Abstract
An important abnormality in Optical Coherence Tomography (OCT) images is Hyper-Reflective Foci (HRF). This anomaly can be interpreted as a biomarker of serious retinal diseases such as Age-related Macular Degeneration (AMD) and Diabetic Macular Edema (DME) or the progression of disease from an early stage to a late one. In this paper, a new method is proposed for the identification of HRFs. The new method divides the OCT B-scan into patches and separately verifies each patch to determine whether or not the patch contains an HRF. The procedure of patch verification contains a texture-based framework which assigns appropriate labels according to intensity changes to each column and row. Then, a feature vector is extracted for each patch based on the assigned labels. The feature vectors are utilized in the training step of well-known classifiers like Support Vector Machine (SVM). Then, the classifiers are used to produce the labels for the test OCT images. The new method is evaluated on a public dataset including HRF labels. The experimental results show that the new method is capable of providing outstanding results in terms of speed and accuracy.
Similar content being viewed by others
Introduction
Retina is an important body organ which is responsible for providing vision for human1. Optical Coherence Tomography (OCT) is a comparatively new imaging modality which is able to capture from light-scattering organs such as retina. Due to its simplicity, accuracy and non-invasiveness, OCT is widely utilized for verifying retinal health status1,2,3,4,5,6.
Hyper-Reflective Foci (HRF) is a kind of lesion appeared in the OCT images in healthy persons or patients with several important retinal diseases7,8,9. Some studies suggested that HRFs have the same characteristics with small aggregates of hard exudates7,8,10. However, in several non-exudative processes, they have been observed, too11,12.
An HRF can be considered as a risk factor for disease progression from intermediate AMD to late AMD. Also, HRFs are from the first symptoms appeared in the OCT images of Diabetic Macular Edema (DME) patients13,14. HRFs appear as bright roundish lesions within retinal layers and they can be considered as lipid extravasation in DME, macrophages15 and microglia16,17,18 in AMD, degenerated photo receptor cells and migrating Retinal Pigment Epithelium (RPE) cells19. In addition, the origin of HRFs can be considered as leukocytes of RPE cells which represent retinal inflammation14. A sample OCT B-scan which contains HRFs is presented in Fig. 1. The HRF regions are shown in Fig. 1 with yellow arrows.
As mentioned in20, HRFs have bright intensities and irregular shapes, varying sizes, blurry boundaries. Also, they are mostly distributed between Retinal Nerve Fiber Layer (RNFL) and Inner/Outer Segment (IS/OS) layers of retina. The location, the number and the size of HRFs can determine the stage of disease. Therefore, the detection of HRFs can facilitate the procedure of disease identification and staging.
Capturing OCT from one subject produces a large volume of information including a number of B-scans. The manual verification and analysis of OCT B-scans is a tedious and time-consuming task which may be prone to error. Therefore, to propose novel methods for automatic or semi-automatic analysis of OCT images is of considerable importance2,3,4,5,6.
In this paper, an innovative method is proposed for automatic detection of HRFs in the OCT images. The new method works based on the analysis of texture in a patch-wise procedure to extract discriminating feature for HRF regions. The main idea for finding the unique feature for HRF regions is that the pixels located at the center of HRFs has the highest intensity value. Thus, a new textural label is defined to characterize the patch columns and rows including such a pixel. A feature vector is formed based on the values of the defined labels and a classifier is trained based on the input feature vectors.
The structure of this paper is as follows. Section II includes the related works performed in the field of HRF detection in OCT images. The novel method is introduced in detail in Section III. Section IV presents the numerical results and finally the concluding remarks are presented in Section V.
Related works
In this section, the existing research works in the field of HRF identification are introduced and explained.
In21 a method is proposed for the visualization, localization and quantification of HRFs in OCT images. In order to do so, it is necessary to segment Inner Limiting Membrane (ILM), Retinal Pigment Epithelium (RPE) and Bruch’s Membrane (BM) layers using the automated software of capturing device. Also, OCT B-scans are converted to Optical Attenuation Coefficients (OAC) maps. Using these maps, the contrast between RPE and its migrating cells observed as HRFs and the background is enhanced and their visualization becomes simpler.
