Abstract
BIVV003 is a gene-edited autologous cell therapy in clinical development for the potential treatment of sickle cell disease (SCD). Hematopoietic stem cells (HSC) are genetically modified with mRNA encoding zinc finger nucleases (ZFN) that target and disrupt a specific regulatory GATAA motif in the BCL11A erythroid enhancer to reactivate fetal hemoglobin (HbF). We characterized ZFN-edited HSC from healthy donors and donors with SCD. Results of preclinical studies show that ZFN-mediated editing is highly efficient, with enriched biallelic editing and high frequency of on-target indels, producing HSC capable of long-term multilineage engraftment in vivo, and express HbF in erythroid progeny. Interim results from the Phase 1/2 PRECIZN-1 study demonstrated that BIVV003 was well-tolerated in seven participants with SCD, of whom five of the six with more than 3 months of follow-up displayed increased total hemoglobin and HbF, and no severe vaso-occlusive crises. Our data suggest BIVV003 represents a compelling and novel cell therapy for the potential treatment of SCD.
Similar content being viewed by others
Introduction
Sickle cell disease (SCD) is a hereditary hemoglobinopathy characterized by inflammation, hemolytic anemia, vaso-occlusive crises, organ damage, and early mortality1,2,3,4. Elevated expression of fetal hemoglobin (HbF) has a well-established beneficial effect on the clinical course of SCD, with decreased disease severity in patients with naturally occurring genetic variants or mutations leading to a persistence of HbF into adulthood5,6,7,8. The ameliorative effect of HbF in patients with SCD has prompted assessment of genetic and pharmacological therapeutic approaches aimed at inducing HbF9,10,11,12.
Allogeneic hematopoietic stem cell (HSC) transplant remains the only curative option for SCD but is limited to the minority of patients with a matched donor and carries a risk of graft failure or rejection and graft-versus-host disease13,14,15,16. Transplantation of autologous, genetically modified HSC has therefore been pursued as an alternative. Several gene-addition and gene-editing strategies using autologous HSC are currently approved or in clinical development in patients with SCD, including approaches to reactivate HbF9,11,12,17,18. Several of these share a common approach of targeting the BCL11A erythroid-specific enhancer (ESE), a key regulatory element in the fetal-to-adult hemoglobin (Hb) switch, to selectively generate erythroid cells with reduced BCL11A expression (and induced HbF production) without affecting non-erythroid cells9,11,12,19,20,21,22. Other gene editing strategies to reactivate HbF include mimicking mutations in the promoter of the gamma-globin gene that lead to high persistence of fetal hemoglobin (HPFH)23,24,25,26.
BIVV003 is a novel gene-edited autologous cell therapy in clinical development for the potential treatment of SCD, in which HSC are genetically modified with mRNA encoding zinc finger nucleases (ZFN) that target and disrupt a specific regulatory GATAA motif in the BCL11A ESE to reactivate HbF27,28. Zinc finger proteins combined with the nuclease domain of the restriction endonuclease FokI create double-strand breaks (DSBs) at precisely defined genomic locations. Repair of the DSBs via non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ) results in target sequence disruption29.
ZFN-mediated gene editing of the BCL11A ESE in HSC from healthy donors and donors with β-thalassemia reactivates HbF in erythroid cells without compromising in vivo engraftment30,31. In these studies, we show that ZFN-mediated editing is highly efficient and produces HSC capable of long-term multilineage engraftment in vivo,independent of mobilization strategy or disease state, and robustly express HbF in the erythroid progeny. Through clonal analysis of edited HSC from healthy donors and donors with SCD, we have also shown enriched biallelic editing with a high frequency of small indels, generating a high efficiency of ZFN gene editing. Together with preliminary clinical studies32,33, our data suggest BIVV003 represents a novel cell therapy for the potential treatment of SCD and support ongoing clinical development of this approach (NCT03653247).
Results
ZFN-mediated gene editing is highly efficient and leads to HbF induction ex vivo independently of mobilization strategy or disease state
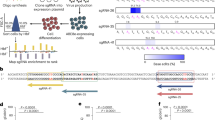
In individuals with β-thalassemia, the combination of granulocyte colony–stimulating factor (G-CSF) with plerixafor is used for stem cell mobilization34,35. However G-CSF is not recommended for hematopoietic stem-progenitor cell (HSPC) mobilization in patients with SCD due to the risk of vaso-occlusive complications associated with leukocytosis, therefore single-agent plerixafor mobilization is utilized36. We characterized ZFN-edited HSPC from healthy donors from single (plerixafor)- or dual-agent (plerixafor plus G-CSF) mobilization. Modification of the BCL11A ESE target site (Fig. 1a) was efficient using both mobilization strategies, with > 75% of alleles modified as measured by frequency of indels (i.e., insertions and deletions) and high post-editing viability (Figs. 1b and 1c).
