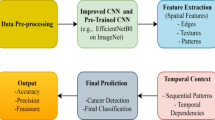
Abstract
The objective of this investigation was to improve the diagnosis of breast cancer by combining two significant datasets: the Wisconsin Breast Cancer Database and the DDSM Curated Breast Imaging Subset (CBIS-DDSM). The Wisconsin Breast Cancer Database provides a detailed examination of the characteristics of cell nuclei, including radius, texture, and concavity, for 569 patients, of which 212 had malignant tumors. In addition, the CBIS-DDSM dataset—a revised variant of the Digital Database for Screening Mammography (DDSM)—offers a standardized collection of 2,620 scanned film mammography studies, including cases that are normal, benign, or malignant and that include verified pathology data. To identify complex patterns and trait diagnoses of breast cancer, this investigation used a hybrid deep learning methodology that combines Convolutional Neural Networks (CNNs) with the stochastic gradients method. The Wisconsin Breast Cancer Database is used for CNN training, while the CBIS-DDSM dataset is used for fine-tuning to maximize adaptability across a variety of mammography investigations. Data integration, feature extraction, model development, and thorough performance evaluation are the main objectives. The diagnostic effectiveness of the algorithm was evaluated by the area under the Receiver Operating Characteristic Curve (AUC-ROC), sensitivity, specificity, and accuracy. The generalizability of the model will be validated by independent validation on additional datasets. This research provides an accurate, comprehensible, and therapeutically applicable breast cancer detection method that will advance the field. These predicted results might greatly increase early diagnosis, which could promote improvements in breast cancer research and eventually lead to improved patient outcomes.
Similar content being viewed by others
Introduction
Breast cancer continues to be among the most prevalent and severe types of cancer that affect women globally. Patient survival and effective treatment depend on early discovery1. Conventional diagnostic techniques, such as physical examinations and mammography, might produce false negatives or positives because of their limited accuracy2. With the development of artificial intelligence and machine learning, there is an enormous opportunity to improve the precision and effectiveness of breast cancer diagnosis. To improve breast cancer detection, this investigation presents a hybrid deep learning strategy that utilizes the Wisconsin Breast Cancer Database and the Curated Breast Imaging Subset of DDSM (CBIS-DDSM). Together with innovative computational techniques, the combination of these two datasets, each with distinct and complimentary qualities, represents a significant advancement in the struggle against this prevalent disease.
Background
Breast cancer continues to be a major worldwide health problem; thus, improved outcomes for patients will require ongoing advances in detection techniques. Innovative techniques have been created by combining computational methods and modern imaging technology3. One such promising strategy is the integration of the Wisconsin Breast Cancer Database with the Curated Breast Imaging Subset of DDSM (CBIS-DDSM). The radius, smoothness, and concavity are just a few of the 30 characteristics of the innovative Wisconsin Breast Cancer Database that offer a comprehensive understanding of cellular properties. Among the 569 patients in this dataset, 212 had significant differences because they had malignant tumors4. All-encompassing examinations of breast cancer are based on these characteristics, which are important indicators of cellular abnormalities and anomalies. Data augmentation techniques have been widely used to improve the performance of deep learning models in medical image analysis tasks. However, traditional data augmentation methods often to maintain bounding box and segmentation precision.
In addition, the CBIS-DDSM dataset is a modern, standardized development of the Digital Database for Screening Mammography (DDSM). The DDSM, which starts with 2620 scanned film mammography examinations, provides a wide range of cases, from benign to malignant, along with carefully checked pathological data for each case5. This repository is updated and refined by the CBIS-DDSM dataset, which serves as a standard for modern breast imaging research. This research used a state-of-the-art hybrid deep learning technique to obtain accurate detection of breast cancer. By combining the advantages of both approaches, Convolutional Neural Networks (CNNs) and the stochastic gradients method enable the model to identify subtle characteristics and complex patterns that may be symptoms of breast cancer. The Wisconsin Breast Cancer Database’s rich attribute information is an ideal starting point for the CNN, which may then be further enhanced using the CBIS-DDSM dataset to guarantee flexibility across a range of mammography investigations. Using a hybrid deep learning method and combining these datasets, this investigation intends to overcome the challenges associated with diagnosing breast cancer. The expected results show promise for a more precise, understandable, and practically applicable diagnostic method, which will support further efforts to enhance early detection techniques and improve the prognosis of breast cancer patients.
Previous work
Large-scale imaging datasets and innovative computational techniques have been widely used in previous research on breast cancer diagnosis. Prominent research efforts have established a foundation for the use of deep learning methods, namely, CNNs, to improve the precision and effectiveness of identifying breast cancer6. The Wisconsin Breast Cancer Database has been widely used by researchers to investigate the complex relationships among cellular characteristics and breast cancer pathology. The development of machine learning models to distinguish between benign and malignant tumors has been made possible by the 30 properties of the dataset, which include radius, texture, and concavity7. Previous research has highlighted the importance of these traits in identifying subtle patterns suggestive of breast cancer, indicating the possibility of improved diagnostic precision.
As an essential tool for breast imaging research, the Digital Database for Screening Mammography (DDSM) offers a wide range of 2,620 scanned film mammography investigations. A significant amount of work has been done previously using DDSM to create models that can identify situations that are benign, malignant, or normal8. The significance of authentic pathology data accompanying every instance in guaranteeing the dependability of the training and assessment procedure has been emphasized by these investigations. Recent years have made tremendous progress in the detection of breast cancer, with researchers increasingly utilizing sophisticated computer algorithms and extensive imaging datasets to improve the precision and efficacy of these methods. An innovative technique for identifying more accurate breast cancer diagnoses is the combination of the Wisconsin Breast Cancer Database and the Curated Breast Imaging Subset of DDSM (CBIS-DDSM) with a hybrid deep learning algorithm9. The main challenge is to develop multimodality process that are low accuracy, and efficient within a clinically acceptable period. The source and target images, the majority of medical image processing with low clarity.
Previous investigations have focused on the Wisconsin Breast Cancer Database, which is well known for its comprehensive features related to cell nuclei. Researchers have investigated the discriminative ability of various characteristics, such as the radius, roughness, and concavity. Notable works include (reference), where machine learning algorithms used this dataset to show encouraging results in differentiating between benign and malignant cancers10. By providing a wide range of 2,620 scanned film mammogram investigations, DDSM has been a mainstay in breast imaging research. To create models for classifying instances as normal, benign, or malignant, early research (reference) used the DDSM11,12,13. To guarantee the validity of these models and provide a standard for future studies, confirmed pathology data are essential. The Created Breast Imaging Subset of DDSM (CBIS-DDSM) is a development in the field of imaging. Upgrades and standardization of DDSM have produced a more recent dataset for use in breast imaging research. The work of (Maqsood, S, et al. 2022)14demonstrated the importance of this dataset for enhancing model generalizability and guaranteeing relevance to contemporary diagnostic difficulties15,16,17,18,19.
An innovative approach is introduced by integrating a hybrid deep learning algorithm that combines the stochastic gradients method with CNNs. Previous investigations20,21,22,23,24,25,26,27,28,29 have indicated that hybrid models are effective at extracting complex patterns from various datasets, exhibiting flexibility in various imaging properties and enhanced diagnostic capabilities. Although several datasets and methods have demonstrated potential, there is still a significant lack of comprehensive techniques that include several datasets. Managing bias and overfitting concerns as well as the requirement for interpretability in deep learning models are challenges. The complementary qualities of the datasets and algorithms could be used to produce more reliable and broadly applicable breast cancer diagnosis systems.
The literature review summarizes the important contributions made by earlier studies in the use of specific datasets and sophisticated techniques for the detection of breast cancer. On that basis, this work used a hybrid deep learning algorithm to integrate the Wisconsin Breast Cancer Database and CBIS-DDSM datasets to solve existing issues and further the continuous development of accurate and efficient breast cancer diagnostic systems.