A method utilizing Morphological Component Analysis (MCA) for detecting HRFs is presented in22. In MCA, an image is considered as a linear combination of morphologically distinct images. Each of these distinct images is a sparse representation in a certain dictionary. For the purpose of HRF detection, known overcomplete dictionaries from curvelet and Daubechies wavelet transforms are used.
An unsupervised learning-based framework for OCT image enhancement and HRF detection is proposed in13. The reconstruction of OCT images is performed using a Restricted Boltzmann Machine (RBM) with a special target function. The reconstructed images have an improved contrast which makes them suitable for detecting HRFs. Gaussian Mixture Models (GMM) are used to cluster HRFs in the reconstructed images. Then, the false positive regions which are located in RPE layer are removed.
For the purpose of quantification and visualization of HRFs in the Outer Nuclear Layer (ONL) of retina, a Convolutional Neural Network (CNN) classifier is employed in23. Firstly, candidate detection is a part of23 where blob detectors are used to find the regions similar to HRFs. Then, feature extraction step extracts a set of features such as intensity, radius and the proximity to a blood vessel to find HRFs. All of the mentioned features are fed to CNN classifier for classifying the patches.
In24 a method including three main phases for pre-processing, layer segmentation and HRF segmentation is proposed for HRF segmentation. A graph search method and Sobel edge algorithm are used for layer segmentation. Finally, HRFs are segmented using a grow-cut algorithm. In25 an algorithm for the detection and quantification of HRFs in DME patients is proposed. In the first step, a U-shaped CNN is developed to segment HRFs. The structure of utilized U-net consists of three down-sampling and up-sampling steps. In order to deal with HRFs with sizes, several convolutional modules in different scales are considered. Moreover, a Channel Attention Module (CAM) is utilized to remove redundant information and guide the model to focus on useful information25.
An automatic method based on deep learning approach for HRF segmentation is suggested in20. In order to improve the accuracy of segmentation in low contrast images, enhanced images are produced using image processing methods. The enhanced images and the denoised image are cascaded to enter 3-D U-net. The structure of 3-D U-net is modified in such a way that the standard convolution is replaced with dilated convolution layer in the last encoder path.
In26 a fully automatic deep learning model is proposed for HRF segmentation. The pre-trained networks such as VGG-16, ResNet50 are utilized for finding the patches which include HRFs. Then, another deep learning approach employing U-net architecture is utilized for segmenting HRFs26.
A segmentation model for HRFs is suggested in27. Firstly, the OCT images are de-noised using non-local means method. Then, the images are divided into patches to solve the problem of imbalanced ratio of HRFs and background. Then, a DBR neural network is employed to receive image patches as inputs. The hidden features in the mentioned patches are extracted by three DBR blocks using a coarse-to-fine architecture.
In28 a deep learning approach is used for the diagnosis of early biomarkers of AMD including HRFs. A CNN is used for the mentioned purpose and its parameters are initialized by transfer learning. Also, a deep learning approach called ReLayNet is utilized for segmenting retinal layers28.
An automatic method based on deep learning is proposed for the classification of Macular Edema (ME) and normal cases in29. In this method, the related biomarkers including HRFs and fluids are segmented. Four CNN architectures are independently trained to identify biomarkers. Pre-processing operations consisting of normalization and augmentation are performed at the starting point.
In contrast to the mentioned research works, in this paper the focus is on the extraction of appropriate manual image processing features for discriminating HRF regions. To the best of our knowledge, the existing works in the field of HRF detection did not pay attention to the discriminating features of HRFs which can be extracted by analyzing the texture of related image regions. Also, the suggested work has interpretability which may not exist in many deep learning methods.
Method
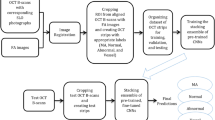
In this section, the novel method is explained in details. The method called Texture-based Label Assignment for HRF (TLA-HRF) detection consists of several steps. The main steps include pre-processing, feature extraction using texture-based label assignment and classification. All the steps are explained in the following. A block diagram for the proposed method is shown in Fig. 2.
