 °C for 30 s, repeated for 40 cycles, followed by melt curve analysis. No PCR product was observed in the no-template control after 40 PCR cycles, and the specificity of the primers was confirmed as a single melt peak and single band when electrophoresed on a 3% agarose gel (not shown). qPCR efficiency calculated from the slope was between 95 and 105% with co-efficiency of reaction R2 = 0.98–0.99.
RNA isolation and qRT‒PCR
For each sample, approximately 50 oocytes were collected in TRIzol reagent (Invitrogen, USA) and homogenized via TissueLyser LT (Qiagen, The Netherlands) for 5 min at 50 Hz. Chloroform was used for phase separation; the aqueous phase was mixed 1:1 with 70% ethanol and applied to RNeasy MinElute spin columns (Qiagen). The washing and elution steps were performed according to the column manufacturer´s instructions. The RNA was quantified via quantitative reverse transcription‒polymerase chain reaction with Pgk1 primers (see below). Integrity was assessed using the RNA 6000 Pico Kit running on a 2100 Bioanalyzer (Agilent, Germany).
Total RNA was reverse transcribed using SuperScript IV (Thermo Fisher Scientific). Mouse phosphoglycerate kinase (Pgk1) served as an endogenous control. Pgk1 cDNA was amplified using the primers Pgk1_c140F (5' GGTGTTGCCAAAATGTCGCT 3') and Pgk1_c186R (5' AACGGACTTGGCTCCATTGT 3'), with a 186 bp amplicon size. Sirt1 cDNA was amplified using the primers Sirt1_c1093F (5’ AGCAGGTTGCAGGAATCCAAA 3’) and Sirt1_c1234R (5’ CACCTAGGGCACCGAGGAA 3’). The expected amplicon size was 142 bp. The Sirt1 primers are located in exons 5 and 6. Cre recombinase removes exons 5, 6, and 7 of the floxed Sirt1 gene in oocytes. The amplification reaction was performed with an Applied Biosystems 7900HT thermal cycler using PowerUp SYBRGreen master mix (Thermo Fisher Scientific). Relative Sirt1 expression was computed using the 2−ΔCt method (wild-type average = 1).
RNA sequencing
The quantity and quality of the isolated RNA were measured using a NanoDrop ND-2000 (NanoDrop Technologies) and analysed with an Agilent 2100 Bioanalyzer (Agilent Technologies). The isolated RNA was processed using the Takara Smarter Stranded Total RNA-seq Kit v2 Pico Input Mammalian (PN:634,413) according to the manufacturer's instructions. Libraries were sequenced on an Illumina NextSeq® 500 instrument using a 75 bp single-end configuration on a high-output flowcell. The read quality was assessed via FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). For subsequent read processing, the bioinformatic pipeline nf-core/RNAseq version 3.10.1 was used50,51. Individual steps included removing sequencing adaptors and low-quality reads with the Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), mapping to the reference genome GRCm39 (Ensembl annotation version 104)52 with STAR53 and quantifying expression at the gene level with Salmon54.
Each quantified gene fragment count served as input for differential expression analysis via negative binomial GLM fitting and the Wald test in the DESeq2 R Bioconductor package55. Only genes whose raw quantified expression was greater than 5 in at least 2 samples were included in the analysis.
We supplied the experimental model, assuming that the KO status (Sirt1null or wt) of the samples was the main effect. Genes exhibiting absolute log2-fold change values of 1 or greater and statistical significance (a false discovery rate < 0.05) between compared groups of samples were considered differentially expressed. The RNA-seq data are available in the ArrayExpress database (https://www.ebi.ac.uk/biostudies/arrayexpress)56 under accession number E-MTAB-13483.
Statistics
The data were analysed using GraphPad Prism 8.1.1 (GraphPad Software Inc., San Diego, CA, USA). Based on the results of normality testing with the Shapiro–Wilk test, differences in quantitative variables were analysed via either a t test or the Mann‒Whitney U test. Categorical variables were evaluated via the chi-square test. P values ≤ 0.05, 0.01, 0.001, and 0.0001 were considered to indicate statistical significance and are indicated with asterisks, as follows: *, **, ***, and ****, respectively. Normally and nonnormally distributed data are expressed as means and medians, respectively.
Data availability
The RNA-seq data are available in the ArrayExpress database under accession number E-MTAB-13483: https://www.ebi.ac.uk/biostudies/arrayexpress. Live-cell imaging data are available in the BioStudies database under accession number S-BSST1217: https://www.ebi.ac.uk/biostudies/studies.
References
Nevoral, J. & Sutovsky, P. Epigenome modification and ubiquitin-dependent proteolysis during pronuclear development of the mammalian zygote: Animal models to study pronuclear development. Animal Models Hum. Reprod. https://doi.org/10.1002/9781118881286.ch17 (2017).