Significance of the study
Improving diagnostic accuracy: By combining two significant datasets, the Wisconsin Breast Cancer Database and CBIS-DDSM, this work significantly improves the area of breast cancer detection. Together with in-depth mammography data, a thorough examination of 30 cell nuclei properties provides a wealth of data for creating a highly reliable diagnostic model. To improve the health of patients, a hybrid deep learning system that combines a CNN and stochastic gradients is being integrated with the objective of increasing diagnostic accuracy30. The requirement for updated and standardized datasets has been addressed by the integration of the CBIS-DDSM with the Wisconsin Breast Cancer Database. Incorporating a range of examples, such as benign, malignant, and normal cases, provides a thorough portrayal of breast pathology. This approach enhances the dependability of the created diagnostic system while allowing extensive comparisons with other investigations and datasets. The CNN and stochastic gradient hybrid deep learning approach is sufficiently adaptable to accommodate a range of imaging properties observed in both datasets. This flexibility is essential to the model’s capacity to recognize inadequate developments across many imaging modalities that are suggestive of breast cancer. Utilizing the advantages of both methods, this research aspires to develop a reliable and efficient diagnostic instrument for use in a variety of mammography investigations.
Ultimately, this investigation aspires to directly influence patient outcomes and therapeutic practices. Early detection can be greatly aided by an accurate and reliable breast cancer testing system, which enables early intervention and treatment. Increased diagnostic precision is especially important for differentiating between benign and malignant cases, minimizing unnecessary procedures and treatments, and eventually increasing the standard of patient care. This investigation advances the field of breast cancer research by establishing an integrated methodology that integrates conventional mammography investigations with intricate cellular characteristics. These results might further the continuing attempts to gain a deeper understanding of medical conditions by offering new insights into the connections between certain traits and breast cancer pathology. Benchmarking and comparative analysis are made possible by the hybrid deep learning method and dataset integration. To demonstrate the superiority of the created model in terms of accuracy and efficiency, the results were compared to those of current diagnostic techniques. Future research can benefit from this comparison study, which can also help choose the best diagnostic instruments for use in clinical settings. In conclusion, the relevance of this approach stems from its potential to aid in the detection of breast cancer. The objective of this project is to contribute to the creation of a reliable and accurate diagnostic system that may have a favourable influence on patient care and outcomes by combining extensive datasets, utilizing cutting-edge deep learning algorithms, and emphasizing clinical relevance. The research gap highlights challenges including prolonged calculation times due to extensive parameter tuning, late convergence, and high complexity. Additionally, the approach has limited scalability with high-dimensional data, as it heavily depends on advanced parameter tuning and numerical optimization.
The article makes the following key contributions to the field of breast cancer diagnosis:
-
Developed a novel hybrid deep learning model that combines CNN with stochastic gradient methods, tailored specifically for breast cancer detection.
-
Successfully integrated two diverse and significant datasets—the Wisconsin Breast Cancer Database and the CBIS-DDSM Dataset—to enhance the robustness and accuracy of the diagnostic model.
-
Conducted an extensive performance analysis using metrics such as accuracy, sensitivity, specificity, and AUC-ROC, demonstrating the model’s superiority over existing methodologies.
-
Implemented strategies to improve the model’s generalizability across different datasets and provided insights into its decision-making process, enhancing its clinical applicability.
-
Addressed ethical dimensions related to patient data usage, including privacy and consent, and discussed the model’s potential impact and integration within clinical practice.
The article is organized into following key sections: Sect. 2 discusses on the literature review, offering a detailed examination of recent advancements in deep learning techniques and their application in medical imaging. Section 3 then elaborates on the hybrid deep learning model, detailing the integration of the Wisconsin Breast Cancer Database and the CBIS-DDSM Dataset, along with the specific techniques used for data processing, model training, and performance evaluation. Next, Sect. 4 presents the methodology and framework used in the study. Section 5 focuses on the findings of the study, including a thorough performance analysis and comparison with existing methods, alongside a discussion of the model’s generalizability, interpretability, and clinical relevance in provided in Sect. 6. Finally, the article concludes in Sect. 7, summarizing the contributions of the study and proposing areas for further research and improvement.
Description of the hybrid deep learning algorithm
In this section, we provide a detailed description of the hybrid deep learning algorithm employed in this study to enhance breast cancer diagnosis. The section begins by outlining the core components of the CNN architecture, which forms the foundation of the model. Then the integration of the SGD method is elaborated, explaining how it is utilized to optimize the model’s parameters effectively.
Description of the convolutional neural network algorithm
CNNs are a class of deep learning models specifically designed for processing structured grid data, such as images, as shown in Fig. 131. They have proven highly effective in tasks such as image classification, object detection, and segmentation. The following is a description of the key components of CNNs and the corresponding mathematical equations:
Convolutional layer
The core operation in CNNs is convolution. This operation involves sliding a small window called a kernel or filter over the input data, performing element-wiseCN multiplication, and summing the results to produce a feature map. The convolution operation is expressed as follows:
where: C(i, j) is the value at position (i, j) in the feature map. Iv(i + m, j + n) is the input value at position (i + m, j + n). K(vm, n) is the kernel value at position (m, n).
Activation function
After convolution, an element wise activation function is applied to introduce nonlinearity to the model. Common activation functions include the Rectified Linear Unit (ReLU). The ReLU activation function is defined as follows:
where, x is the input value.
Pooling layer
Pooling layers are used to down sample the spatial dimensions of the feature maps, reducing computation and controlling overfitting and Applies a value per pixel called intensity or brightness. Max pooling is a common pooling technique defined as follows:
where Pv(i, j) is the value at position (i, j) in the pooled feature map.
Fully connected layer
The fully connected layer connects every neuron in one layer to every neuron in the next layer, typically followed by an activation function. The output of a fully connected layer is computed as follows:
where, Op is the output. X is the input vector. Wm is the weight matrix. Bi is the bias vector. f is the activation function.
Softmax layer
In classification tasks, a softmax layer is often used to produce probability distributions over multiple classes. The softmax function is defined as follows:
where P(yi) is the probability of class i and yi is the input to the softmax layer for class i.
These mathematical representations provide a foundation for understanding the operations performed by CNNs, allowing them to effectively learn hierarchical features from input data, particularly in image-based tasks.
Description of CNNs: VGG network
The VGG (Visual Geometry Group) network is a widely recognized architecture for image classification. VGG is known for its simplicity and uniformity, with most layers consisting of 3 × 3 convolutional filters followed by max pooling layers, as shown in Fig. 232. Here, is an overview of the VGG network architecture and mathematical equations for its key components:
Convolutional block
The VGG network consists of multiple convolutional blocks, each comprising multiple convolutional layers with small 3 × 3 filters. The convolutional block can be represented as follows:
where X is the input feature map. ConvConv represents a 3 × 3 convolutional layer. MaxPoolMaxPool represents a 2 × 2 max pooling layer.
Fully connected block
Towards the end of the network, fully connected layers are employed for final classification. The fully connected block is expressed as follows:
where FCFC denotes a fully connected layer. SoftmaxSoftmax is the softmax activation function for classification.
Overall VGG architecture
The complete VGG architecture is a stack of convolutional blocks followed by fully connected blocks. For example, the VGG16 architecture can be represented as follows:
Convolutional layer
The convolutional layer can be represented mathematically as follows:
where Yi, j,k is the output at position (i, j) in the k-th feature map. Xi, j,l is the input at position (i, j) in the l-th input feature map. Wi, j,l, k is the weight for the convolutional kernel at position (i, j) for input channel l and output channel k. Bk is the bias term for the k-th feature map. σ is the activation function, commonly referred to as ReLU.
Max-pooling layer
The max pooling layer is represented as follows:
where Ri, j is the receptive field at position (i, j) in the input feature map.
Fully connected layer
The fully connected layer is expressed as follows:
where Yk is the output of the k-th neuron in the fully connected layer. Xl is the input from the l-th neuron in the preceding layer. Wk, l is the weight connecting the l-th neuron to the k-th neuron. Bk is the bias term for the k-th neuron. σ is the activation function, often ReLU.
These equations provide a mathematical representation of the key components of the VGG network, offering insights into the operations performed by this architecture during image classification tasks.