Pre-processing
In this step, the OCT B-scan is de-noised with the help of some noise reduction method. The method which is utilized here is median filtering due to its simplicity and speed. Also, retinal segmentation of the OCT B-scans for the determination of RNFL and IS/OS borders is executed. The reason is that according to the studies, the HRFs are usually located between the borders of RNFL and IS/OS layers. The software utilized for segmenting the retinal layers is Caserel30.
Primary idea
Here, the primary idea for the feature extraction step is described. In order to clearly explain the main idea, two sample HRFs are presented in parts (a) and (g) of Fig. 3. As can be observed in the figure, the rectangular windows including HRFs are considered. Also, the related masks produced by the opinion of experts are shown in parts (b) and (h). It is obvious that the pixels located near the center of HRFs have the highest intensity levels. In fact, we have an incremental trend in the intensity levels when one moves from far to the center of HRF. Also, a decremental trend is observed when one moves from the center to the outside of an HRF. These trends are presented in the horizontal and vertical directions. Parts (c) and (d) present the trend of intensity changes in the horizontal direction in the third and sixth rows of the rectangular window in part (a). Also, parts (e) and (f) show the trend of changes in the intensity values in the fifth and sixth column of the same rectangular window. In addition, parts (i) and (j) present the changes trend in the fourth and fifth rows of the rectangular window in part (g). Furthermore, the trends of changes in the fourth and fifth columns of the same rectangular window are presented in parts (k) and (l), respectively.
(a, g) Two rectangular windows from the OCT image containing HRF and (b, h) the corresponding related masks, (c, d) the changes trend in the second and third rows of part (a), (e, f) the changes trend in the second and third columns of part (a), (i, j) the changes trend in the second and third rows of part (g), (k, l) the changes trend in the third and fourth columns of part (g).
Feature extraction
In this section, the procedure for feature extraction is explained. It should be noted that the focus is on the features which represent the main idea described in the previous section. Let I denote the OCT B-scan with m rows and n columns. The whole image is divided into p*p rectangular windows, RWs, which may or may not contain an HRF. The feature extraction phase is performed on a patch-wise basis. For each column and row of the rectangular window, a separate verification procedure is performed. For each column or row, it is verified whether or not it contains a pixel with local maximum intensity value. In fact, it is verified that the intensity incremental trend and decremental trend are observed in each column or row.
Let pi, j denote the pixel located at (i,j) in cartesian coordinates. Also, Xi, j denotes the intensity value of pi, j. The pseudo-code for the feature extraction and forming feature vectors is presented in Fig. 4. Let \(\:{r}_{j}^{RW}\) and \(\:{c}_{i}^{RW}\) denote the pixel sets in the jth column of RW and in the ith row of RW, respectively. Also, \(\:{L}_{j}^{C},1\le\:j\le\:p\) and \(\:{L}_{i}^{R},1\le\:i\le\:p\) denote the label of jth column and ith row in the RW, respectively. The process of forming feature vector is summarized in the assignment of appropriate labels to the columns and rows of RW. In fact, each column (row) is separately verified to receive a suitable label. The conditions verified for each column are indicated in lines 1 to 12 of the pseudo-code. For every pixel located in the column, several parameters including dk, k = 0, 1, …, 3 are computed. If d0 and d1 have positive values, pi, j is a local maximum. However, such conditions are not sufficient for our proposed method. The reason is that some pixels may accidently satisfy the mentioned conditions due to noise or other artefacts. Therefore, it is necessary for d2, d3 to be positive (line 5). If these conditions are true, the label assigned to the column is equal to Xi, j which is the intensity value of the local maximum (line 6). If the mentioned conditions are not true, the zero value in the related column is assigned as a label for the column (line 9). The similar procedure is performed in each row to assign the proper label to each row (lines 13 to 24). This label assignment procedure highlighting the primary idea described in the previous section. The final feature vector is formed by concatenating the labels assigned to the columns and rows (line 25).