ZFN-mediated gene editing is highly efficient and leads to HbF induction independently of mobilization strategy or disease state. (a) Schematic of zinc-finger nucleases (ZFN) targeting the BCL11A erythroid-specific enhancer. The FokI domains dimerize over the core GATAA motif to induce double-strand breaks. Arrows represent the ZFN-nucleotide contacts. (b) Indel frequency and (c) cell viability using acridine orange/propidium iodide in single- (n = 5) and dual-mobilized (n = 4) CD34 + HSPC 2 days after ZFN editing of the BCL11A erythroid enhancer. (d) Gamma-globin (γ-globin) protein levels measured by reverse phase ultra-performance liquid chromatography (RP-UPLC) at day 21 of erythroid differentiation (single-mobilized n = 4, dual-mobilized n = 4). (e) Fraction of HbF+ cells in enucleated glycophorin A–positive erythroid cells derived from single- and dual-mobilized CD34 + HSPC (left; single-mobilized n = 3, dual-mobilized n = 2). A representative flow cytometry plot of HbF+ cell frequency is shown on the right. (f) Indel levels and (g) cell viability in HSPC derived from healthy (n = 4) and SCD (n = 3) donors 2 days post-transfection of ZFNs. (h) γ-globin protein levels measured by RP-UPLC at terminal erythroid differentiation (day 18). Data expressed as mean ± standard deviation (*P < 0.05; **P < 0.01; ***P < 0.001). G + P, granulocyte colony-stimulating factor plus plerixafor; HbF, fetal hemoglobin; HD, healthy donor; HSC, hematopoietic stem cell; P, plerixafor; RP-UPLC, reversed-phase ultra-performance liquid chromatography; SCD, sickle cell disease donor; SSC, side scatter; UT, untreated; ZFN, zinc finger nuclease.
For both mobilization strategies, we observed approximately 3–fold increases in γ-globin protein and the frequency of HbF+ cells after erythroid differentiation of ZFN-edited HSPC, compared with unedited control cells (Figs. 1d and 1e).
ZFN transfection in HSPC from four healthy donors and three participants with SCD (all from single-agent plerixafor mobilization) resulted in comparable BCL11A ESE target site editing in a majority of both healthy and SCD donor HSPC (75.3% and 64.2%, respectively; p > 0.05; Fig. 1f), with high post-editing viability (Fig. 1g). Editing led to approximate 2–threefold increases in γ-globin levels in erythroid progenies from healthy and SCD donor HSPC at day 18 of differentiation (Fig. 1h).
Owing to unexpected differential growth kinetics between SCD donors and premature cell death of unknown cause in two SCD donor HSPC preparations, we were only able to evaluate HbF+ cell frequency in HSC derived erythroid cells from one SCD donor. We found comparable increases in HbF+ cell frequency after editing in healthy (n = 1) and SCD (n = 1) donor HSPC derived erythroid cells (Figure S1a). Red blood cell (RBC) sickling from the same SCD donor under hypoxic conditions was lower after editing (Figure S1b-d).
ZFN-mediated gene editing does not compromise the function or progenies of HSC and results in stable editing levels after engraftment in in vivo mouse models
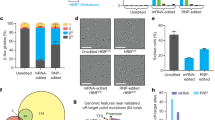
We next measured colony-forming unit (CFU) production in single-agent mobilized HSPC from healthy donors in vitro. CFU composition was similar between edited and unedited cells 2 days post-ZFN editing (Fig. 2a; n = 3). Furthermore, there were no significant differences in frequency of enucleated cells after 20 days of in vitro erythroid differentiation (n = 4, p > 0.05; Fig. 2b).