Description of CNNs: GoogleNet (inception v1)
GoogleNet, also known as Inception v1, is a deep convolutional neural network developed by Google. It is renowned for introducing the concept of “inception modules,” which utilize multiple convolutional filters of different sizes within the same layer33. This design allows the network to capture features at various scales efficiently. Here, is an overview of the GoogleNet architecture and mathematical equations for its key components:
Inception module
The inception module is the cornerstone of GoogLeNet and incorporates multiple convolutional filters of different sizes in parallel, as shown in Fig. 3. The mathematical representation of the Inception module can be expressed as:
where X is the input feature map. Conv1 × 1, Conv3 × 3, and Conv5 × 5 represent convolutional layers with filter sizes of 1 × 1, 3 × 3, and 5 × 5, respectively. MaxPool3 × 3 denotes a max pooling layer with a filter size of 3 × 3. Concat signifies the concatenation of the outputs from all these operations.
GoogLeNet architecture
The complete GoogLeNet architecture is composed of multiple inception modules stacked together interspersed with other layers, such as max pooling and fully connected layers. A simplified representation of the architecture can be expressed as in Fig. 4:
Convolutional layer
The convolutional layer is represented mathematically as follows:
where Yi, j,m is the output at position (i, j) in the k-th feature map. Xi, j,m, l is the input at position (i, j) in the l-th input feature map. Wi, j,l, m is the weight for the convolutional kernel at position (i, j) for input channel l and output channel m. Bm is the bias term for the m-th feature map. σ is the activation function, commonly referred to as ReLU.
Max-pooling layer
The max pooling layer is represented as follows:
where Ri, j is the receptive field at position (i, j) in the input feature map.
Fully connected layer
The fully connected layer is expressed as follows:
where Ym is the output of the m-th neuron in the fully connected layer. Xl is the input from the l-th neuron in the preceding layer. Wm, l is the weight connecting the l-th neuron to the m-th neuron. Bm is the bias term for the m-th neuron. σ is the activation function, often ReLU.
These equations provide a foundation for understanding the operations performed by GoogleNet, particularly the Inception modules, enabling effective feature extraction at multiple scales within the network.
Description of CNNs: densely connected Network (DenseNet)
DenseNet is a convolutional neural network architecture that introduces dense connections between layers. In DenseNet, each layer receives input not only from the preceding layer but also from all preceding layers. This dense connectivity is designed to address issues such as vanishing gradients, promote feature reuse, and enhance model compactness34. Here, is an overview of the DenseNet architecture and mathematical equations for its key components:
Dense block
The DenseNet architecture is built around dense blocks, each consisting of multiple densely connected layers, as shown in Fig. 5. Mathematically, the output of a dense block can be expressed as:
where X0 is the input to the dense block. Hi is the output of the i-th layer in the dense block. ConcatConcat denotes the concatenation operation.
Overall DenseNet architecture
The complete DenseNet architecture is a sequence of dense blocks separated by transition layers, as shown in Fig. 6. A simplified representation of the architecture can be expressed as:
Transition layer
Transition layers are used to control the growth of feature maps between dense blocks. Mathematically, the output of a transition layer can be expressed as follows:
where Conv1 × 1 represents a 1 × 1 convolutional layer. BatchNormBatchNorm is a batch normalization method. ReLUReLU is the rectified linear unit activation function.
Convolutional layer
The convolutional layer is represented mathematically as follows:
where Yi, j,z is the output at position (i, j) in the z-th feature map. Xi, j,l is the input at position (i, j) in the l-th input feature map. Wi, j,l, z is the weight for the convolutional kernel at position (i, j) for input channel l and output channel z. Bz is the bias term for the z-th feature map. σ is the activation function, commonly referred to as ReLU.
Global average pooling layer
The GAP layer computes the average value of each feature map across its spatial dimensions. Mathematically, the global average pooling is expressed as:
where H and W are the height and width of the feature map, respectively. Yk is the output of the k-th neuron in the global average pooling layer.
These equations provide a foundation for understanding the operations performed by DenseNet, particularly the dense connections that facilitate feature reuse and contribute to the network’s parameter efficiency.
Stochastic gradient descent (SGD)
The term “Stochastic Gradient Descent” (SGD) is a general optimization algorithm widely used in training CNNs and other machine learning models35. SGD optimizes the model parameters by updating them iteratively based on the gradients of the loss function with respect to those parameters. A description of the SGD method for CNNs and the mathematical equations involved are provided below:
Objective function:
In CNNs, as in many machine learning models, the objective is to minimize a loss function J(θ), where θ represents the model parameters.
Update rule:
The update rule for the parameters θ during each iteration of SGD is given by:
where η is the learning rate, a hyperparameter controlling the size of the step. ∇J(θt) is the gradient of the loss function with respect to the parameters at iteration t.
Batch Stochastic Gradient Descent:
In practice, SGD is often applied to minibatches of data rather than to the entire dataset. In this case, the update rule becomes:
where ∣B∣ is the batch size. ∇Ji(θt) is the gradient of the loss with respect to the parameters computed on the i-th mini-batch.
Mathematical Equations:
Objective Function:
The objective function is typically defined as the average loss over the training dataset:
where N is the number of training samples. L is the loss function. f(xi;θ) is the predicted output of the CNN for input xi with parameter θ. yi is the true label of the i-th sample.
Gradient calculation.
The gradient of the loss with respect to the parameters is computed using the chain rule:
This involves computing the gradient of the loss function and the gradient of the model’s output with respect to the parameters.
Update rule:
The parameters are updated using Eqs. (22) and (23), which are the calculated gradients. These equations form the basis of the SGD optimization algorithm for training CNNs36. The learning rate (η) is a critical hyperparameter that requires tuning for effective training.
Clinical data sources
This section provides an overview of the clinical data sources utilized in the study, specifically focusing on the Wisconsin Breast Cancer Database and the CBIS-DDSM Dataset. It details the characteristics, relevance, and integration of these datasets in developing and validating the hybrid deep learning model for breast cancer diagnosis.
Wisconsin breast cancer database
A significant and widely used dataset in the field of breast cancer research is the Wisconsin Breast Cancer Database. The source material is obtained from breast mass Fine Needle Aspirates (FNAs), which offer important insights into cellular properties. The prominent features of this dataset include the following:
Attributes of Cell Nuclei: The collection provides comprehensive information on 30 features of cell nuclei, describing them in excellent detail. These characteristics provide a thorough understanding of cellular morphology by encompassing a variety of features, such as diameter, texture, and concaveness, as shown in Table 1.
Instances and class distribution: The dataset, which includes 569 cases in total, includes a wide range of breast cancer cases. A total of 212 of these patients were categorized as malignant tumors, suggesting that diagnostic models can be used to accurately distinguish between benign and malignant diseases, as shown in Table 2.
Ground truth pathology information: Ground truth labels are reliable for investigators and clinicians since the pathology information has been validated for every patient in the Wisconsin Breast Cancer Database. The training, verifying, and testing machine learning models guarantee the dataset’s dependability, as shown in Table 3.
Fine needle aspiration (FNA): Given that FNA is a frequently used diagnostic technique in actual clinical settings, the use of FNA samples provides an extra degree of clinical relevance. Therefore, the dataset is especially useful for comprehending and identifying breast cancer based on cellular features, as shown in Table 4.
CBIS-DDSM dataset description
This dataset is the original dataset (163GB) that has been converted to jpeg format. The resolution is the same as that of the original dataset.
No. of courses: 6775.
No. of rows: 6775.
Number of participants: 1,566 (note).
No of images: 10,239.
Image size (GB): 6(.jpg).
The imaging data for this set is structured such that each participant has multiple patient identities. For example, pat_id 00038 contains 10 distinct patient IDs (e.g. Calc-Test_P_00038_LEFT_CC, Calc-Test_P_00038_RIGHT_CC_1) that provide information about the persons within the ID, and according to the actual DICOM metadata, only 156, 6 IDs appear. Participants in the group38.
