Abstract
Group B Streptococcus (GBS) is a major cause of pneumonia, sepsis, and meningitis in infants younger than 3 months of age. Furthermore, GBS infection in pregnant women is associated with stillbirths and pre-term delivery. It also causes disease in immunocompromised adults and the elderly, but the highest incidence of the disease occurs in neonates and young infants. At this time, there are no licensed vaccines against GBS. Complete GBS genome sequencing has helped identify genetically conserved and immunogenic proteins, which could serve as vaccine immunogens. In this study, in silico reverse vaccinology method were used to evaluate the prevalence and conservation of GBS proteins in invasive and colonizing isolates from South African infants and women, respectively. Furthermore, this study aimed to predict potential GBS vaccine targets by evaluating metrics such as antigenicity, physico-chemical properties, subcellular localization, secondary and tertiary structures, and epitope prediction and conservation. A total of 648 invasive and 603 colonizing GBS isolate sequences were screened against a panel of 89 candidate GBS proteins. Ten of the 89 proteins were highly genetically conserved in invasive and colonizing GBS isolates, nine of which were computationally inferred proteins (gbs2106, SAN_1577, SAN_0356, SAN_1808, SAN_1685, SAN_0413, SAN_0990, SAN_1040, SAN_0226) and one was the surface Immunogenic Protein (SIP). Additionally, the nine proteins were predicted to be more antigenic than the SIP protein (antigenicity score of > 0.6498), highlighting their potential as GBS vaccine antigen targets.
Similar content being viewed by others
Introduction
Invasive Group B Streptococcus (GBS) disease in infants younger than 90 days of age has a case fatality rate of 5 to 23% 1–4. Children who survive invasive GBS disease may develop neurodevelopmental challenges in 27.6% of cases5,6,7,8,9. Maternal recto-vaginal GBS colonization is a major risk factor for the development of early-onset disease (EOD; i.e. 0–6 days of age)10,11. Furthermore, invasive GBS disease in infants between 7 and 89 days of age (i.e. late onset disease [LOD]) can be contracted from the mother (including via breast milk) or from surrounding environmental sources12,13. The high incidence rate of invasive GBS disease, estimated at 0.49 per 1000 live births (95% confidence interval, 0.43–0.56)14, warrants innovative strategies for its prevention, including the possibility of maternal vaccination15.
Early efforts at developing a GBS vaccine mainly focused on targeting the capsular polysaccharides (CPS) that surround GBS bacterial cells. An inverse association of maternally derived serotype-specific anti-capsular immunoglobulin G (IgG) and risk of homotypic serotype invasive GBS disease in young infants is evident14. GBS expresses 10 distinct serotypes of CPS (i.e. Ia, Ib, II-IX), necessitating development of a multivalent polysaccharide-based vaccine16,17,18. Furthermore, immunization against only selected serotypes may induce immune pressure which could result in capsular switching to serotypes not targeted by the vaccine19,20. This supports the need for novel approaches to identify vaccine candidates that can confer broad protection against all GBS serotypes, including surface proteins.
GBS commonly expresses surface proteins with limited sequence diversity that play a virulence role in the pathogenesis of invasive GBS disease21,22,23. Surface proteins may be alternate vaccine targets to CPS epitopes against GBS16. Also, protein-based vaccines are likely to be cheaper to manufacture than polysaccharide-protein conjugate vaccines24 and would induce predominantly IgG1 sub-class responses, which are more efficiently transferred across the placenta than the predominantly IgG2 subclass responses that is induced by polysaccharide antigens vaccines25. An alternative strategy for developing a GBS vaccine involves the identification and targeting of surface expressed proteins that are genetically conserved and immunogenic, allowing for precise targeting of conserved GBS antigens through protein-based vaccines.
Reverse vaccinology has been applied to the design of viral, parasitic, and bacterial vaccines26. In silico approaches have been used in the development of Neisseria meningitidis and COVID-19 vaccines27,28 Several studies have been undertaken to identify GBS protein vaccine antigens using multiple genome screening29,30,31, but less so using reverse vaccinology. Reverse vaccinology utilizes bioinformatics and enables the identification of highly conserved and immunogenic antigen targets32,33. By integrating reverse vaccinology with serological testing, animal models and epidemiological studies can enhance the process of identifying potential vaccine candidates33,34. This study aimed to identify GBS proteins with promising vaccine attributes, hence providing an initial step toward their potential inclusion as vaccine candidates. Proteins exhibiting suitable characteristics for vaccine candidacy were subjected to validation through flow cytometry to confirm their surface expression on GBS clinical isolates.
Results
GBS protein candidate distributions among GBS clinical isolates
A total of 89 GBS surface proteins were evaluated across the whole genome sequences of 1251 GBS isolates from infant invasive disease (n = 648) and recto-vaginal colonizing (n = 603) isolates from women in South Africa. The proteins were obtained from translations of DNA sequences available in public genomic databases. Sixty-one (68.5%) of the 89 proteins were present in greater than 95% of all the GBS isolates (Table 1). Out of the 61 highly prevalent proteins, 59 are newly described proteins, while two proteins—SIP (present in 99% [1239/1251] of isolates) and LrrG (present in 97.6% [1221/1251] of isolates)—are well-established in GBS research. Notably, the prevalence of other extensively studied proteins, including Alp-1, Alp-2/3, AlpC, Rib, C5a, PI-1, PI-2a, PI-2b, and Srr2, was found to be less than 95% in the study isolates (21.8% [273/1251], 5.1% [64/1251], 9.7% [121/1251], 60.0% [750/1251], 85.5% [1070/1251], 70.2% [878/1251], 41.7% [522/1251], 54.0% [676/1251], and 12.5% [157/1251] respectively). Regarding the Alp family of proteins, one of the Alp family proteins was detected in 96.1% (1202/1251) of the isolates. Furthermore, it was revealed that 98.7% (1235/1251) of the isolates exhibited the presence of at least 1 type of Pilus Island protein and the majority of the isolates (53.6% [671/1251]) had a combination of PI-1 and PI-2b proteins. Moreover, the 61 proteins that were present in greater than 95% of the study isolates had full, non-truncated sequences and were considered for further analysis to assess for potential vaccine targets.