ZFN-mediated gene editing does not adversely affect HSC progenies. (a) Number of CFU-derived colonies of HSPC 2 days post-ZFN editing from plerixafor cells. Shown are the contribution of CFU-E, BFU-E, CFU-G/M/GM, and CFU-GEMM (n = 3). (b) Frequency of enucleated cells at Day 20 of in vitro differentiation (n = 4). (c) Proportion of human CD45+ (hCD45) cells in NSG mouse PB. Data from 9–10 mice per donor and condition at Weeks 8 and 12 and from 4–5 mice at Weeks 16 and 19. Error bars correspond to the standard error of the mean. (d) Proportion of hCD45+ cells in mouse BM at Week 19. (e and f) Frequencies of HSPC (Lin-CD34highCD38-), committed progenitors (Lin-CD34highCD38 +), myeloid (CD33 +), erythroid (CD71 +), lymphoid B (CD19 +), lymphoid T (CD3 +), and natural killer (CD56 +) within the hCD45 + population within the total bone marrow cells are shown for two healthy donors at 19 weeks after engraftment. Data expressed as mean ± standard deviation unless specified (*P < 0.05; **P < 0.01; ***P < 0.001). BFU-E, burst-forming unit-erythroid; CFU, colony-forming unit; CFU-E, CFU-erythroid; CFU-G/M/GM, CFU-granulocyte–macrophage; CFU-GEMM, CFU-granulocyte-erythrocyte-monocyte-megakaryocyte; CP, committed progenitor; HD, healthy donor; HSC, hematopoietic stem cell; HSPC, hematopoietic stem and progenitor cell; NK, natural killer; UT, untreated; ZFN, zinc finger nuclease.
To determine the effect of ZFN editing on the function of HSC and engraftment potential, we transplanted ZFN-edited and unedited dual-agent mobilized control HSPC from two healthy donors into immune-deficient NOD scid gamma (NSG) mice. Mice treated with edited or unedited HSPC had similar chimerism (Figs. 2c and 2d). We observed a small increase in HSPC and committed progenitors in the bone marrow of mice engrafted with the first donor between edited and unedited cells after engraftment in NSG mice (p < 0.05; Fig. 2e). No significant difference was observed in the bone marrow of mice engrafted with cells from the second donor (Fig. 2f).
Comparable indel levels were observed in both healthy donor HSPC in the cell input and after transplantation at Week 19 in peripheral blood (Fig. 3a). No significant difference in indel levels were measured in BCL11A-dependent (CD19+ B cells; CD34+CD38+ primitive progenitors) and BCL11A-independent (CD33+ myeloid cells) lineages at Week 19 after engraftment of HSC from both donors (p > 0.05; Fig. 3b). Human in vitro differentiated erythroid cells derived from engrafted HSPC showed similar indel levels to the other lineages (p > 0.05; Fig. 3b). In addition, no significant differences in terminal erythroid differentiation in vitro were observed based on the proportion of enucleated cells in recipients treated with edited or unedited HSPC at Week 19 post-engraftment (p > 0.05; Fig. 3c). A significant increase in γ-globin in the erythroid progeny was observed in edited versus unedited HSPC (Fig. 3d).
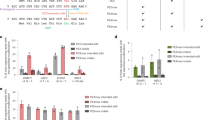
ZFN-mediated gene editing does not compromise the functionality or progenies of HSPC and results in stable editing levels after engraftment in vivo. (a) PB indel levels at input (Week 0, n = 2 donors) and after engraftment (For each donor, n = 9–10 mice for week 8 and 12 post engraftment, and n = 4–5 mice for week 16 and 19). (b) Indel levels in human bone marrow (BM) as well as cell subpopulations within the hCD45 + compartment sorted using antibodies recognizing the indicated lineage markers at input (Week 0) and after engraftment (Week 19 post engraftment). No significant difference was observed between lineages at week 19 using a multivariate ANOVA controlling for donor differences (P > 0.05, excluding input). BM refers to the total human CD45 + cell population in the bone marrow. (c) Frequency of enucleated cells in terminal erythroid differentiation in vitro from BM-derived HSPC at Week 19. (d) Globin protein levels were analyzed at Day 16 of in vitro erythroid differentiation by UPLC. Average γ-globin protein levels for each group are plotted as percentage of all β-like globin protein (A-γ-, G-γ-, δ-, and adult β-globin protein) on the left, and average γ-globin protein levels as percentage of α-globin levels are plotted on the right. (e) Read counts of the 10 most frequent sequences identified by next-generation sequencing at input (Week 0) in healthy donor 1 (HD 1) and healthy donor 2 (HD 2). The unedited (wild type) sequence is shown in bold, with a rectangle highlighting the core GATAA site and lines representing ZFN contacts. (f) Fraction of total indels that are likely derived from MMEJ. MMEJ patterns represent those identified by the RGEN Microhomology-Predictor tool (see Methods). (g) Frequency of indels by size of insertion (> 0) or deletions (< 0). Data expressed as mean ± standard deviation (*P < 0.05; **P < 0.01; ***P < 0.001). HD, healthy donor; HSC, hematopoietic stem cell; MMEJ, microhomology-mediated end-joining; n.s., not significant; PB, peripheral blood; BM, bone marrow; UPLC; ultra-performance liquid chromatography; UT, untreated; ZFN, zinc finger nuclease.