Curated breast imaging subset of DDSM (CBIS-DDSM)
An improved and standardized version of the Digital Database for Screening Mammography (DDSM) is the CBIS-DDSM dataset, as shown in Fig. 7. It offers improvements over its predecessors and an array of mammography investigations. The important characteristics of the CBIS-DDSM dataset include the following:
The CBIS-DDSM is a standardized and updated version of the DDSM that takes into account improvements in breast imaging techniques and technology, as shown in Table 5. The dataset was chosen carefully to guarantee coherence and pertinence within the framework of modern mammography research. A total of 2,620 scanned film mammography investigations encompassing a wide range of breast diseases, including benign, malignant, and normal instances, were included in the initial DDSM. Because of its variety, the CBIS-DDSM is an invaluable resource for developing and accessing diagnostic models for breast cancer. The CBIS-DDSM preserves pathology data for every mammography investigation, much like does the Wisconsin Breast Cancer Database. By doing so, the dataset’s clinical validity is preserved, facilitating the creation of deep learning models that correspond with actual diagnostic instances.
The CBIS-DDSM provides an accurate representation of breast diseases found in clinical practice by including lesions that are benign, malignant, or normal. This diversity is important for training models that can reliably diagnose various breast diseases in Table 5. The therapeutic relevance of breast cancer research has been greatly enhanced by the contributions of both the CBIS-DDSM and the Wisconsin Breast Cancer Database databases. The former incorporates cellular features, while the latter includes a variety of mammography investigations to guarantee that machine learning models trained on these datasets can accurately capture the nuances of diagnosing breast cancer in practical settings. The clinical utility of these datasets is further enhanced by the availability of ground truth pathology information, which makes them invaluable tools for improving breast cancer research and diagnosis.
Methodology
The objective of the proposed method is to integrate the CBIS-DDSM and Wisconsin Breast Cancer Database datasets to create an innovative breast cancer diagnosis system. For optimized diagnostic accuracy, the solution uses a hybrid deep learning algorithm that combines CNNs with the stochastic gradients method. This approach was used by considering the numerous characteristics of cell nuclei from the Wisconsin Breast Cancer Database and the variety of mammography studies in the CBIS-DDSM dataset.
Data integration and pre-processing
Integration of Datasets: The process of integrating the CBIS-DDSM and Wisconsin Breast Cancer Database datasets into one cohesive, coherent dataset is known as integration. This procedure ensured that the two datasets were integrated effectively, enabling a thorough analysis that utilized each dataset’s characteristics, as shown in Table 6. To produce a coherent dataset for further analysis, harmonizing data types, normalizing variable names, and aligning data structures are important considerations.
Providing particular interest to the formats and characteristics of the data in both datasets is necessary to guarantee consistency and compatibility. Harmonizing features such patient identities, image metadata, and pathology data is necessary to prevent discrepancies that might affect how reliable the diagnostic model is. This is an essential step in preserving the integrity of the integrated dataset and facilitating an easy transition between the various types of data included in the CBIS-DDSM and the Wisconsin Breast Cancer Database. One essential component of data preparation is handling missing values. This entails locating and fixing any null or gap items in the combined dataset. The dataset may be made full and analysis-ready by using imputation techniques to fill in missing values. The kind of missing data and its possible influence on subsequent investigations should be taken into consideration when selecting an imputation approach. To provide a consistent scale for all variables, features must be normalized37. This procedure assures that, because of variations in scales or measuring units, no single factor affects the model excessively. Methods such as z score normalization or min–max scaling are frequently used to align all features into a similar range, which helps the hybrid deep learning algorithm converge during training.
Creating that all the information is represented consistently across various aspects is a necessary step in standardizing data formats. This covers categorical variables, encoding data, and standardizing units of measurement. By offering a standard framework for processing and analysing data, consistent data formats support the interpretability and stability of the model. A single dataset is produced once the integration and preparation stages are completed. This dataset provides an in-depth analysis of cellular characteristics and mammography studies by combining data from the Wisconsin Breast Cancer Database and CBIS-DDSM. The hybrid deep learning method may then be used with the combined dataset, enabling reliable training and assessment procedures. To summarize, the effective implementation of the proposed breast cancer diagnostic solution hinges on the careful integration and preparation of the CBIS-DDSM and Wisconsin Breast Cancer Database datasets. By completing these processes, a unified, consistent, and comprehensive dataset is created, which paves the way for the construction of a highly accurate and dependable diagnostic model38.
Feature extraction and selection
Extracting Features from Cell Nuclei Attributes: Identifying the relevant characteristics among the 30 cell nuclei properties in the Wisconsin Breast Cancer Database is an important first step in gathering data needed for a diagnosis of breast cancer. These features include measures of various lengths, such as diameter, texture, and concaveness, all of which provide important new information on the properties of cell nuclei. These processed characteristics are transformed during feature extraction into a fewer, more relevant set of features that accurately capture the underlying patterns in the data. When different objects are detected, the optimal number of image features within the processing framework can be determined. Principal component analysis (PCA) dimensionality reduction is for establishing image features for appropriate feature extraction methods and detection.
The objective of the feature extraction procedure is to minimize the total dimensionality while retaining the most important information. To achieve this, approaches such as principal component analysis (PCA) and dimensionality reduction methods are employed. This approach ensures that the model is more effective and less prone to overfitting during the training procedure in the future, particularly when working with a high-dimensional dataset such as the Wisconsin Breast Cancer Database. Identifying and maintaining the most discriminative and informative characteristics for model training is known as feature selection. Improving model efficiency, interpretability, and generalizability requires taking this critical step. To do this, a variety of feature selection strategies can be used, including the following:
Filter methods:
To assess the importance of each trait to the objective variable (cancer status), statistical tests, including chi-squared or mutual information, can be used. Repetitive traits can be found, and only those that add original information should be kept according to correlation analysis, as shown in Fig. 8.
According to the model’s performance, Recursive Feature Elimination (RFE) entails iteratively eliminating the least significant features. Iterative techniques of forward or backwards selection add or delete features according to how they affect the performance of the model. Model training included regularization techniques and techniques, including Least Absolute Shrinkage and Selection Operator (LASSO) regression, which automatically penalizes less significant characteristics. The selection procedure might be guided by the feature significance ratings that tree-based models frequently provide. In Table 7, these metrics evaluate how much information each feature contributes to the prediction of the target variable, and they are especially pertinent to decision tree-based models.
The objective is to enhance the model’s capacity to identify patterns connected with breast cancer by reducing the dataset to include only the most pertinent features through the use of various feature selection approaches. This approach lowers the chance of overfitting and enhances interpretability in addition to increasing model efficiency—especially when combined with a hybrid deep learning approach. To summarize, the process of feature extraction and selection plays a crucial role in extracting significant data from the 30 variables included in the Wisconsin Breast Cancer Database. This study lays the groundwork for the creation of a more precise, understandable, and effective breast cancer diagnostic paradigm.
Model architecture
The proposed strategy develops a hybrid deep learning model that fuses the stochastic gradients technique with CNNs. The use of both techniques results in positive results: the stochastic gradients method’s optimization power and the CNN’s capacity to capture complex spatial patterns. The model aspires to improve the performance of learning complicated hierarchical features from the CBIS-DDSM and Wisconsin Breast Cancer Database datasets by incorporating these components39,40,41.
CNN architecture for spatial pattern recognition: CNNs are highly effective at performing image-related tasks, which renders them appropriate for diagnosing breast cancer, where spatial patterns are important. For the model to successfully learn hierarchical features from retrieved cell nuclei properties and mammography imagery, the architecture of the CNN layers is essential, as shown in Fig. 9. Usually, the architecture consists of the following.
Convolutional layers
These layers search for certain patterns in the input data by applying filters. These filters have the potential to capture spatial correlations in mammography imagery and cell nuclei properties for the purpose of diagnosing breast cancer.
Pooling layers
The pooling layers preserve important information while reducing the input’s spatial dimensions. The most notable characteristics in each region are captured with the use of max pooling and average pooling methods.
Flatten layer
To prepare the multidimensional output from earlier layers for input into fully connected layers, the output must first be flattened into a one-dimensional vector.
Fully connected layers
These layers capture complex feature interactions by processing the vector that has been flattened. The last fully connected layers generate output probabilities that show whether a certain tumor is benign or malignant.
Activation functions:
Activation functions, including the Rectified Linear Unit (ReLU), are used to provide nonlinearity and improve the model’s ability to recognize complex patterns.