Antigenic potential
Ten out of the 61 proteins that were present in more ≥ 95% of all study isolates were predicted to be highly antigenic (Table 2). SAN_1577 had the highest antigenicity score (1.1756), followed by gbs2106 (0.8361). The antigenicity score of SIP using Vaxijen, which was used as a reference cut-off value, was 0.6498.
Distribution of dominant and antigenic GBS proteins among clinical isolates by GBS serotype
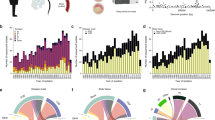
Further analysis was restricted to 10 proteins with an antigenic score greater than 0.6498 and which were prevalent in greater than 95% of invasive and colonizing isolates. The 10 selected proteins were present in greater than 95% of each of the six most invasive disease-causing capsular serotypes (i.e. Ia, Ib, II, III, IV, V) (Fig. 1). Furthermore, when compared with other GBS proteins candidates (such as the Alp family, C5a, PI-1, PI-2a, PI-2b, and Srr2) that have been considered as potential vaccine antigen targets, the 10 highly abundant and antigenic proteins identified in our analysis were more frequently present in both invasive disease and colonizing isolates as well as being present across the different GBS serotypes (Fig. 1.)
Heatmap illustrating the frequency of highly antigenic GBS surface proteins present in South African GBS clinical isolates and stratified according to disease phenotype: Infant invasive and non-invasive (Maternal Colonization) isolates. Heat map created using GraphPad Prism Version 7.0 (GraphPad Software Inc. California USA).
Characterization of GBS proteins
In silico analysis of proteins was performed to deduce whether the 10 selected proteins had suitable vaccine characteristics (Table 3). The molecular weight varied between 15 to 58 kDa, while the number of amino acids varied between 142 and 518. The largest protein was SAN_0226 (58 kDa) whereas the smallest protein was SAN_1577 (15 kDa). Six out of the 10 proteins (i.e. SAN_1577, gbs2106, SAN_1685, SAN_0356, SAN_1040, and SIP) were predicted to be extracellular; 2 proteins (SAN_1808 and SAN_0226) were predicted to be on the cell wall; and 2 proteins (SAN_0413, and SAN_0990) were predicted to be cytoplasmic proteins. The proteins SAN_1577, gbs2106, SAN_0356, SAN_1808, SAN_1685, SAN_0413, SAN_0990, SAN_1040, SAN_0226, and SIP exhibited molecular weights less than 110 kDa. These proteins were predicted to not have transmembrane regions and did not have any homology to human proteins. Moreover, SAN_1577, SAN_0356, and SAN_1808 were predicted to be unstable, as suggested by their high instability index (67.92, 41.17, and 45.19, respectively) which indicates their propensity for rapid degradation or denaturation. Furthermore, SAN_0413 and SAN_0990 were predicted to be allergens based on computational analysis that identified structural and sequence features indicative of allergenic potential.
B-cell epitope prediction
The five protein targets (gbs2106, SAN_1685, SAN_1040, SAN_0226, and SIP) that were predicted to be stable and non-allergens were evaluated to predict their B-cell epitopes, using epitopes with a length of greater than six amino acids for antigenicity prediction (Table 4). Gbs2106 had five predicted B-cell epitopes, all of which were characterized as being antigenic. SAN_1685 had three B-cell epitopes, two of which were antigenic. Five B-cell epitopes were predicted for SAN_1040, four of which were antigenic. Seven B-cell epitopes were predicted for SAN_0226, six of which were antigenic. Ten B cell epitopes were predicted for SIP, nine of which were antigenic. Three out of five predicted B-cell epitopes for gbs2106 (0.9341–0.9916 vs. 0.8361), two out three epitopes for SAN_1685 (1.3514 and 1.6488 vs. 0.7336), four out of five epitopes of SAN_1040 (0.6813–1.9689 vs. 0.6757) and seven out of ten epitopes of SIP (0.6529–1.2648 vs. 0.6498) exhibited higher antigenicity compared with the overall antigenicity of their respective proteins.
Protein secondary structure analysis and identification of potential vaccine targets
The analysis of the proteins’ secondary structure showed that gbs2106 had a high proportion of exposed residues spanning the peptide chain from AA 1- 116, which contained a predicted antigenic linear B-cell epitope (Supplementary Fig. S1A). Conversely, the remaining portions of the protein exhibited buried peptides that were presented as α-helix. Similarly, SIP demonstrated a large portion of exposed residues, also containing predicted linear B-cell epitopes (Supplementary Fig. S1E). In contrast, SAN_1685, SAN_1040, and SAN_0226 exhibited a high proportion of buried residues (Supplementary Fig. S1B–D).
Two proteins (gbs2106 and SIP) were selected for 3D modelling (Fig. 2) due to their prominence among GBS colonizing and invasive isolates, high antigenicity predictions, suitable vaccine characteristics and the presence of predicted B-cell epitopes that are anticipated to be exposed on the surface of their respective proteins. The detailed 3D models of the gbs2106 and SIP proteins and their predicted antigenic linear and conformational B-cell epitopes are shown in Supplementary Figs. S2–S5. Significant portions of the predicted antigenic B-cell epitopes are prominently located on their surfaces. It is worth noting that the linear B-cell epitopes are highly conserved among colonizing and invasive isolate sequences (Supplementary Table S1). One B-cell epitope on gbs2106 (96TYRPAQHQT103) showed a substitution of amino acids at position 98, while lysine (K) substituted for bulkier arginine (R) residue in 52% (313/603) of colonizing isolates and 48% (311/648) of invasive isolates exhibited an arginine residue (R). Additionally, an amino acid change in SIP at position 163 (159EQVSPAPVKS168) was observed where 72% (432/603) of colonizing and 68% (441/648) of invasive isolates presented with a Proline (P), while 28% (171/603) of colonizing and 32% (208/648) of invasive isolates exhibited a threonine (T). Despite the variations, there was no discernible impact on the predicted antigenicity of gbs2106 and SIP.
The flowchart illustrates the systematic selection of two proteins; SIP and gbs2106 for 3D modelling and protein expression analysis using flow cytometry. These proteins were selected using bioinformatic tools and the selection criteria included; high protein frequency among GBS clinical isolates, high antigenicity, favourable vaccine characteristics and surface exposed B-cell epitopes.