Preparing classifier
For the classification purpose, it is necessary to train a classifier such as SVM. For training SVM, all HRFs are extracted from the masks existing for the HRFs in each OCT B-scan of the dataset. From each extracted HRF in the ground truth, we found the core patch. The procedure for finding the core patch inside the HRF includes finding the center of HRFs. Since intensity values are larger in the center of HRFs and core patch is located at the center of HRF, it is reasonable to find the maximum intensity value in the mask related to each HRF. Then, a window around that pixel is considered as the core patch. The dimensions of core patch are 3*3 and 5*5 with the center of the pixel discussed above.
The number of all HRFs in the dataset is equal to 4999 and therefore the number of core patches is equal to 4999.
In order to sufficiently train the classifier, the core patches are increased through augmentation methods. The augmentation methods which are utilized here include rotation, horizontal and vertical flip and brightness adjustment. For each core patch, 8 augmented patches are generated. Thus, the number of core patches for training reaches to 4999*9 = 44,991. Since some patches are located at the margins of images and it is not possible to compute the feature vector for them, they are removed and 44,802 square patches are used for training SVM.
For training SVM, it is also necessary to extract normal patches which do not include HRFs. In order to do so, we have used the segmented OCT B-scans in which the borders of RNFL and IS/OS layers are determined. From each OCT B-scan, all the normal patches which are located in the mentioned ROI are selected. The number of such patches is equal to 825,631. From these normal patches, 44,802 patches are randomly selected and utilized for training SVM. In fact, 44,802 normal and 44,802 abnormal patches including HRFs are used for training SVM to provide balance in the training process. The feature vector which is computed for each normal or abnormal patch is a 1*10 vector (for 5*5 core patches) or a 1*6 vector (for 3*3 core patches). This vector includes 2*p features for a p*p core patch. From each row and each column, one feature is calculated according to the pseudo code of Fig. 4.
From all normal (abnormal) patches, 70% and 30% are used for training and testing purposes. The accuracy values obtained on the testing patches are equal to 90% and 88% for 5*5 and 3*3 dimensions of core patch, respectively. Therefore, the dimensions of 5*5 are considered for core patches in the simulations. After training SVM, it is necessary to traverse all the OCT B-scan and label all patches. In order to do so, we divide the whole OCT B-scan into p*p square patches. It should be noticed that only the patches which belong to the region between RNFL and IS/OS layers are considered and others are removed. The trained classifier is employed to label the mentioned patches.
Experimental results
In this section, the results obtained from evaluating the TLA-HRF are presented. In order to evaluate the performance of the proposed method, a public dataset31 is utilized. This dataset includes 210 OCT B-scans with masks which are related to HRFs. These masks are annotated manually by the experts.
Visual and numerical results
Figure 5 presents several sample rectangular windows which include and do not include HRFs.
Table 1 summarizes the value of sensitivity, specificity and accuracy for the TLA-HRF method. As can be observed.
It should be noted that our proposed method is a patch-based method and the sensitivity, specificity and accuracy values are presented in Table 1 were computed using a patch-based approach. To the best of our knowledge, the methods focusing on identifying HRFs are limited and no work has evaluated its performance using a patch-based approach.