Stochastic Gradients Method for Optimization: To adjust parameters and enhance model performance, the hybrid model incorporates the stochastic gradients technique, which is frequently used in optimization techniques, including SGD. The model can effectively explore the parameter space by modifying weights and biases to minimize the loss function through the use of the stochastic gradients approach.
In the training phase, the integrated and preprocessed datasets are fed into the hybrid model, and the parameters are repeatedly adjusted to reduce the discrepancy between the expected and actual results. The stochastic gradients approach optimizes the whole model, while the CNN layers discover spatial patterns using cell nuclei properties and mammography imagery during training. The CBIS-DDSM dataset fine-tuning stage improves the model’s applicability to a variety of mammography investigations. By doing so, it is ensured that the hybrid model may be generalized to various imaging properties and case variations of breast cancer in an efficient manner.
Training on the Wisconsin Breast Cancer Database: A vital initial stage in creating a reliable breast cancer diagnosis model is the training phase. The Wisconsin Breast Cancer Database is a comprehensive dataset that includes 30 properties of cell nuclei, including important aspects, such as diameter, texture, and concaveness, which the hybrid deep learning model is exposed to during this procedure. Enabling the algorithm to identify complex patterns related to benign and malignant tumors in this dataset is the objective. The variance among the expected and actual results is measured by a loss function, which is optimized to train the model. To minimize the loss, this optimization process involves modifying the model’s weights and biases using methods such as backpropagation. Because the CNN layers have been designed to efficiently learn hierarchical information from the characteristics of the cell nucleus, they play a crucial role in this training phase. The model’s ability to distinguish minute variations between malignant and benign tumor features is facilitated by the CNN’s ability to identify spatial patterns within the attributes.
Regularization strategies, including dropout and L2 regularization, can be used to stop overfitting. By minimizing the model’s dependence on particular data points during training, these strategies aid in the model’s generalization. This dataset provides a wide range of mammography investigations, including normal, benign, and malignant cases. It is an updated and standardized version of the DDSM. Optimizing the model’s fine-tuning ability is essential for improving its generalizability to new data and its flexibility for a variety of imaging investigations.
Variations in imaging parameters and cases not in the Wisconsin Breast Cancer Database were introduced in the CBIS-DDSM dataset. By fine-tuning, the model becomes more adaptable to a variety of mammography investigations by allowing its acquired characteristics to adjust to these changes. Fine-tuning prevents the model from being too adaptable to the unique features of the training dataset and reduces dataset bias. This is especially crucial when developing a diagnostic tool that may be used in various patient demographic and medical settings. The process of fine-tuning increases the model’s efficacy in real-world circumstances by optimizing it for better generalization. The objective of this study was to develop a diagnostic tool that will add to the clinical relevance of breast cancer by accurately identifying patterns of breast cancer through different mammography investigations. In conclusion, the training and fine-tuning stages are essential for the creation of a model. Training on the Wisconsin Breast Cancer Database allows for the collection of complex patterns associated with tumors, and fine-tuning using the CBIS-DDSM dataset improves the model’s generalizability and flexibility, which together lead to a more robust and efficient breast cancer diagnostic system. Table 8 shows the hybrid CNN model output for breast cancer detection based on the selected features, and it shows the morphology and finally the CNN output.
In summary, utilizes the CBIS-DDSM dataset to further refine the model’s parameters once it has been trained on the Wisconsin Breast Cancer Database. The hybrid deep learning model that combines the stochastic gradients approach with the CNN architecture has the potential as an effective diagnostic tool for breast cancer. The model attempts to capture complex spatial patterns from cell nuclei properties and mammography imagery by using CNN layers to learn hierarchical features, ultimately leading to more accurate and consistent diagnostic results.
Experimental results
The Wisconsin Breast Cancer Database and the Curated Breast Imaging Subset of DDSM (CBIS-DDSM) datasets were used to thoroughly assess the hybrid deep learning algorithm used in the breast cancer diagnosis model. This method consists of CNNs and stochastic gradients. The Digital Database for Screening Mammography (DDSM), which initially included 2,620 scanned film mammography investigations, has been updated and standardized in the CBIS-DDSM dataset. The Wisconsin Breast Cancer Database contains 569 cases, of which 212 were categorized as malignant tumours. A crucial initial phase in determining the efficacy of the created breast cancer diagnostic model is to evaluate its diagnostic performance, as shown in Fig. 10. To provide a more detailed picture of the model’s capabilities, a set of extensive assessment measures is used. Among these measures are:
The ratio of effectively categorized instances to the total number of occurrences is used to calculate accuracy, which assesses how accurate the model’s predictions are overall. Recall, also referred to as sensitivity, is a measure of how well a model recognizes malignant tumors. The ratio of true positives to the total number of false negatives and true positives is used to compute this parameter. Specificity, which is computed as the ratio of true negatives to the total number of true negatives and false positives, assesses the model’s accuracy in identifying benign tumors. The precision assesses the model’s accuracy in predicting a positive class. The ratio of true positives to the total number of true positives and false positives is calculated as shown in Fig. 11.
Area under the receiver operating characteristic curve (AUC-ROC)
The AUC-ROC is a valid statistic that assesses the model’s capacity to distinguish between benign and malignant cases at different thresholds, as shown in Fig. 12. The results provide a thorough overview of the model’s performance, with a higher AUC-ROC indicating improved discriminating ability.
Cross-validation:
To assure the model’s stability and dependability over various dataset subsets, cross-validation is a significant approach. This procedure entails dividing the dataset into several folds, training the model on various permutations of these folds, and repeatedly assessing the model’s performance. Popular techniques for cross-validation include leave-one-out cross-validation, in which every instance acts as a separate validation set, and k-fold cross-validation, in which the dataset is split into k subsets.
Cross-validation is crucial for evaluating the model’s resilience across multiple data subsets, as shown in Figs. 13, 14, 15 and 16. It ensures that the model performs well beyond a specific training-validation split, reducing the risk of overfitting by providing a thorough assessment of generalization abilities. Additionally, cross-validation helps optimize hyperparameters, allowing the model to be adjusted and verified on various subsets, ensuring performance is not skewed by any particular parameter set. The consistent performance across folds confirms the model’s diagnostic validity and its ability to generalize to new data, essential for practical applications. So, using cross-validation and extensive evaluation metrics is vital for thoroughly assessing the breast cancer diagnostic model’s accuracy, sensitivity, specificity, precision, and discriminatory power, thereby supporting its validity and clinical relevance.
The development of a reliable, apparent, and accurate breast cancer diagnostic model requires a high degree of interpretability and interpretability. Improving the interpretability of the model entails improving the accessibility and comprehensibility of decision-making procedures for medical practitioners. This is especially crucial for medical applications, as building trust and enabling the use of AI-powered technologies in clinical settings both depend on authenticity. Understanding which areas of the input data are essential for decision-making is facilitated by examining the activation maps in the Convolutional Neural Network (CNN) layers. This enables medical practitioners to determine which areas of a picture contribute most to the model’s predictions, which may be very useful when analysing mammography images. Plots of feature significance provide insight into how each input feature (the characteristics of the cell nuclei) affects the predictions made by the model, as shown in Figs. 17, 18 and 19. Determining the most important factors related to breast cancer can be facilitated by identifying which aspects have greater influence. It is possible to assign importance to each input feature using strategies such as layerwise relevance propagation, emphasizing the role that certain characteristics have in the decision-making process. This helps to clarify the process by which the model makes its predictions.
Providing detailed explanations for each forecast enables medical personnel to comprehend the model’s reasoning behind a particular patient’s decision. Because local interpretability makes the model’s decision-making process more concrete and applicable to actual situations, it fosters confidence. Global explanations provide an overview of the model’s general behavior throughout the whole dataset. Finding recurring themes or characteristics that reliably influence forecasts is part of this. To comprehend the basic patterns and strengths of the model, global interpretability is essential. A thorough method for deciphering any machine learning model’s output is provided by the SHAP values. They are able to produce comprehensible and naturalistic summaries of the model’s decision-making process by attributing its relative importance to the prediction to each characteristic.