Major histocompatibility complex class I (MHC I) and class II (MHCII) epitope prediction
Several antigenic human MHC-I epitopes were predicted for gbs2106 and SIP (Table 5). For gbs2106, three MHC-I epitopes (135STWEHIIAR143, 13KVRVAKKSK21, and 22MTKATSKSK30) were predicted to be highly antigenic, had low IC50 values, a high score (> 0.80), and percentile rank between 0.01 and 0.02, which indicates strong binding between epitope and MHC-I alleles. For SIP, five MHC-I epitopes (132KTYSSAPALK141, 60YPETTLTVTY69, 221ASAKVVTPK229, 38EAMSIDMNV46, and 72KSHTATSMK80), met the pre-specified criteria for strong binding between epitopes and MHC-I alleles. Three antigenic MHC-II epitopes were predicted for gbs2106, while none were predicted for SIP (Table 6). The gbs2106 epitope 176QVNSAIKAYRAQGLS190 had the highest antigenicity, despite having a predicted IC50 value of 14.93. The majority of the predicted MHC-I and MHC-II epitopes for gbs2106 and SIP were highly conserved among the invasive and colonizing GBS isolate sequences. Nevertheless, amino acid polymorphism was observed in the predicted MHC-I epitope 221ASAKVVTPK229 on SIP. Specifically, 52% (312/603) of colonizing isolates and 53% (343/648) of invasive isolates having an alanine (A) at amino acid position 223, and 48% (291/603) of colonizing isolates and 47% (306/648) of invasive isolates showing a valine (V) on the same amino acid position 223 (Supplementary Table S2). Similar to the B-cell epitopes that showed variation, there was no discernible reduction in antigenicity when the alanine at position 223 was substituted with valine.
Docking analysis
Due to the limited MHC-I and MHC-II allele variation on the Protein Bank database, not all probable epitopes could be docked to their respective MHC-I and MHC-II alleles. Accordingly, the MHC-II epitope 176QVNSAIKAYRAQGLS190 on gbs2106 was docked to HLA-DRB1*15:01. Among the models predicted on Cluspro, the model with the lowest energy score of −1180.6, which indicates good binding affinity, was selected (Fig. 3).
Protein surface expression
SIP and gbs2106 displayed high prevalence in both invasive and colonizing GBS isolates. Along with their high antigenicity, favorable vaccine characteristics, and the presence of surface-exposed highly antigenic B-cell epitopes, SIP and gbs2106 are promising candidates for surface expression analysis using flow cytometry. Surface expression of gbs2106 was evident in 86.2% (50/58) and 89.2% (107/120) of invasive and colonizing isolates, respectively. Furthermore, the gene for gbs2106 was present in 94.4% (34/36) and 100% (81/81) of invasive and colonizing isolates tested, respectively. The gene for SIP was present in 97.2% (35/36) and 100% (81/81) of invasive and colonizing isolates tested, respectively; and surface expression of SIP was evident in 93.1% (54/58) and 97.5% (117/120) of the respective isolates (Fig. 4).
The bar charts illustrate the percentage of South African GBS clinical isolates that contain gbs2106 and SIP genes (shown by the black bars) determined using in silico analysis, and the percentage of isolates that express gbs2106 and SIP on their surface (shown by the grey bars) determined using flow cytometry.
Discussion
Using in silico methods, gbs2106 and SIP were identified as having potential to be used as GBS vaccine targets. Both proteins were highly prevalent in infant invasive and maternal colonizing GBS isolates. Additionally, gbs2106 and SIP had conserved B-cell epitopes which were predicted to be highly antigenic, enhancing their potential as vaccine antigen targets.
Reverse vaccinology plays a pivotal role in vaccine development by enabling in silico analysis of pathogen genomes to identify potential vaccine antigen targets. While this approach has shown promise across diverse pathogens, it is essential to acknowledge that the validation and translation of identified vaccine targets require experimental validation. Exploratory research has been conducted across diverse pathogens has proven to be instrumental in identification of potential vaccine targets, exemplified by the discovery of two Streptococcus pneumoniae surface proteins (pavB and spuA35), two Moraxella catarrhalis outer membrane proteins (BamA and LptD36), the Actinetobacter baumannii Outer membrane protein A (OmpA)37, the SARS-CoV-2 Nucleocapsid protein27, and the GBS Spy0416 protein38. It is important to highlight that although these proteins have been identified as potential vaccine targets, they have not yet been successfully utilized to develop effective vaccines. Therefore, their validation and translation into vaccines that induce protective immune responses require further experimental investigation and evaluation. Reverse vaccinology offers valuable insights into vaccine development and provides a strategic advantage by guiding preliminary investigations before initiating wet lab studies.
Bioinformatic analysis performed in this study helped identify 61 out of 89 proteins that were present in greater than 95% of the invasive and colonizing isolates. SIP (99.0% [1239/1251]) and LrrG (97.6% [1221/1251]) exhibited high prevalence, aligning with their common occurrence in North American (100%)29,39 and European (99.5% and 100%)39,40 isolates. Despite its high prevalence, LrrG protein was predicted to be non-antigenic (Supplementary Table S3). Conversely, the well-established GBS protein Srr2 showed lower prevalence (12.5% [157/1251]) compared to the USA (96.4%)31 and was absent in serotype Ia isolates, a common invasive disease-causing serotype41,42,43 diminishing its potential as a vaccine candidate. 92% of invasive and 99.8% of colonizing isolates had one of the Alp family protein sequences. The prevalence of Alp-like proteins in this study’s isolates was similar compared with that reported in the USA (99.2%)31. Nearly all isolates (98.7% [1236/1251]) had at least one type of Pilus Island protein, with the most predominant combination being PI-1 and PI-2b (53.6% [167/1251]. The data aligns with reports from the USA31 and South Africa44,45, but, notably, this study revealed the prevalence of the Pilus Island proteins was higher compared to previous studies44,45. Gbs2106, a potential protein vaccine target, was found to be highly prevalent (98.2% [1230/1251]) in both invasive and colonizing isolates, prompting further analysis, as gbs2106 may potentially induce protective immune responses and provide broad protection against both invasive and colonizing GBS strains.