Sha, Q. Q. et al. Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat. Commun. https://doi.org/10.1038/s41467-020-18680-6 (2020).
Abe, K. I. et al. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. U. S. A. 115, E6780–E6788 (2018).
Schulz, K. N. & Harrison, M. M. Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234 (2019).
Vastenhouw, N. L., Cao, W. X. & Lipshitz, H. D. The maternal-to-zygotic transition revisited. Development https://doi.org/10.1242/dev.161471 (2019).
Landry, J. et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U. S. A. 97, 5807 (2000).
Rodgers, A. B., Morgan, C. P., Leu, N. A. & Bale, T. L. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. 112, 13699–13704 (2015).
Li, J. et al. Metabolic control of histone acetylation for precise and timely regulation of minor ZGA in early mammalian embryos. Cell. Discov. https://doi.org/10.1038/s41421-022-00440-z (2022).
Jęśko, H. & Strosznajder, R. P. Sirtuins and their interactions with transcription factors and poly(ADP-ribose) polymerases. Folia Neuropathol. 3, 212–233 (2016).
Ma, R. et al. Sirt1/Nrf2 pathway is involved in oocyte aging by regulating Cyclin B1. Aging 10, 2991–3004 (2018).
Vaquero, A. et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes. Dev. 20, 1256–1261 (2006).
Adamkova, K. et al. SIRT1-dependent modulation of methylation and acetylation of histone H3 on lysine 9 (H3K9) in the zygotic pronuclei improves porcine embryo development. J. Anim. Sci. Biotechnol. https://doi.org/10.1186/s40104-017-0214-0 (2017).
Vaquero, A. et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 16, 93–105 (2004).
Liu, Y., Zhao, L. W., Shen, J. L., Fan, H. Y. & Jin, Y. Maternal DCAF13 Regulates Chromatin Tightness to Contribute to Embryonic Development. Sci. Rep. https://doi.org/10.1038/s41598-019-42179-w (2019).
Taylor, G. C. A., Eskeland, R., Hekimoglu-Balkan, B., Pradeepa, M. M. & Bickmore, W. A. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 23, 2053 (2013).
McBurney, M. W. et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23, 38–54 (2003).
Coussens, M., Maresh, J. G., Yanagimachi, R., Maeda, G. & Allsopp, R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One 3, e1571 (2008).
Iljas, J. D., Wei, Z. & Homer, H. A. Sirt1 sustains female fertility by slowing age-related decline in oocyte quality required for post-fertilization embryo development. Aging Cell https://doi.org/10.1111/acel.13204 (2020).
Mouchiroud, L. et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 154, 430 (2013).
Mahlknetch, U. & Voelter-Mahlknecht, S. Chromosomal characterization and localization of the NAD+-dependent histone deacetylase gene sirtuin 1 in the mouse. Int. J. Mol. Med. 23, 245–252 (2009).
Lewandoski, M., Wassarman, K. M. & Martin, G. R. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7, 148–151 (1997).
Nevoral, J. et al. Epigenetic and non-epigenetic mode of SIRT1 action during oocyte meiosis progression. J. Anim. Sci. Biotechnol. 10, 67 (2019).
Adamkova, K. et al. SIRT1-dependent modulation of methylation and acetylation of histone H3 on lysine 9 (H3K9) in the zygotic pronuclei improves porcine embryo development. J. Anim. Sci. Biotechnol. 8, 83 (2017).
Iniesta-Cuerda, M., Havránková, J., Řimnáčová, H., García-Álvarez, O. & Nevoral, J. Male SIRT1 insufficiency leads to sperm with decreased ability to hyperactivate and fertilize. Reprod. Domest. Anim. https://doi.org/10.1111/rda.14172 (2022).
Shao, G. B., Ding, H. M. & Gong, A. H. Role of histone methylation in zygotic genome activation in the preimplantation mouse embryo. In Vitro Cell. Dev. Biol. Anim. 44, 115–120 (2008).
Matsuoka, S., Huang, M. & Elledge, S. J. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282, 1893–1897 (1998).
Hori, Y. S., Kuno, A., Hosoda, R. & Horio, Y. Regulation of FOXOs and p53 by SIRT1 modulators under oxidative stress. PLoS One 8, e73875 (2013).
Di Emidio, G. et al. SIRT1 signalling protects mouse oocytes against oxidative stress and is deregulated during aging. Hum. Reprod. 29, 2006–2017 (2014).
Zhang, W. et al. SIRT1 modulates cell cycle progression by regulating CHK2 acetylation-phosphorylation. Cell Death Differ. 27, 482–496 (2020).
Rasti, G. et al. SIRT1 regulates DNA damage signaling through the PP4 phosphatase complex. Nucleic Acids Res. https://doi.org/10.1093/NAR/GKAD504 (2023).
Benc, M. et al. Enucleolation and nucleolus transfer in mammalian oocytes and zygotes. Int. J. Dev. Biol. 63, 253–258 (2019).