However, in order to make the evaluations complete, we chose the method of13 for comparison. This method called RBM has been introduced in section II. RBM presented its performance metrics in a pixel-wise approach. Therefore, it is necessary to compute the segmentation results for our proposed method to make the comparison possible. In order to do so, for each patch correctly labeled as the patch with HRF, we perform some computations to identify the pixels which belong to HRFs. In these patches, we look for the pixels having the local maximum intensity. In fact, if for a pixel \(\:{p}_{i,j}\) which belongs to a HRF patch, \(\:{X}_{i,j}\ge\:{X}_{i,j-1}\), \(\:{X}_{i,j}\ge\:{X}_{i,j+1}\), \(\:{X}_{i,j-1}\ge\:{X}_{i,j-2}\), and \(\:{X}_{i,j+1}\ge\:{X}_{i,j+2}\) are true, all \(\:{p}_{i,j-2}\), \(\:{p}_{i,j-1}\), \(\:{p}_{i,j}\), \(\:{p}_{i,j+1}\), and \(\:{p}_{i,j+2}\) are labeled as HRF pixels. In addition, if for a pixel \(\:{p}_{i,j}\) which belongs to a HRF patch, \(\:{X}_{i,j}\ge\:{X}_{i-1,j}\), \(\:{X}_{i,j}\ge\:{X}_{i+1,j}\), \(\:{X}_{i+1,j}\ge\:{X}_{i+2,j}\), and \(\:{X}_{i-1,j}\ge\:{X}_{i-2,j}\) are true, all \(\:{p}_{i-2,j}\), \(\:{p}_{i-1,j}\), \(\:{p}_{i,j}\), \(\:{p}_{i+1,j}\), and \(\:{p}_{i+2,j}\) are labeled as HRF pixels. The results related to pixel-wise evaluation of our proposed-method and comparison with13 are presented in Table 2. The results show that all parameters have improvement compared to RBM. It should be also noticed that contrast enhancement is necessary in RBM which imposes extra computations.
Figure 6 presents several rectangular windows including HRFs which are correctly identified by TLA-HRF method.
Figure 7 presents several rectangular windows without HRFs which are correctly identified as true negatives by TLA-HRF method.
Regarding the speed of the proposed method. It is worth pointing out that the required computations for labeling the rectangular patches are very simple. For each rectangular patch, only several comparisons between the intensity values of each row and each column are necessary for labeling. If each patch contains p*p dimensions, the number of comparisons for each patch is a multiplicator of 2*p. Also, it should be noted that although the number of patches used for training is large, the training process is performed only one time and it does not affect the processing time for labeling a sample OCT B-scan. The required time for labeling a sample OCT image is around 4.6 s which is really low.
Discussion on patch selection
It should be noted that in this research work, all the HRFs in a big important dataset have been verified and evaluated. It is true that HRFs do not have the same shapes and sizes. However, it is possible to consider rectangular bounding boxes around them. Using the mentioned bounding boxes, it is possible to count the number of columns and rows existing in each HRF. Therefore, it is possible to cover HRFs with different and irregular shapes using the bounded boxes. Table 3 shows the percentage of HRFs with different rows and columns.
A comprehensive verification on the HRFs of the utilized dataset has been performed. Our evaluations show that although different HRFs may have various sizes, it is possible to consider a core patch for all HRFs. In fact, in order to localize the HRFs, it is sufficient to identify the core patch for HRFs. The core patch is a small patch which can be found in the center of all HRFs. As mentioned before, the number of small HRFs in the utilized dataset is considerable. Our verifications show that around 23% of HRFs in the utilized dataset have less than 4 rows and less than 4 columns. Therefore, it is reasonable to consider the small dimensions such as 3*3 and 5*5 for the core patch.
Limitations
It should be mentioned that the proposed method requires pre-processing steps including noise-reduction and also retinal layer segmentation. With respect to the noisy nature of OCT B-scans and using the intensity-based feature, a step for reducing noise before the main processing is necessary. Also, in order to train the classifier more accurately, it is necessary to extract ROI by segmenting the borders of two retinal layers. Although the pre-processing steps help in the improvement of accuracy results, they affect the processing time. However, it is planned to employ simple noise reduction and layer segmentation methods.
Moreover, it should be mentioned that it is necessary to train the classification model with a sufficient number of patches. The more the number of patches for training, the more accuracy can be obtained. In fact, it is necessary to utilize the trained classifier for labeling all the image patches. Therefore, the classifier should be trained with different kinds of normal and abnormal patches and consequently the number of patches for training purpose should be large. It is interesting that if the classifier is trained with 1000 or 40,000 patches, the accuracy value on the utilized patches may be the same. However, the accuracy value obtained on one sample test image is not the same. Thus, the training process is time-consuming. However, training process is performed only one time and does not repeat during the labeling process of images.