With GBS having diverse invasive and colonizing serotypes, it is important to develop a vaccine that can target all relevant serotypes to ensure broad coverage and protection. Ten proteins, including gbs2106, were highly prevalent among the global leading invasive GBS serotypes (Ia, Ib, II, III, and V)46,47, suggesting a vaccine that contains the prevalent proteins may confer broad protection against various GBS serotypes5354. It is worth noting that this study did not assess the presence of these proteins in the less common GBS serotypes (IV, VI, VII, VIII and IX). Recognizing the potential temporal changes and evolutionary changes in serotype prevalence over time, it is important to investigate the prevalence of these proteins in the less common GBS serotypes ( IV, VI, VII, VIII and IX), so as to develop an effective vaccine strategy48.
Evaluating potential protein vaccine targets by identifying suitable vaccine characteristics is a crucial aspect of vaccine development. Using reverse vaccinology, 10 highly prevalent proteins that had high antigenicity scores (greater than 0.6) were identified, any of which have the potential to elicit protein-specific antibodies in a vaccinated mother that can be transplacentally transferred to the fetus to confer antibody mediated immunity49. Gbs2106, SAN_1685, SAN_1040, SAN_0226, and SIP proteins had desirable protein-based vaccine candidate characteristics, including stable surface proteins with high antigenicity, molecular weights of less than 110 kDa, less than or equal to 1 transmembrane helix, no homology to human proteins, and non-allergenic properties50. The five identified proteins—gbs2106, SAN_1685, SAN_1040, SAN_0226, and SIP—were predicted to be expressed on the surface of GBS, rendering them accessible to the host’s immune system. The proteins exhibited molecular weights of less than 110 kDa, which facilitates efficient antigen processing and presentation to the immune system50,51. Furthermore, the proteins had no transmembrane helices, suggesting they are more likely to be accessible to circulating antibodies and potentially induce a robust immune response50,52. Based on the proteins’ characteristics, they would be appropriate for further vaccinology studies that would provide a strategic foundation for the advancement of efficacious vaccine development.
While the focus of many studies on GBS protection and vaccine development is predominantly on antibodies and B-cell responses, it is crucial to recognize the contribution of T-cell mediated immunity in preventing the establishment and spread of a GBS infection. T cells, especially CD4 + and CD8 + T-cells play a role in immune responses against various bacterial infections53,54. It has been elucidated that the production of interferon-gamma (IFN-γ) by CD4 + T cells is a key cytokine in controlling GBS infections55. Additionally, Toll-like receptor 13 (TLR13) has been identified as a critical receptor in the response to Streptococci bacteria in myeloid cells56, implicating the involvement of T-cell activation pathways in the host’s response against GBS. The exploration of T-cell epitopes in this study represents a novel aspect as majority of existing literature on GBS focuses on B-cells and antibodies. By predicting MHC-I and MHC-II epitopes aids in determining the potential involvement of T-cell medicated immunity in GBS, thus broadening the scope of vaccine antigen target identification. For an effective GBS vaccine, elicitation of both B-cell and T-cell responses could contribute to protection of the host from future re-infections. Several antigenic B-cell epitopes were identified on gbs2106 and SIP proteins, with potential to stimulate humoral immunity57,58. Additionally, gbs2106 and SIP proteins have several highly antigenic MHC-I epitopes that have the potential to activate CD8 + T cells. Furthermore, gbs2106 has a highly antigenic MHC-II epitope that could potentially stimulate CD4 + T cells59, while SIP had no predicted MHC-II epitopes In this study a potentially conserved epitope (ETPAPVAK) was identified on SIP which overlapped with another epitope on SIP identified by Zhang et al., (AAETPAPVAKVAPVRTVAAPRVA) with slight amino acid variations flanking it. The latter epitope showed higher antigenicity compared to our identified epitope60 Moreover, the predicted epitopes of gbs2106 and SIP were highly conserved among the colonizing and invasive GBS isolates, with the amino acid variation in a few epitopes not predicted to attenuate the antigenicity of the epitopes, illustrating the robustness of the antigenic properties of these two proteins. Nevertheless, the minor antigenic variation could influence the immunogenicity of vaccines targeting the identified epitopes65.
Given the predicted antigenic B-cell, MHC-I and MHC-II epitopes identified on both gbs2106 and SIP as alternative vaccine design strategy of a multi-epitope subunit vaccine should be pursued. In this strategy, a combination of multiple highly antigenic epitopes from different GBS proteins may lead to a synergistic effect that targets the most immunogenic regions of GBS proteins, thereby eliciting a robust immune response and enhancing the efficacy of a GBS vaccine60,61. Utilizing multiple epitopes in vaccine designs, a strategy used for pathogens such as Plasmodium62, not only enhances efficacy but also minimizes use of non-essential or less immunogenic components in the vaccine design63.
In silico prediction alone is not sufficient to validate prevalence, protein expression and vaccine candidates. Therefore, surface expression of two proteins that displayed suitable vaccine characteristics and highly conserved B-cell, epitopes, SIP, and gbs2106, was confirmed in clinical isolates by flow cytometry experiments. The SIP and gbs2106 proteins were highly expressed among invasive and colonizing GBS isolates, further supporting their putative potential as vaccine antigen targets.
Despite the findings of this study, there are some limitations. While in silico analysis provides valuable insights in potential vaccine candidates, it is important to recognize that these predictions have not been validated through pre-clinical studies. Moreover, the protective efficacy of immune responses elicited by these predicted epitopes have not been demonstrated experimentally. Biological complexity, such as interactions of the immune system, can present a challenge for accuracy in silico modelling. This highlights the need for future preclinical studies to validate the immunogenicity and protective capacity of these epitopes against GBS disease. Furthermore, not all epitopes predicted in this study could be analyzed for protein-protein interactions by docking methods, due to the lack of crystalized protein 3D structures. Further research into the exploration of protein-protein interactions is needed to fully understand the functional significance and potential of epitopes identified in this study for GBS vaccine development29,30,64. Additionally, predictive accuracy is subject to limitations, as there is a potential for false positives or negatives, emphasizing the need for experimental validation such as crystallography61,62. Furthermore, the study focused on gbs2106 and SIP based on their prevalence, predicted antigenicity and the presence of favorable vaccine characteristics, and the study did not consider the Alp-family proteins which are included in the most recent advanced protein vaccine which may represent a limitation.