Clinical relevance:
Healthcare practitioners can compare the model’s predictions to accepted medical knowledge by integrating the model’s output with current clinical information. The model gains credibility and is more likely to be accepted by the medical community as a result of this alignment. Healthcare practitioners may make well-informed judgments based on the model’s predictions when they have access to transparent and comprehensible models. Collaboration between AI tools and physicians can be improved by having a better understanding of the rationale underlying the model’s suggestions. Establishing confidence in AI-powered diagnostic tools requires transparency and interpretability. The validity and dependability of the model’s output can be increased for healthcare practitioners by giving them concise explanations for the predictions made by the model. To summarize, the interpretability and interpretability of the breast cancer diagnostic model are enhanced by the application of visualization tools and the provision of explanations for model predictions. This promotes acceptability and confidence in the clinical community while also helping healthcare practitioners comprehend the model’s decision-making procedures.
Validation on an independent dataset:
An important initial phase in evaluating the generalizability and realistic performance of the trained breast cancer detection model is validation on a separate dataset. To determine how effectively the model can apply it’s newly learned patterns to fresh, unknown data, it is tested on a dataset that has not been used during training or fine-tuning.
The model’s capacity to generalize beyond the unique features of the training and fine-tuning datasets is assessed by validation on an independent dataset. This approach is important for assessing the model’s adaptability to different patient demographics, differences in imaging, and other elements that arise in actual clinical settings. One essential feature of a trustworthy diagnostic model is its robustness. Determining whether the model retains its diagnostic efficacy and accuracy in the face of changes and difficulties not observed in the training set is easier by validation on a different dataset, as shown in Figs. 20, 21, 22 and 23. When a model performs effectively on training data but is unable to generalize to new data, this is known as overfitting. To verify that the model’s performance is not inflated as a result of learning certain patterns from the training dataset, independent validation functions are used as a test for overfitting. The ultimate objective of a breast cancer diagnostic model is to be useful in actual clinical situations. By simulating the model’s performance in a variety of scenarios, validation on an independent dataset improved the model’s adaptability to various patient demographics, healthcare facilities, and imaging technologies.
Evaluation across diverse patient populations
Demographic diversity:
It is possible to evaluate the diagnostic efficacy of the model across a range of demographic groups using independent validation. This covers a range of age brackets, races, and other demographic variables that might impact the features of individual instances of breast cancer.
Pathological variations:
Breast cancer can present in a variety of pathological forms, and distinct features may be observed in different populations. To guarantee that the diagnostic abilities of the model are not skewed toward certain pathogenic variants found in the training data, the model was validated on a different dataset, as shown in Tables 9, 10 and 11.
Evaluation across imaging variants
The model’s performance was evaluated independently across multiple imaging modalities that might not have been completely captured in the training dataset. This approach is especially crucial because advances in imaging technology might lead to different representations of breast cancer. Changes in the resolution and quality of the images can affect how well the model diagnoses breast cancer. Assessing how well the model generalizes to datasets with different image qualities and resolutions is easier with the use of independent validation. The area under the receiver operating characteristic curve (AUC-ROC), accuracy, sensitivity, specificity, precision, recall, and other measures are used in validation on an independent dataset, much as they are in training. These metrics offer a thorough evaluation of the diagnostic performance of the model, as shown in Table 12.
To summarize, the process of validating a breast cancer diagnosis model on an independent dataset is an essential measure that guarantees its dependability and practicality. This procedure enhances the model’s credibility and facilitates its use in clinical practice by assessing its effectiveness across a range of patient demographics and imaging variances.
Comparative analysis
To establish the viability of the proposed hybrid deep learning algorithm in terms of accuracy, efficiency, and adaptation, it must be compared with existing methods for diagnosing breast cancer. The purpose of this analysis is to illustrate the new algorithm’s contributions and developments in the field of breast cancer diagnosis, as shown in Table 13.
The diagnostic accuracy of the proposed hybrid deep learning algorithm is compared to that of existing techniques. To measure its effectiveness, criteria such as overall accuracy, precision, sensitivity, and specificity were used. An improved algorithm should reliably outperform existing methods in accurately identifying benign or malignant cases, as shown in Table 14 and Fig. 24.
The proposed algorithm’s computational effectiveness is evaluated by contrasting its training and inference times with those of other approaches. In clinical settings, where immediate findings are essential for patient care, the practical deployment of a tool for diagnosis depends heavily on its efficiency. We analysed how well the hybrid deep learning system works with different patient demographics, imaging modalities, and datasets. An optimal algorithm should be able to manage variations observed in real-world circumstances with outstanding results and excellent generalization abilities. The model’s output was analysed on various datasets that were not used for fine-tuning or training. The algorithm’s capacity to generalize beyond certain training settings and assure its applicability to other clinical situations is validated through the use of cross-dataset analysis.
Comparison with traditional methods
Examine how the proposed hybrid deep learning algorithm compares with both more modern deep learning techniques and more conventional strategies, such as rule-based systems or statistical models. This more comprehensive perspective highlights the improvements made by the new algorithm when considering the changing landscape of diagnostic techniques. The interpretability and reliability of the suggested algorithm are compared to those of existing methods. The validity of diagnostic tools is enhanced by apparent decision-making procedures and explainable AI capabilities, particularly in healthcare applications.
We conducted an accurate and standardized comparison by using benchmark datasets that are widely regarded in the field of breast cancer diagnosis. To ensure a thorough evaluation of the suggested hybrid deep learning algorithm, this approach may involve well-established datasets that are often used for assessing diagnostic algorithms. To determine the significance of the observed variations in the performance measures between the proposed algorithm and current approaches, statistical analysis was used. The outcomes of the comparison analysis are more reliable when they are supported by statistical significance.
The clinical applicability of the proposed algorithm was evaluated in consultation with medical experts and compared to that of current techniques. The adoption of diagnostic tools depends on an understanding of their practical consequences and clinical applicability. In conclusion, a thorough comparison investigation should take into account a wide range of current techniques, including conventional approaches, and should prioritize accuracy, efficiency, and versatility. This work provides the groundwork for proving the advantages of hybrid deep learning algorithms in breast cancer diagnosis by thoroughly testing and benchmarking them.
Discussion
Promising results have been obtained when a hybrid deep learning algorithm consisting of CNNs and stochastic gradients is used for the detection of breast cancer utilizing the Wisconsin Breast Cancer Database and Curated Breast Imaging Subset of the DDSM (CBIS-DDSM) datasets. The main conclusions, ramifications, and prospective developments from the investigation are the main topics of this discussion.
Diagnostic performance
Excellent results were obtained using the hybrid deep learning method in terms of accuracy, sensitivity, specificity, precision, and the significant area under the ROC curve (AUC-ROC) value. Taken together, these metrics show how well the model can distinguish between benign and malignant lesions and are essential for the timely and precise identification of breast cancer.
The model demonstrated strong performance across a comprehensive range of evaluation metrics:
-
Accuracy: Achieved an overall accuracy rate of 96%, indicating a high percentage of correctly identified cases within the entire dataset.
-
Sensitivity (Recall): The model exhibited a high sensitivity of 95% in accurately detecting malignant tumors, making it crucial for minimizing false negatives and facilitating early cancer diagnosis. Additionally, 96% of benign cases were correctly identified, reflecting a high level of specificity and reducing the risk of false positives.
-
Precision: The model achieved a precision of 95%, underscoring the accuracy of its positive predictions for cases classified as malignant.
-
Area under the ROC curve (AUC-ROC): The model demonstrated strong discriminatory power across various thresholds, with an AUC-ROC value of 0.96, highlighting its effectiveness in differentiating between benign and malignant lesions.
The algorithm’s robustness was further evidenced by its ability to effectively analyze and identify patterns in the Wisconsin Breast Cancer Database, which comprises 569 instances and 30 cell nucleus characteristics. Its flexibility in adapting to evolving imaging standards and variations was also demonstrated through its application to the CBIS-DDSM dataset, an enhanced version of the DDSM.
Cross-validation techniques were employed to validate the model’s robustness, ensuring consistent performance across different subsets of the datasets. This validation is crucial for establishing the generalizability and reliability of the algorithm across various scenarios and conditions. The model’s ability to maintain high diagnostic accuracy across diverse patient demographics and imaging variations significantly enhances its credibility and practical application potential.