We determined that the most common indel sequences at input resulting from editing in both donors were comparable, with 15-, 13-, and 2-bp deletions the most frequently observed (Fig. 3e). The frequencies of indel patterns and sizes of insertions or deletions after 19 weeks of engraftment were similar between the two donors (Fig. 3g), with no difference between the fractions of total indels likely derived from microhomology-mediated end-joining (MMEJ; Fig. 3f). The frequency of -13 and -15 MMEJ indels dropped significantly after engraftment in both donors, likely reflecting loss of short-term HSC and committed progenitors (Fig. 3f and 3g, Figure S2). Insufficient cell preparations were available to conduct these experiments for HSPC from sickle cell donors.
ZFN-mediated gene editing leads to enriched biallelic-edited cells and allele-additive increases in HbF
We sorted single-agent mobilized, ZFN-edited HSPC from healthy (n = 5) and SCD (n = 3) donors into single cells and performed erythroid differentiation on each resulting colony (Fig. 4)37. We genotyped 1,175 single-cell–derived HSPC colonies by next-generation sequencing (Table S1a). Similar levels of editing, up to 95%, were observed after erythroid differentiation in single cells from healthy and SCD donor HSPC (Fig. 4a). Indel levels in single-cell derived colonies were generally higher than in bulk HSPC or in vitro differentiated erythroid cells (Figure S3). The most observed indel patterns in healthy donor and SCD donor samples were comparable, with the 15-bp, 2-bp, and 13-bp deletions the most frequent (Fig. 4b).
ZFN-mediated gene editing leads to enriched biallelic edited cells and allele-additive increases in HbF. (a) Total editing levels in erythroid colonies derived from single-cell CD34 + HSPC from healthy and SCD donors. (b) Frequency of the most common indel patterns in single-cell derived colonies. The unedited (wild type) sequence is shown in bold, with a rectangle highlighting the core GATAA site and lines representing ZFN contacts. (c and d) Frequency of wild-type, monoallelic, and biallelic-edited genotypes derived from single-cell clonal analysis of plerixafor-mobilized HSC from (c) 5 healthy and (d) 3 SCD donors. The distribution of genotype was significantly skewed towards biallelic clones. The expected distribution of clones was calculated using an equation analogous to the Hardy–Weinberg equilibrium (Online Methods). P-values from χ2 tests comparing the observed and expected number of wild-type, monoallelic, and biallelic clones for each sample are reported on the figure. Significant P-values reflect a skew of the distributions towards wild-type and biallelic clones. (e and f) Association of γ-globin levels measured by reversed-phase ultra-performance liquid chromatography with genotypes in (e) healthy donors (linear regression P = 8 × 10−84; beta = 14.9; standard error = 0.7) and (f) SCD donors (linear regression P = 7 × 10−8; beta = 14.1; standard error = 2.5). Mean differences between biallelic-edited and unedited clones were 32% (HD 1), 38% (HD 2), 28% (HD 3), 30% (HD 4), 21% (HD 5), 28% (SCD 1), 32% (SCD 2), and 20% (SCD 3). (g) Effect of indels on gamma-globin expression in healthy donor cells, stratified by whether they disrupt the core GATAA site (GATAA-, left) or not (GATAA + , right). For (e, f, and g), beta and P-values were calculated by fitting linear regression models of HbF (gamma-globin) against genotype in all clones, adjusted for the different donors (HbF ~ genotype + donor). The beta coefficient for the genotype effect is reported. HbF, fetal hemoglobin; HD, healthy donor; HSC, hematopoietic stem cell; SCD, sickle cell disease; WT, wild type.