Conclusions
In this paper, a new method for localizing HRFs in the retinal OCT images is proposed. The new method focuses on the texture of HRF regions in the retinal OCT images. The primary idea for finding HRFs is that the pixels located near the center of HRFs have the highest intensity value. In fact, when one moves from the center of an HRF to outside of HRF, the intensity values decrease. Based on this point, a label assignment framework is designed to allocate a proper label to each row and each label in a patch-based procedure. Each row or column which includes a pixel having the primary idea receives the maximum intensity value as a label. The row or columns not satisfying the primary idea receive the minimum intensity vale as a label. Then, the labels assigned to columns and rows form a feature vector for each patch. The feature vectors related to a considerable number of sample patches are fed to SVM classifier as well as their labels. The experimental results show the effectiveness of the proposed feature and method in the localization of HRFs in the retinal OCT images.
Data availability
The utilized dataset is available in the following link: https://github.com/yeisonlegarda/focisdukemarkeddataset.
References
Fujimoto, J. G., Drexler, W., Schuman, J. S. & Hitzenberger, C. K. Optical coherence tomography (OCT) in ophthalmology: Introduction. Opt. Express. 17(5), 3978–3979 (2009).
Monemian, M. & Rabbani, H. Analysis of a novel segmentation algorithm for optical coherence tomography images based on pixels intensity correlations. IEEE Trans. Instrum. Meas.70, 1–12 (2021).
Monemian, M. & Rabbani, H. Mathematical analysis of texture indicators for the segmentation of optical coherence tomography images. Optik. 219(165227). (2020).
Monemian, M. & Rabbani, H. A new texture-based segmentation method for optical coherence tomography images. EMBC. (2019).
Mousavi, N. et al. Cyst identification in retinal optical coherence tomography images using hidden Markov model. Sci. Rep.13(12) (2023).
Monemian, M., Irajpour, M. & Rabbani, H. A review on texture-based methods for anomaly detection in retinal optical coherence tomography images. Optik. 288, 171165, (2023).
Torm, M. E. W. et al. Characterization of hyperreflective dots by structural and angiographic optical coherence tomography in patients with diabetic retinopathy and healthy subjects. J. Clin. Med.11, 6646 (2022).
Bolz, M. et al. Optical coherence tomographic hyperreflective foci: A morphologic sign of lipid extravasation in diabetic macular edema. Ophthalmology. 116, 914–920 (2009).
Schreur, V. et al. Retinal hyperreflective foci in type 1 diabetes mellitus. Retina. (2019).
Coscas, G. et al. Hyperreflective dots: A new spectral-domain optical coherence tomography entity for follow-up and prognosis in exudative age-related macular degeneration. Ophthalmologica. 229, 32–37 (2012).
Vujosevic, S. et al. Hyperreflective Intraretinal spots in diabetics without and with Nonproliferative Diabetic Retinopathy: An in vivo study using spectral domain OCT. J. Diabetes Res., 491835. (2013).
Kuroda, M. et al. Intraretinal hyperreflective foci on spectral-domain optical coherence tomographic images of patients with retinitis pigmentosa. Clin. Ophthalmol.8, 435–440 (2014).
Mansooreh Ezhei, G., Plonka & Rabbani, H. Retinal optical coherence tomography image analysis by a restricted Boltzmann machine. Biomed. Opt. Express. 13, 4539–4558 (2022).
Framme, C., Schweizer, P., Imesch, M., Wolf, S. and Wolf-Schnurrbusch, U. Behavior of SD-OCT-detected Hyperreflective Foci in the retina of anti-VEGF-treated patients with diabetic macular edema. Invest. Ophthalmol. Vis. Sci.53(9), 5814–5818 (2012).
Omri, S. et al. Microglia/Macrophages Migrate through retinal epithelium barrier by a Transcellular Route in Diabetic Retinopathy: role of PKCζ in the Goto Kakizaki Rat Model. Am. J. Pathol.179, 942–953 (2011).
Ling, E. A. A light microscopic demonstration of amoeboid microglia and microglial cells in the retina of rats of various ages. Arch. Histol. Jpn. 45, 37–44 (1982).