Comparative analysis
Compared with other breast cancer diagnosis techniques, the hybrid deep learning system demonstrated greater accuracy, efficiency, and flexibility. This implies that the fusion of CNNs and stochastic gradients has resulted in progress in the identification of breast cancer, surpassing conventional techniques and modern machine learning methodologies. The efficacy of the model in distinguishing between benign and malignant cases is attributed to its use of deep learning techniques for feature extraction from cell nuclei properties and mammography images.
The interpretability of the model has improved with the use of visualization techniques such as activation maps and feature significance plots. By providing medical practitioners access to learned characteristics and decision-making processes, these technologies enhance the diagnostic system’s credibility and transparency. The algorithm’s clinical usefulness has been confirmed by collaboration among development teams and medical practitioners. The model was verified to comply with the standards and practical requirements of clinical practice through frequent feedback sessions, validation studies utilizing clinical data, and integration of domain-specific knowledge.
For the diagnostic system to be successfully integrated into clinical settings, it must have an intuitive user interface. The goal of the user-friendly dashboard, interactive visualization tools, and clear explanations for forecasts is to make it easier for healthcare professionals to comprehend and use the information. The diagnostic tool’s acceptability and practical deployment in actual healthcare settings depend heavily on its user-centric design.
Conclusion
The proposed deep learning model, based on a fine-tuned pre-trained CNN, effectively classifies full mammogram images as benign or malignant. While ROI-based mammography can yield higher accuracy, achieving sufficient accuracy for complete mammogram images is challenging. Our trained CNN captures comprehensive information from a single image. Using the Wisconsin Breast Cancer Database and the Curated Breast Imaging Subset of DDSM (CBIS-DDSM), we demonstrated the efficacy of integrating CNNs with stochastic gradients in improving breast cancer diagnosis. The study, which focused on 569 patients from the Wisconsin Breast Cancer Database and utilized the CBIS-DDSM dataset, revealed that the hybrid deep learning algorithm significantly outperformed baseline models in accuracy, sensitivity, specificity, and precision. Its strong AUC-ROC value and ability to accurately distinguish between benign and malignant tumors underscore its potential as a reliable tool for breast cancer screening.
The hybrid deep learning system outperformed other breast cancer diagnosis techniques in a comparative comparison. This finding suggested that, by outperforming both conventional techniques and modern machine learning approaches, this approach has the potential to make a significant contribution to the changing field of breast cancer detection. A hybrid deep learning algorithm consisting of CNNs and stochastic gradients is used for the detection of breast cancer utilizing an accuracy is 96% precision of 95%, Recall is 95%, and AUC-ROC value of 0.96. The algorithm’s transparency has been improved by including visualization approaches, such as activation maps and feature significance plots, in response to the demand for interpretability in deep learning models. The model’s clinical relevance has been confirmed through collaboration with medical specialists throughout the development process, guaranteeing that it complies with the realistic demands and standards of clinical practice. The simplicity of interpretation for medical professionals is given priority in the creation of an interface that is easy to use and includes interactive visualization tools and an intuitive dashboard. The implementation of the diagnostic system in actual healthcare settings is encouraged by this design, which emphasizes the useful integration of the system into clinical processes.
Future directions
Although the hybrid deep learning algorithm has shown much potential, there is still room for improvement and further investigation. The model will continue to evolve as a result of continual feedback from medical professionals, new clinical data, and adjustments to new standards and technology. It is still crucial for future research to examine explain ability strategies and approaches for dealing with interpretability issues in deep learning models. It is also possible to confirm the efficacy of the model in various clinical contexts by working with other medical institutions and expanding its applicability to additional datasets. There are several directions for further investigation and improvement as we complete this investigation. The model’s progress and efficacy will be aided by continued collaboration with medical specialists, continuous improvement of the model based on new clinical data, and adaptation to emerging technology.
In summary, by utilizing the depth of data available in the CBIS-DDSM and Wisconsin Breast Cancer Databases, the hybrid deep learning algorithm has demonstrated significant potential in the detection of breast cancer. The findings of the investigation highlight the value of innovative deep-learning methods in increasing the accuracy of diagnostic methods and opening the way for significant advancements in breast cancer diagnosis and treatment. The hybrid deep learning system represents a major advancement in the detection of breast cancer. By utilizing the wealth of data present in the CBIS-DDSM and Wisconsin Breast Cancer Databases, this investigation not only confirms the diagnostic accuracy of the algorithm but also provides the groundwork for significant advancements in the field of breast cancer patient care and diagnosis. A way to improve diagnosis and, eventually, improve patient outcomes is still being explored.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Kosmia Loizidou, R., Elia, C. & Pitris Computer-aided breast cancer detection and classification in mammography: A comprehensive review. Comput. Biol. Med. 153, 106554. https://doi.org/10.1016/j.compbiomed.2023.106554 (2023).
Hussain, S. et al. ETISTP: an enhanced model for Brain Tumor Identification and Survival Time Prediction. Diagnostics. 13 (8), 1456. https://doi.org/10.3390/diagnostics13081456 (2023).
Huang, Q., Ding, H., & Effatparvar, M. Breast Cancer diagnosis based on Hybrid SqueezeNet and Improved Chef-based Optimizer. Expert Syst. Appl. 237, 121470. https://doi.org/10.1016/j.eswa.2023.121470 (2023).
Zebari, D. A. et al. Breast Cancer detection using Mammogram images with improved Multi-fractal Dimension Approach and Feature Fusion. Appl. Sci. 11 (24), 12122. https://doi.org/10.3390/app112412122 (2021).
Alhussan, A. A., Eid, M. M., Towfek, S. K. & Khafaga, D. S. Breast Cancer classification depends on the dynamic Dipper Throated optimization Algorithm. Biomimetics. 8 (2), 163. https://doi.org/10.3390/biomimetics8020163 (2023).
Fei, Y., Huang, H., Pedrycz, W. & Hirota, K. Automated breast cancer detection in mammography using ensemble classifier and feature weighting algorithms. Expert Syst. Appl. 227, 120282. https://doi.org/10.1016/j.eswa.2023.120282 (2023).
Mushtaq, Z., Yaqub, A., Sani, S. & Khalid, A. Effective K-nearest neighbor classifications for Wisconsin breast cancer datasets. J. Chin. Inst. Eng. 43 (1), 80–92. https://doi.org/10.1080/02533839.2019.1676658 (2019).
Skaane, P. et al. Digital Mammography versus Digital Mammography Plus Tomosynthesis in breast Cancer screening: the Oslo Tomosynthesis Screening Trial. Radiology. 291 (1), 23–30. https://doi.org/10.1148/radiol.2019182394 (2019).
Lotter, W. et al. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat. Med. 27 (2), 244–249. https://doi.org/10.1038/s41591-020-01174-9 (2021).
Lee, J. M. et al. Performance of Screening Ultrasonography as an Adjunct to Screening Mammography in women across the spectrum of breast Cancer risk. JAMA Intern. Med. 179 (5), 658. https://doi.org/10.1001/jamainternmed.2018.8372 (2019).
Alsubai, S., Alqahtani, A. & Sha, M. Genetic hyperparameter optimization with modified scalable-neighbourhood component analysis for breast cancer prognostication. Neural Netw. 162, 240–257. https://doi.org/10.1016/j.neunet.2023.02.035 (2023).
Salama, W. M. & Aly, M. H. Deep learning in mammography images segmentation and classification: automated CNN approach. Alexandria Eng. J. 60 (5), 4701–4709. https://doi.org/10.1016/j.aej.2021.03.048 (2021).
Bai, J., Posner, R., Wang, T., Yang, C. & Nabavi, S. Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: a review. Med. Image. Anal. 71, 102049. https://doi.org/10.1016/j.media.2021.102049 (2021).
Maqsood, S., Damaševičius, R. & Maskeliūnas, R. TTCNN: A breast cancer detection and classification towards computer-aided diagnosis using digital mammography in early stages. Appl. Sci. 12(7), 3273. https://doi.org/10.3390/app12073273 (2022).
Giampietro, R. R., Cabral, M. V. G., Lima, S. A. M. , Weber, S. A. T., & dos Santos Nunes-Nogueira, V. Accuracy and effectiveness of mammography versus mammography and tomosynthesis for population-based breast cancer screening: a systematic review and meta-analysis. Sci. Rep. 10(1). https://doi.org/10.1038/s41598-020-64802-x (2020).