The distribution of wild-type (WT), monoallelic-, and biallelic-edited genotypes was highly skewed towards biallelic-edited cells in both healthy donor and SCD donor HSPC, representing > 90% and 88% of edited clones, respectively, and indicating that ZFN-edited cells contained edits at both alleles (Fig. 4c and 4d; Table S1b and S1c). Background HbF protein levels varied across donors, which were processed in batches, ranging from 7 to 22% in WT clones from healthy donors and from 22 to 51% for WT clones from SCD donors. Nevertheless, we observed a similar increase in HbF across donors: HbF protein measurements indicated that each edited allele contributed additively to an increase of γ/(γ + β) ratio of ~ 15% (Figs. 4e and 4f). Biallelic-edited clones from healthy donors and SCD donors had a 29% and 27% increase, respectively, of γ/(γ + β) ratio, compared with basal levels in unedited clones.
To understand the contribution of different indel patterns to HbF induction, we separated indels according to whether they disrupted the internal GATAA motif. We observed a similar additive increase in HbF in clones with GATAA-disrupting indels in both healthy (n = 5) and SCD (n = 3) donor cells compared with WT, with biallelic clones expressing ~ 34% more HbF than WT (Fig. 4g, Figure S4). In contrast, biallelic-edited clones with both GATAA motifs intact expressed ~ 13% more HbF compared with WT, suggesting that non–GATAA-disrupting indels contribute to HbF up-regulation, albeit to a lower extent (Fig. 4g).
To determine the effect of ZFN editing on erythroid function, we examined the frequency of cell enucleation based on genotype and whether the indel pattern disrupted the internal GATAA motif. In healthy donor HSC (n = 3), biallelic-edited cells showed a slight decrease in enucleation frequency compared with WT (Figure S5a). However, biallelic-edited cells with intact GATAA sites (“WT/WT”) did not show differences in enucleation compared with biallelic-edited cells with disrupted GATAA sites (Figure S5b), suggesting that the observed differences in enucleation may be due to the process of transfection and editing rather than specific effects of BCL11A knockdown. No differences in enucleation frequency were observed among various genotypes or indel patterns within GATAA sites in SCD donor HSPC (n = 3; Figures S5c and S5d).
ZFN-mediated gene editing is characterized by high frequency of small indels
It was possible that biallelic-edited homozygous clones (N = 162) and clones that failed to genotype (N = 16 with visible colonies and good HbF detection) harbored larger deletions at the target sites in one of the alleles, encompassing the primer sites used for the amplicon, and thus were not captured by sequencing. Therefore, we further genotyped these clones from healthy donors (n = 4) using an amplicon design that included both the target site and an informative heterozygous single nucleotide polymorphism that allowed the discrimination of both alleles. We subsequently sequenced clones that remained indeterminate using a larger 12 kb Nextera XT amplicon assay centered on the target site (Fig. 5a). Overall, the majority (89.7%) of edited alleles were small indels (1–50 bp; N = 963; Fig. 5b). Large deletions and insertions > 1 kb accounted for less than 1% and 0.3%, respectively. Similarly, we observed a very small number of complex events involving both insertions and deletions (0.5%). The largest deletion was 5.9 kb within the BCL11A ESE (Fig. 5c). However, this was an outlier considering that the second largest deletion was ~ 2.5 kb and the median size of large deletions (> 1 kb) was 1.4 kb. The largest insertion was estimated to be 2.9 kb. All large insertions mapped to other genomic loci. Overall, between 79.9% and 86.6% of total indels disrupted the GATAA motif (Fig. 5d).