Conclusion
SIP and gbs2106 have the potential to elicit robust immune responses and provide protection against the majority of GBS serotypes prevalent in the South African population. The observed high conservation of these proteins and their associated predicted B-cell, MHC-I and MHC-II epitopes among majority of the GBS invasive and colonizing isolates suggests the potential of using multiple proteins in combination to develop a vaccine, or a multi-epitope subunit vaccine, which could elicit a broader spectrum of protection against invasive and colonizing GBS serotypes. Further studies are warranted to investigate the immunogenicity and efficacy of the proteins identified in this study as potential vaccine targets in animal models. This research pathway is crucial for the development of potential GBS vaccines.
Methods
Clinical GBS isolates
Available sequences of GBS isolates from previous case-control and surveillance studies conducted at Chris Hani Baragwanath Academic Hospital, Soweto, South Africa, between 2004 and 2016 were assessed46,65,66. Briefly, the sequences were from invasive GBS isolates obtained from blood and cerebral spinal fluid (CSF) samples from infants and colonizing GBS isolates obtained from vaginal and rectal swabs from pregnant women67. All GBS isolates were serotyped using latex agglutination45,46. Genomic DNA was extracted, quantified, and sequenced as previously described67. All the GBS isolate sequences (n = 1251) are available at the Streptococcus agalactiae multilocus sequencing typing website PUBMLST (http://pubmlst.org/sagalactiae)67.
Protein sequences conservation and prevalence
The deoxyribonucleic acid (DNA) and amino acid sequences of 89 candidate GBS proteins were analyzed. These include 77 sequences provided by Boston Children’s Hospital (BCH), 1 sequence (gbs2106) contributed by The University of the Witwatersrand Vaccine and Infectious Diseases Analytics Research Unit (WITS-VIDA) (Supplementary Table S4), and 11 reference proteins. These reference proteins include Alp1 (Accession number U33554)68, Alp2 and Alp3-which are identical over the first half of their DNA length-(Accession number AF245663.1)68, AlpC (Accession number MN725039)68, Rib (Accession number MN725044)68, C5a (Accession number AF189004.2)69, LrrG (Accession number AY909605.1)39, the backbone protein of PI-1 (Accession number EU929860.1)70, the backbone protein of PI-2a (Accession number EU929968.1)70, the backbone protein of PI-2b (Accession number EU929123.1)70, SIP (Accession number AF151359)29 and Srr2 (Accession number AY669067.1)71. These candidate proteins’ sequences were mapped against the GBS clinical isolates using the genome comparator tool on Public databases for molecular typing and microbial genome diversity database (PUBMLST), with parameters set at 90% minimum identity, 70% minimum alignment, and 90% core threshold. The alignments were performed using MAFFT72 and viewed with the ALIVIEW73 and JALVIEW74 software. Candidate proteins that were found to be highly prevalent (greater than or equal to 95%) in our invasive and colonizing GBS isolates (n = 1251) were shortlisted as potential vaccine antigen targets.
Antigenicity analysis
The predicted antigenicity of the candidate GBS proteins was determined using the web-based tool Vaxijen (v.20) (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html)75. The threshold value was set at greater than 0.4 Vaxijen Probability score. Candidate proteins with a predictive score of greater than or equal to 0.6, which is the predictive score of the highly conserved and antigenic SIP protein, were shortlisted as potential vaccine antigen targets.
Protein characteristics
The structural and functional characteristics of the candidate GBS proteins were determined by performing Protein BLAST (Basic local alignment search tool [BLASTp]) analysis. Additionally, to confirm that these proteins will not cause autoimmune reactions, homology with human proteins was evaluated using BLASTp analysis using human proteome (taxid:9606) as reference (Parameters: e-value 10e−5, identity greater than 30%, query coverage greater than or equal to 70%50. The localization of selected GBS proteins and the prediction of their surface exposure was determined using Subcellular Localization Prediction Tool (PSORTb v3.0)50,76. Protein sequences were further screened to have not more than one transmembrane helix. A high number of these transmembrane helices can pose challenges in the cloning and expression of the proteins during vaccine development. The bioinformatic tool TransMembrane prediction using Hidden Markov Models (TMHMM) (https://services.healthtech.dtu.dk/services/TMHMM-2.0/) was used to detect the presence of transmembrane helices in the selected GBS proteins. Potential protein targets were also assessed for their allergenic potential using the AllerTOP V.2.0 server (https://www.ddg-pharmfac.net/AllerTOP/)77 Additionally, the Protparam server (https://web.expasy.org/protparam/) was used to predict the proteins’ molecular weight, instability index, and stability prediction (https://www.psort.org/psortb/)50,76,77.
B-cell epitope mapping
B-cell epitope mapping was performed on the B Cell Epitope Prediction Tools server (http://tools.iedb.org/bcell/) with the threshold set at 0.978. The antigenicity of the predicted B-cell epitopes was confirmed using Vaxijen (v.2.0). Predicted epitopes of less than 5 amino acids were eliminated.
Prediction of human major histocompatibility Class-I (MHC-I) and class II (MHC-II) epitopes
The prediction of MHC-I and MHC-II epitopes were performed on the Immune Epitope Database & Tools (IEDB ) server. For MHC-I epitopes prediction, the IEDB-recommended 2020.09 (NetMHCpan EL 4.1) and NetMHCpan BA 4.1 prediction methods were used and the default HLA allele reference set, which consists of 54 HLA alleles, was utilized for the analysis. For the MHC-II epitopes prediction, the IEDB-recommended 2023.05 (NetMHCIIpan 4.1EL) and NetMHCIIpan 4.1 BA prediction methods were used, and the 7-allele HLA reference set was used for this analysis. The antigenicity of the predicted epitopes was determined using Vaxijen 2.0. Highly antigenic epitopes, with an IC50 of less than 10, score of greater than 0.80, and percentile rank of 0.01–0.02 were considered to have strong binding to the MHC alleles and were selected for docking analysis.