Zhang, Y. et al. Repopulating retinal microglia restore endogenous organization and function under CX3CL1-CX3CR1 regulation. Sci. Adv.4, eaap8492 (2018).
De Benedetto, U., Sacconi, R., Pierro, L., Lattanzio, R. & Bandello, F. Optical coherence tomographic hyperreflective foci in early stages of diabetic retinopathy. Retina. 35, 449–453 (2015).
S. Fragiotta, S. Abdolrahimzadeh, R. D. Marco, Y. Sakurada, O. Gal-Or, G. Scuderi, Significance of Hyperreflective Foci as an Optical Coherence Tomography Biomarker in Retinal diseases: Characterization and clinical implications. J. Ophthalmol. (2021).
Xie, S. et al. Fast and automated hyperreflective Foci Segmentation based on image enhancement and improved 3D U-Net in SD-OCT volumes with Diabetic Retinopathy. Trans. Vis. Sci. Tech.9(2), 21 (2020).
Hao Zhou, J. et al. Depth-resolved visualization and automated quantification of hyperreflective foci on OCT scans using optical attenuation coefficients. Biomed. Opt. Express. 13, 4175–4189 (2022).
Mokhtari, M., Kamasi, Z. G. & Rabbani, H. Automatic detection of hyperreflective foci in OCT B-scans using morphological component analysis. in 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (2017).
Schmidt, M. F. et al. Automated detection of hyperreflective foci in the outer nuclear layer of the retina. Acta Ophthalmol.101, 200–206 (2023).
Okuwobi, I. P. et al. Automated segmentation of hyperreflective foci in spectral domain optical coherence tomography with diabetic retinopathy. J. Med. Imaging (Bellingham). 5(1), 014002 (2018).
Huang, H. et al. Algorithm for detection and quantification of Hyperreflective dots on Optical Coherence Tomography in Diabetic Macular Edema. Front. Med. (Lausanne). 8, 688986 (2021).
Goel, S., Sethi, A., Pfau, M., Munro, M., Chan, R.V.P., Lim, J.I., Hallak, J., Alam, M. Automated region of interest selection improves deep learning-based segmentation of hyper-reflective foci in optical coherence tomography images. J. Clin. Med. 11, 7404 (2022).
Wei, J. et al. Automatic segmentation of Hyperreflective Foci in OCT images based on Lightweight DBR Network. J. Digit. Imaging. 36, 1148–1157 (2023).
Saha, S., Nassisi, M. & Wang, M. et al. Automated detection and classification of early AMD biomarkers using deep learning. Sci. Rep. 9, 10990 (2019).
Padilla-Pantoja, F.D., Sanchez, Y.D., Quijano-Nieto, B.A., Perdomo, O.J. & Gonzalez, F.A. Etiology of macular edema defined by deep learning in optical coherence tomography scans. Trans. Vis. Sci. Tech. 11(9), 29 (2022).
Teng, P. Caserel—An open source software for computer-aided segmentation of retinal layers in optical coherence tomography images. Zenodo Sep.15, (2013).
Y. D. Sanchez, B. Nieto, F. D. Padilla, O. Perdomo, F. A. G. Osorio, Segmentation of retinal fluids and hyperreflective foci using deep learning approach in optical coherence tomography scans. in The 16th International Symposium on Medical Information Processing and Analysis, Lima, Peru (2020).
Author information
Authors and Affiliations
Contributions
M.M. designed/implemented the final method and wrote the main manuscript. P.D. implemented the related codes for some parts of the final method. S.R. contributed in summarizing several research works with deep learning approaches. H.R. modified the main method and evaluated the final results. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Monemian, M., Daneshmand, P.G., Rakhshani, S. et al. A new texture-based labeling framework for hyper-reflective foci identification in retinal optical coherence tomography images. Sci Rep 14, 22933 (2024). https://doi.org/10.1038/s41598-024-73927-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-73927-2
This article is cited by
-
Learning feature dependencies for precise tumor region detection and segmentation in optical coherence tomography images
International Ophthalmology (2025)