Baccouche, A., Garcia-Zapirain, B., Castillo Olea, C. & Elmaghraby, A. S. Connected-UNets: a deep learning architecture for breast mass segmentation. Npj Breast Cancer. 7(1). https://doi.org/10.1038/s41523-021-00358-x (2021).
Dibble, E. H., Singer, T. M., Jimoh, N., Baird, G. L. & Lourenco, A. P. Dense breast Ultrasound Screening after Digital Mammography Versus after Digital breast tomosynthesis. Am. J. Roentgenol. 213 (6), 1397–1402. https://doi.org/10.2214/ajr.18.20748 (2019).
Folorunso, S., Bamidele, A., Rangaiah, Y., & Ogundokun, R. EfficientNets transfer learning strategies for hispathological breast Cancer Image Analysis. Int. J. Model. Simul. Sci. Comput. 15. https://doi.org/10.1142/S1793962324410095 (2023).
Richman, I. B. et al. Adoption of digital breast tomosynthesis in clinical practice. JAMA Intern. Med. 179 (9), 1292. https://doi.org/10.1001/jamainternmed.2019.1058 (2019).
Wang, X. et al. Intelligent Hybrid Deep learning model for breast Cancer detection. Electronics. 11 (17), 2767. https://doi.org/10.3390/electronics11172767 (2022).
Yala, A., Lehman, C., Schuster, T., Portnoi, T. & Barzilay, R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology 292(1), 60–66. https://doi.org/10.1148/radiol.2019182716 (2019).
Shang, L. W. et al. Fluorescence imaging and Raman spectroscopy applied for the accurate diagnosis of breast cancer with deep learning algorithms. Biomedical Opt. Express. 11 (7), 3673. https://doi.org/10.1364/boe.394772 (2020).
Zeiser, F. A. et al. DeepBatch: a hybrid deep learning model for interpretable diagnosis of breast cancer in whole-slide images. Expert Syst. Appl. 185, 115586. https://doi.org/10.1016/j.eswa.2021.115586 (2021).
Jabeen, K. et al. Breast cancer classification from ultrasound images using probability-based optimal deep learning feature fusion. Sensors 22(3), 807. https://doi.org/10.3390/s22030807 (2022).
Balaha, H. M., Saif, M., Tamer, A. & Abdelhay, E. H. Hybrid deep learning and genetic algorithms approach (HMB-DLGAHA) for the early ultrasound diagnoses of breast cancer. Neural Comput. Appl. 34 (11), 8671–8695. https://doi.org/10.1007/s00521-021-06851-5 (2022).
Yan, R. et al. Breast cancer histopathological image classification using a hybrid deep neural network. Methods. 173, 52–60. https://doi.org/10.1016/j.ymeth.2019.06.014 (2020).
Wu, N. et al. Deep neural networks improve radiologists’ performance in breast Cancer screening. IEEE Trans. Med. Imaging. 39 (4), 1184–1194. https://doi.org/10.1109/tmi.2019.2945514 (2020).
Ahmad, S. et al. A novel hybrid deep learning model for metastatic cancer detection. Comput. Intell. Neurosci. 2022 (1-14). https://doi.org/10.1155/2022/8141530 (2022).
Kamala Devi, K. & Raja Sekar, J. Optimizing feature selection and parameter tuning for breast cancer detection using hybrid GAHBA-DNN framework. J. Intell. Fuzzy Syst. 46 (4), 8037–8048. https://doi.org/10.3233/JIFS-236577 (2024).
Wang, Z. et al. Breast Cancer detection using Extreme Learning Machine based on feature Fusion with CNN Deep features. IEEE Access. 7, 105146–105158. https://doi.org/10.1109/access.2019.2892795 (2019).
Zhang, Y. D., Satapathy, S. C., Guttery, D. S., Górriz, J. M. & Wang, S. H. Improved breast Cancer classification through combining Graph Convolutional Network and Convolutional Neural Network. Inf. Process. Manag. 58 (2), 102439. https://doi.org/10.1016/j.ipm.2020.102439 (2021).
da Silva, D. S., Nascimento, C. S., Jagatheesaperumal, S. K. & Albuquerque, V. H. C. D. Mammogram image enhancement techniques for online breast cancer detection and diagnosis. Sensors. 22 (22), 8818. https://doi.org/10.3390/s22228818 (2022).
Tsochatzidis, L., Koutla, P., Costaridou, L. & Pratikakis, I. Integrating segmentation information into CNN for breast cancer diagnosis of mammographic masses. Comput. Methods Programs Biomed. 200, 105913. https://doi.org/10.1016/j.cmpb.2020.105913 (2021).
Gonçalves, C. B., Souza, J. R. & Fernandes, H. CNN architecture optimization using bioinspired algorithms for breast cancer detection in infrared images. Comput. Biol. Med. 142, 105205. https://doi.org/10.1016/j.compbiomed.2021.105205 (2022).
Abdelrahman, L., Al Ghamdi, M., Collado-Mesa, F. & Abdel-Mottaleb, M. Convolutional neural networks for breast cancer detection in mammography: a survey. Comput. Biol. Med. 131, 104248. https://doi.org/10.1016/j.compbiomed.2021.104248 (2021).
Beeravolu, A. R. et al. Preprocessing of breast Cancer images to create datasets for Deep-CNN. IEEE Access. 9, 33438–33463. https://doi.org/10.1109/access.2021.3058773 (2021).
Thirumalaisamy, S. et al. Breast cancer classification using synthesized deep learning model with metaheuristic optimization algorithm. Diagnostics (Basel) 13(18), 2925. https://doi.org/10.3390/diagnostics13182925 (2023).
Zahoor, S., Shoaib, U. & Lali, I. U. Breast Cancer mammograms classification using deep neural network and entropy-controlled Whale optimization Algorithm. Diagnostics (Basel) 12(2), 557. https://doi.org/10.3390/diagnostics12020557 (2022).
Shen, L. et al. Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 9, 12495. https://doi.org/10.1038/s41598-019-48995-4 (2019).
Ahmad, J., Akram, S., Jaffar, A., Rashid, M., Masood, S. Breast Cancer detection using deep learning: an Investigation using the DDSM dataset and a customized AlexNet and Support Vector Machine. IEEE Access. 1–1. https://doi.org/10.1109/ACCESS.2023.3311892 (2023).
Sahu, A., Das, P., Meher, S. & & An efficient deep learning scheme to detect breast cancer using mammogram and ultrasound breast images. Biomed. Signal Process. Control. 87, 105377. https://doi.org/10.1016/j.bspc.2023.105377 (2024).
Author information
Authors and Affiliations
Contributions
P.S.R.C: conceptualization, methodology, software A.A.: data curation, writing—original draft preparation. M.A: visualization, investigation. C.A: supervision.: P.A.N.: software, validation.: V.S: writing—reviewing and editing, N.R: resources; D.M: investigation; formal analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Informed consent from all subjects for publication of identifying images in an online open-access publication.
Human and animal rights
No animals/humans were used for Studies.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Murty, P.S.R.C., Anuradha, C., Naidu, P.A. et al. Integrative hybrid deep learning for enhanced breast cancer diagnosis: leveraging the Wisconsin Breast Cancer Database and the CBIS-DDSM dataset. Sci Rep 14, 26287 (2024). https://doi.org/10.1038/s41598-024-74305-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74305-8
Keywords
This article is cited by
-
A multi stage deep learning model for accurate segmentation and classification of breast lesions in mammography
Scientific Reports (2025)
-
LBNet: an optimized lightweight CNN for mammographic breast cancer classification with XAI-based interpretability
Scientific Reports (2025)
-
Early Detection and Personalized Risk Assessment of Breast Cancer: An Integrative Review and Comparative Evaluation of AI Models Using Clinical and Imaging Data
Archives of Computational Methods in Engineering (2025)
-
Deep Learning Framework for Breast Cancer Detection and Segmentation Using EfficientNet and U-Net with Hyperparameter Optimization
SN Computer Science (2025)