3D protein modelling and docking analysis
Protein modelling of selected protein sequences was performed using I-TASSER server (https://zhanggroup.org/I-TASSER/). The 3D models of the HLA alleles were obtained from the Protein Data bank (https://www.rcsb.org/). Protein models were docked to their specific MHC-I and MHC-II allele receptor using the ClusPro Server (https://cluspro.bu.edu/login.php). Docked models that had the lowest energy values were considered as having the strongest binding affinity. The PyMOL software was used to visualize the 3D structures and map the interacting residues between the protein’s epitopes and the MHC-I and MHC-II ligands.
Surface expression profiling
A preliminary assessment of surface exposure was determined by predicting the protein secondary structure based on the protein sequence and was done using the NetSurfP v.2.0 server (https://services.healthtech.dtu.dk/services/NetSurfP-2.0/)79.
Surface expression of two selected proteins that exhibited suitable vaccine characteristics, gbs2106 and SIP, was validated using flow cytometry. This was assessed on maternal colonizing and infant invasive GBS clinical isolates by determining antibody binding to whole bacterial cells using flow cytometry. Briefly, GBS clinical isolates were cultured on Strep B colorex plates and incubated overnight at 37 °C. A single colony was selected and inoculated into Todd Hewitt Yeast broth followed by an overnight incubation at 37 °C, while rotating at 220 rpm. The bacteria culture was subcultured in Todd Hewitt broth (1:100 dilution) until it reached an optical density at 600nm of 1. Bacteria were harvested, washed, and re-suspended with PBS before heat-killing at 58 °C for 1 hour. The heat-killed bacteria were then spun down and re-suspended in PBS/1% BSA and incubated overnight while rotating at 150 rpm at 4 °C. The next day, a volume of 200µl of bacteria suspension was spun down and then incubated with a 1:200 dilution of either pre-immune or post immune rabbit anti-gbs2106 protein antisera(provided by WITS-VIDA) in PBS-1% Tween-20. The post immune rabbit antisera utilized were collected from rabbits seven days after the third dose of gbs2106-protein. After washing twice with PBS-Tween-20/ 1%BSA, the bacteria were incubated with 1:100 dilution of R-phycoerythrin-conjugated F(ab’)2 goat anti-rabbit immunoglobulin (Jackson ImmunoResearch, USA) in PBS-Tween-20/1% BSA and incubated for 1 h. After incubation and washing with PBS-Tween-20/1%BSA, the bacteria were fixed in PBS/4% paraformaldehyde. The samples were acquired by a BD LSRII Fortessa flow cytometer and data analysed using FlowJo software (v10.8.1 < Becton Dickinson). Surface expression of the gbs2106 protein was defined as a greater than or equal to two-fold increase in median fluorescence intensity (MFI) between the pre-immunization and post immunization serum. Surface expression of the protein was further confirmed by observation of a clear shift in signal intensity between the pre-immunization and post-immunization histograms created using FlowJo software v10.8.1.
Statistical analysis
Data was analyzed using Excel and STATA software Version 13.0 (Stata-Corp, Tx USA) and graphs were generated using Graphpad Prism Version 7.0 (GraphPad Software Inc, California USA).
Data availability
All the raw reads are available at the NCBI sequence Read Active, BioProject ID: PRJNA479604, SRA accession number: SRP159611. All GBS protein sequences are available on European Nucleotide Archive, Project Accession number PRJEB76418.
References
Seale, A. C. et al. Estimates of the burden of group B streptococcal disease worldwide for pregnant women, stillbirths, and children. Clin. Infect. Dis.65, S200–S219 (2017).
Edmond, K. M. et al. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet379, 547–556 (2012).
Mynarek, M. et al. Mortality and neurodevelopmental outcome after invasive group B streptococcal infection in infants. Dev. Med. Child Neurolhttps://doi.org/10.1111/dmcn.15643 (2023).
Dangor, Z., Seale, A. C., Baba, V. & Kwatra, G. Early-onset group B streptococcal disease in African countries and maternal vaccination strategies. Front. Public Health11 (2023).
Ku, L., Boggess, K. & Cohen-Wolkowiez, M. Bacterial meningitis in the infant. Clin. Perinatol.42, 29–45 (2015).
Pintye, J., Saltzman, B., Wolf, E. & Crowell, C. S. Risk factors for late-onset group B streptococcal disease before and after implementation of universal screening and intrapartum antibiotic prophylaxis. J. Pediatr. Infect. Dis. Soc.5, 431–438 (2016).
Steer, P. J., Bedford, A., Kochhar, S., Cox, P. & Plumb, J. Group B streptococcal disease in the mother and newborn—A review. Eur. J. Obstet. Gynecol. Reprod. Biol.252, 526–533 (2020).
Dangor, Z. et al. Burden of invasive group B Streptococcus disease and early neurological sequelae in South African infants. PLoS One10, e0123014 (2015).
Paul, P. et al. Neurodevelopmental and growth outcomes after invasive Group B Streptococcus in early infancy: A multi-country matched cohort study in South Africa, Mozambique, India, Kenya, and Argentina. eClinicalMedicine47, 101358 (2022).
Dangor, Z. et al. Burden of invasive group B Streptococcus disease and early neurological sequelae in South African infants. PLoS One10, 1–13 (2015).
Stevens, D. & Kaplan, E. Molecular pathogenesis of Group B streptococcal disease in newborns. in Streptococcal Infections: Clinical aspects, Microbiology and Molecular pathogenesis, vol. 180 (Oxford University Press, 2000).
Dangor, Z. et al. The association between breast milk group B streptococcal capsular antibody levels and late-onset disease in young infants. Clin. Infect. Dis.70, 1110–1114 (2020).
Shabayek, S. & Spellerberg, B. Group B streptococcal colonization, molecular characteristics, and epidemiology. Front. Microbiol.9, 1–14 (2018).
Madrid, L. et al. Infant Group B streptococcal disease incidence and serotypes worldwide: systematic review and meta-analyses. Clin. Infect. Dis.65, S160–S172. https://doi.org/10.1093/cid/cix656 (2017).
Seale, A. C. et al. Vaccines for maternal immunization against Group B Streptococcus disease: WHO perspectives on case ascertainment and case definitions. Vaccine37, 4877–4885. https://doi.org/10.1016/j.vaccine.2019.07.012 (2019).
Nuccitelli, A., Rinaudo, C. D. & Maione, D. Group B Streptococcus vaccine: state of the art. Ther. Adv. Vaccines3, 76–90 (2015).