Benc, M. et al. Improving the Quality of Oocytes with the Help of Nucleolotransfer Therapy. Pharmaceuticals (Basel) https://doi.org/10.3390/ph14040328 (2021).
Wang, Q. & Latham, K. E. Requirement for protein synthesis during embryonic genome activation in mice. Mol. Reprod. Dev. 47, 265–270 (1997).
Zhang, H. et al. Stable maternal proteins underlie distinct transcriptome, translatome, and proteome reprogramming during mouse oocyte-to-embryo transition. Genome Biol. 24, 166 (2023).
Liu, X. et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 17, 349–366 (2016).
Hu, Z. et al. N-acetyltransferase NAT10 controls cell fates via connecting mRNA cytidine acetylation to chromatin signaling. Sci. Adv. https://doi.org/10.1126/sciadv.adh9871 (2024).
Drazic, A., Myklebust, L. M., Ree, R. & Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta Proteins Proteom. 1864, 1372–1401 (2016).
Ooga, M. et al. Parental competition for the regulators of chromatin dynamics in mouse zygotes. Commun. Biol. https://doi.org/10.1038/s42003-022-03623-2 (2022).
Ou, X. et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood 117, 440–450 (2011).
Samata, M. et al. Intergenerationally maintained histone H4 lysine 16 acetylation is instructive for future gene activation. Cell 182, 127-144.e23 (2020).
Yuan, Y. et al. Quadrupling efficiency in production of genetically modified pigs through improved oocyte maturation. Proc. Natl. Acad. Sci. U. S. A. 114, E5796–E5804 (2017).
Jílek, F., Hüttelová, R., Petr, J., Holubová, M. & Rozinek, J. Activation of pig oocytes using calcium ionophore: Effect of protein synthesis inhibitor cycloheximide. Anim. Reprod. Sci. 63, 101–111 (2000).
Yoshioka, K., Suzuki, C., Tanaka, A., Anas, I. M. K. & Iwamura, S. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol. Reprod. 66, 112–119 (2002).
Jílek, F., Hüttelová, R., Petr, J., Holubová, M. & Rozinek, J. Activation of Pig Oocytes using Calcium Ionophore: Effect of the Protein Kinase Inhibitor 6-dimethyl aminopurine. Reprod. Domest. Anim. 36, 139–145 (2001).
Blengini, C. S. et al. Aurora kinase A is essential for meiosis in mouse oocytes. PLoS Genet. 244, 110 (2021).
Knoblochova, L. et al. CHK1-CDC25A-CDK1 regulate cell cycle progression and protect genome integrity in early mouse embryos. EMBO Rep. https://doi.org/10.15252/embr.202256530 (2023).
Sarkans, U. et al. The BioStudies database-one stop shop for all data supporting a life sciences study. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx965 (2018).
Thouas, G. A., Korfiatis, N. A., French, A. J., Jones, G. M. & Trounson, A. O. Simplified technique for differential staining of inner cell mass and trophectoderm cells of mouse and bovine blastocysts. Reprod. Biomed. Online 3, 25–29 (2001).
Malik, A. N., Czajka, A. & Cunningham, P. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion 29, 59–64 (2016).
Tommaso, D. I. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Ewels, P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2020).
Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res. 50, D988–D995 (2022).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Kolesnikov, N. et al. ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 43, D1113–D1116 (2015).
Acknowledgements
We would like to thank Stepanka Jansova, Gabriela Pribanova, Kristyna Popelkova, and Vaclav Rucka for their help with animal breeding, histology, genotyping, and proteomics.
Funding
This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic (the Cooperatio Programme, research area MED/DIAG of Charles University; Grant SVV 260 651; MSTC in the Danube Region, grant No. 8X23009), the Ministry of Health of the Czech Republic (Conceptual Development of Research Organization 00064165, General University Hospital in Prague), the Ministry of Agriculture of the Czech Republic (MZE-RO0723), the Technology Agency of the Czech Republic (grant No. QL24010123), the Grant Agency of the Czech Republic (grant No. 23-07532S), and the Slovak Research and Development Agency under Contract no. DS-FR-22–0003 and VEGA 1/0270/24 in Slovakia.
Author information
Authors and Affiliations
Contributions
J.N., D.D., and M.B. designed experiments and wrote the manuscript. J.N., D.D., M.V., M.B., F.L., I.V., S.S., J.K., J.H., M.S., L.M., M.I.C., L.M., and T.Z. performed experiments. J.N., D.D., M.V., M.B., F.L., J.K., and P.H. analyzed data. F.S., M.K., and J.P. conceived the study and proofread the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nevoral, J., Drutovic, D., Vaskovicova, M. et al. Dynamics and necessity of SIRT1 for maternal–zygotic transition. Sci Rep 14, 21598 (2024). https://doi.org/10.1038/s41598-024-72595-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-72595-6