Carreras-Abad, C., Ramkhelawon, L., Heath, P. T. & Doare, K. L. A vaccine against group b streptococcus: Recent advances. Infect. Drug Resist.13, 1263–1272. https://doi.org/10.2147/IDR.S203454 (2020).
Madhi, S. A. & Dangor, Z. Prospects for preventing infant invasive GBS disease through maternal vaccination. Vaccine35, 4457–4460 (2017).
Bellais, S. et al. Capsular switching in group B streptococcus CC17 hypervirulent clone: A future challenge for polysaccharide vaccine development. J. Infect. Dis. 206, 1745–1752 (2012).
Martcheva, M., Bolker, B. M. & Holt, R. D. Vaccine-induced pathogen strain replacement: What are the mechanisms?. J. R. Soc. Interface 5, 3–13 (2008).
Lindahl, G., Stålhammar-Carlemalm, M. & Areschoug, T. Surface proteins of Streptococcusagalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18, 102–127. https://doi.org/10.1128/CMR.18.1.102-127.2005 (2005).
Paoletti, L. C. & Kasper, D. L. Surface structures of group B Streptococcus important in human immunity. Microbiol. Spectr. 7 (2019).
Shabayek, S. & Spellerberg, B. Group B streptococcal colonization, molecular characteristics, and epidemiology. Front. Microbiol.https://doi.org/10.3389/fmicb.2018.00437 (2018).
Lagousi, T., Basdeki, P., Routsias, J. & Spoulou, V. Novel protein-based pneumococcal vaccines: Assessing the use of distinct protein fragments instead of full-length proteins as vaccine antigens. Vaccineshttps://doi.org/10.3390/vaccines7010009 (2019).
Clements, T. et al. Update on transplacental transfer of IgG subclasses: impact of maternal and fetal factors. Front. Immunol 11, 1920 (2020).
Chong, L. C. & Khan, A. M. Vaccine target discovery. in Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics vols. 1–3 241–251 (Elsevier, 2018).
Can, H. et al. In silico discovery of antigenic proteins and epitopes of SARS-CoV-2 for the development of a vaccine or a diagnostic approach for COVID-19. Sci. Rep. 10, 22387 (2020).
Christodoulides, M. & Heckels, J. Novel approaches to Neisseriameningitidis vaccine design. Pathog. Dis. 75, 1–16 (2017).
Brodeur, B. R. et al. Identification of Group B streptococcal sip protein, which elicits cross-protective immunity. Infect. Immun. 68, 5610–5618 (2000).
Maione, D. et al. Identification of a universal group B streptococcus vaccine by multiple genome screen. www.sciencemag.org/cgi/content/full/309/5731/148/DC1
McGee, L. et al. Multistate, population-based distributions of candidate vaccine targets, clonal complexes, and resistance features of invasive group B streptococci within the United States, 2015–2017. Clin. Infect. Dis. 72, 1004–1013 (2021).
Kanampalliwar, A. M., Soni, R., Girdhar, A. & Tiwari, A. Reverse vaccinology: Basics and applications. J. Vaccines Vaccin. 4, 194 (2013).
Sette, A. & Rappuoli, R. Reverse vaccinology: Developing vaccines in the era of genomics. Immunity 33, 530–541. https://doi.org/10.1016/j.immuni.2010.09.017 (2010).
Masignani, V., Pizza, M. & Moxon, E. R. The development of a vaccine against Meningococcus B using reverse vaccinology. Front. Immunol. 10, 1–14. https://doi.org/10.3389/fimmu.2019.00751 (2019).
Talukdar, S., Zutshi, S., Prashanth, K. S., Saikia, K. K. & Kumar, P. Identification of potential vaccine candidates against Streptococcuspneumoniae by reverse vaccinology approach. Appl. Biochem. Biotechnol. 172, 3026–3041 (2014).
Soltan, M. A. et al. In silico prediction of a multitope vaccine against Moraxellacatarrhalis: Reverse vaccinology and immunoinformatics. Vaccines (Basel) 9, 669 (2021).
Mehdinejadiani, K. et al. In silico design and evaluation of Acinetobacterbaumannii outer membrane protein A (OmpA) antigenic peptides as vaccine candidate in immunized mice. Iran. J. Allergy Asthma Immunol. 18, 655–663 (2020).
Rodríguez-Ortega, M. J. et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 24, 191–197 (2006).
Seepersaud, R. et al. Characterization of a novel leucine-rich repeat protein antigen from group B streptococci that elicits protective immunity. Infect. Immun. 73, 1671–1683 (2005).
Meehan, M. et al. Genomic epidemiology of group B streptococci spanning 10 years in an Irish maternity hospital, 2008–2017. J. Infect. 83, 37–45 (2021).
Madzivhandila, M. et al. Serotype distribution and invasive potential of group B streptococcus isolates causing disease in infants and colonizing maternal-newborn dyads. PLoS One 6, e17861 (2011).
Russell, N. J. et al. Risk of early-onset neonatal group B streptococcal disease with maternal colonization worldwide: systematic review and meta-analyses. Clin. Infect. Dis. 65, S152–S159 (2017).
Martins, E. R. et al. Dominance of serotype Ia among group B streptococci causing invasive infections in nonpregnant adults in Portugal. J. Clin. Microbiol. 50, 1219–1227 (2012).
Madzivhandila, M., Adrian, P. V., Cutland, C. L., Kuwanda, L. & Madhi, S. A. Distribution of pilus islands of group B streptococcus associated with maternal colonization and invasive disease in South Africa. J. Med. Microbiol. 62, 249–253 (2013).
Kwatra, G. et al. Serotype-specific acquisition and loss of group B Streptococcus recto-vaginal colonization in late pregnancy. PLoS One 9, 1–9 (2014).
Dangor, Z. et al. Temporal changes in invasive group B Streptococcus serotypes: implications for vaccine development. PLoS One 11, 1–12 (2016).
Madzivhandila, M. et al. Serotype distribution and invasive potential of group B streptococcus isolates causing disease in infants and colonizing maternal-newborn dyads. PLoS One 6, 2–7 (2011).
Dangor, Z. et al. Temporal changes in invasive group B streptococcus serotypes: Implications for vaccine development. PLoS One 11, e0169101 (2016).
Langel, S. N., Blasi, M. & Permar, S. R. Maternal immune protection against infectious diseases. Cell Host Microbe 30, 660–674. https://doi.org/10.1016/j.chom.2022.04.007 (2022).
Rawal, K. et al. Identification of vaccine targets in pathogens and design of a vaccine using computational approaches. Sci. Rep. 11, 17626 (2021).
Blum, J. S., Wearsch, P. A. & Cresswell, P. Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473. https://doi.org/10.1146/annurev-immunol-032712-095910 (2013).
Kar, T. et al. A candidate multi-epitope vaccine against SARS-CoV-2. Sci. Rep. 10, 10895 (2020).
Shepherd, F. R. & McLaren, J. E. T cell immunity to bacterial pathogens: Mechanisms of immune control and bacterial evasion. Int. J. Mol. Sci. 21, 1–32. https://doi.org/10.3390/ijms21176144 (2020).
Wagner, C. et al. T lymphocytes in acute bacterial infection: Increased prevalence of CD11b+ cells in the peripheral blood and recruitment to the infected site. Immunology 125, 503–509 (2008).
Clarke, D. et al. Group B streptococcus induces a robust IFN-γ response by CD4+ T cells in an in vitro and in vivo model. J. Immunol. Res. 2016, 5290604 (2016).
Kolter, J. et al. Streptococci engage TLR13 on myeloid cells in a site-specific fashion. J. Immunol. 196, 2733–2741 (2016).
Sharon, J., Rynkiewicz, M. J., Lu, Z. & Yang, C. Y. Discovery of protective B-cell epitopes for development of antimicrobial vaccines and antibody therapeutics. Immunology 142, 1–23 (2014).
Ding, P. et al. Nanoparticle orientationally displayed antigen epitopes improve neutralizing antibody level in a model of porcine circovirus type 2. Int. J. Nanomed. 12, 5239–5254 (2017).
Dissanayake, S. K., Tuera, N. & Ostrand-Rosenberg, S. Presentation of endogenously synthesized MHC class II-restricted epitopes by MHC class ii cancer vaccines is independent of transporter associated with Ag processing and the proteasome 1. J. Immunol. 174, 1811–1819 (2005).
Zhang, Y. et al. Development and evaluation of a multi-epitope subunit vaccine against group B Streptococcus infection. Emerg. Microbes Infect. 11, 2371–2382 (2022).
Rashidi, S. et al. Bioinformatics analysis for the purpose of designing a novel multi-epitope DNA vaccine against Leishmania major. Sci. Rep. 12, 18119 (2022).
Tuju, J., Kamuyu, G., Murungi, L. M. & Osier, F. H. A. Vaccine candidate discovery for the next generation of malaria vaccines. Immunology 152, 195–206. https://doi.org/10.1111/imm.12780 (2017).
Curtiss, R. Vaccine design: innovative approaches and novel strategies. Expert Rev. Vaccines 10, 1385–1387 (2011).
Cheng, S. et al. The correlation between expression of sip protein in different serotypes of group B streptococcus and diagnosis. Heliyon 5, e01899 (2019).
Dangor, Z. et al. Infant serotype specific anti-capsular immunoglobulin G antibody and risk of invasive group B Streptococcal disease. Vaccine 39, 6813–6816 (2021).
Cutland, C. L. et al. Increased risk for group b streptococcus sepsis in young infants exposed to HIV, Soweto, South Africa, 2004–2008. Emerg. Infect. Dis. 21, 638–645 (2015).
Lukhele, S. T. et al. Investigation of possible nosocomial-associated invasive group B streptococcus disease using whole-genome sequencing: a report of 3 cases. J. Pediatr. Infect. Dis. Soc.https://doi.org/10.1093/jpids/piab042 (2021).
Bobadilla, F. J., Novosak, M. G., Cortese, I. J., Delgado, O. D. & Laczeski, M. E. Prevalence, serotypes and virulence genes of Streptococcusagalactiae isolated from pregnant women with 35–37 weeks of gestation. BMC Infect. Dis. 21, 73 (2021).
Harris, T. O., Shelver, D. W., Bohnsack, J. F. & Rubens, C. E. A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J. Clin. Investig. 111, 61–70 (2003).
Margarit, I. et al. Preventing bacterial infections with pilus-based vaccines: The group B streptococcus paradigm. J. Infect. Dis. 199, 108–115 (2009).
Seifert, K. N. et al. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (ε) associated with a virulent lineage of serotype III Streptococcusagalactiae. Microbiology (N Y) 152, 1029–1040 (2006).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability article fast track. Mol. Biol. Evol. 30, 772–780 (2013).
Larsson, A. Aliview: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30, 3276–3278 (2014).
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Doytchinova, I. A. & Flower, D. R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 7, 1–7 (2007).
Yu, N. Y. et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 (2010).
Dimitrov, I., Bangov, I., Flower, D. R. & Doytchinova, I. AllerTOP vol 2—A server for in silico prediction of allergens. J. Mol. Model. 20, 2278 (2014).
Jespersen, M. C., Peters, B., Nielsen, M. & Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 45, W24–W29 (2017).
Klausen, M. S. et al. NetSurfP-20: Improved prediction of protein structural features by integrated deep learning. Proteins Struct. Funct. Bioinform. 87, 520–527 (2019).
Acknowledgements
We would like to thank Prof Pascaline Fru for the use of the Flow cytometry at the University of the Witwatersrand, School of Surgery. Anna Seale from Bill & Melinda Gates Foundation who enthusiastically supported our work, offered advice and guidance throughout the study.
Author information
Authors and Affiliations
Contributions
VG, ND, ZD, SAM and GK designed the study. Y.L. and RM provided the protein sequences. S.L. performed whole genome sequencing. V.G performed the in-silico analysis and protein expression. V.G, ND, ZD, SAM, GK analysed the data. VG prepared the 1st draft. All authors have reviewed and had final responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
The study was approved by the Human Research Ethics Committee of the University of the Witwatersrand (HREC140203/M170740).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gent, V., Lu, YJ., Lukhele, S. et al. Surface protein distribution in Group B Streptococcus isolates from South Africa and identifying vaccine targets through in silico analysis. Sci Rep 14, 22665 (2024). https://doi.org/10.1038/s41598-024-73175-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-73175-4