ZFN-mediated gene editing is highly specific and characterized by high frequency of on-target small indels. (a) Experimental strategy to confirm homozygote clones and identified large deletions. Left: With the original 140 bp sequencing assay, clones with only 1 indel pattern may represent homozygote clones or clones with an uncaptured deletion spanning a primer site. Center: 12 homozygous clones could be confirmed by a sequencing assay that captures an informative SNP in 2 healthy donors. Right: For samples with no informative SNPs or uncaptured events, a 12 kb region around the target site was sequenced. Example shows a heterozygous sample with a large deletion that was not captured by the small 140 bp amplicon and therefore ruled out as a homozygote by the subsequent analysis. (b) Frequency of identified allele lengths in clones (N = 1175) derived from healthy donors (n = 4). A 12 kb region was sequenced around the BCL11A enhancer in clones with alleles potentially missed by short amplicon sequencing. Indels > 1 kb represent less than 1% of events. Unknown events include 3 insertions for which the length could not be determined. (c) Depiction of the largest deletion observed (5.9 kb). The deletion is contained within the BCL11A erythroid super-enhancer, itself located within the 77 kb intron 2 of BCL11A. (d) Total fraction of indels that disrupt the core GATAA site. (e) Enrichment of MMEJ indels with large deletions. The figure shows the frequency of small indels (MMEJ or NHEJ) as the second allele of clones carrying a large deletion (> 50 bp), a large insertion (> 50 bp), a complex indel, or a NHEJ indel. MMEJ indels are over-represented as the second allele of clones with deletions > 50 bp (Fisher exact test P = 0.008). Complex indels and insertions show over- and under-representation of large deletions (P = 0.02 and 0.03, respectively). Data from healthy donor cells (n = 4). MMEJ patterns represent those identified by the RGEN Microhomology-Predictor tool (methods). bp, base pairs; HD, healthy donor; MMEJ, microhomology-mediated end joining; NHEJ, non-homologous end joining; SNP, single nucleotide polymorphism.
To understand the effect of indel size on HbF, we correlated the mean indel length per clone with HbF using a linear model accounting for sample differences. There was a significant effect of mean size on HbF, although this was attenuated when accounting for GATAA-disrupting indels (Figure S6a and S6b). There was no significant correlation between edit length and enucleation rate, suggesting that larger indels do not negatively impact erythroid function (Figure S6c). Finally, we hypothesized that deletions > 50 bp may be more frequent in either committed progenitors or HSC. We observed an enrichment of MMEJ indel patterns in the second allele of clones harboring deletions > 50 bp compared with clones with NHEJ small indels (Fig. 5e) and observed an increased frequency of microhomologies around deletions > 50 bp compared to smaller deletions (< 50 bp; p < 0.00005). Since MMEJ mediated repair is less frequent in long-term engrafting HSC, these results suggest that large deletions are less likely to arise in this population.
Interim results from the phase 1/2 PRECIZN-1 study
To show how the preclinical research data shown above translate to the clinical setting, we are summarizing the current results from the PRECIZN-1 study, which was conducted following extensive safety studies.
Participant disposition
At the cutoff date (August 15, 2023), seven participants with SCD had received infusions of BIVV003 (Table S2)32,33. Group 1 comprised four participants administered BIVV003 using the original manufacturing process, each with up to 30 months of follow-up as of the cutoff. Participants in Group 2 received BIVV003 manufactured according to an improved process designed to increase the number of long-term progenitors present at the end of the process without affecting their editing frequency. One participant in Group 2 had 1 month of follow-up at the cutoff. Six of 13 enrolled participants who underwent mobilization and apheresis at least once did not collect sufficient CD34 + HSPC on the first attempt to manufacture the minimal dose of BIVV003 and discontinued from the study without being dosed (Table S3). Four of 13 (31%) participants underwent two cycles of mobilization and apheresis; two collected sufficiently through the repeat cycle to manufacture a dose of BIVV003 and subsequently received the infusion.
Hematopoietic reconstitution and engraftment of edited HSPC
Seven participants underwent busulfan conditioning and infusion of autologous, ZFN-edited HSPC. The mean hemoglobin on the day of BIVV003 infusion was 9.9 g/dL. Median times for neutrophil and platelet recovery were 21 days and 29 days, respectively. Participants received RBC transfusion support up to Day 21 post-transplant. After hematopoietic recovery, all participants were transfusion-free except for Participant 3, who required sporadic transfusions.
The indel frequency in the BIVV003 drug product infused ranged from 56–78% (Table S4). At Week 26, indel frequency ranged from 17–34% in unsorted bone marrow in all four Group 1 participants, and 37% and 33% in two Group 2 participants. The corresponding values for Week 52 were 17–32%. While both input indel percentages and indel percentages at equilibrium tended to be slightly lower in the clinical study, with no strict correlation between the two values for individual samples, the indel patterns in the patient samples mirror those observed after engraftment of ZFN treated human HSC in mice (Fig. 3) with a steep reduction of the percentage of the MMEJ derived 15 and 13 bp deletions compared to the drug product and a high degree of complexity with no specific indel dominant but the 2 bp deletion as the most common indel observed post-infusion in the bone marrow or PBMC derived samples from all patients (data not shown).
Hb fractionation following BIVV003 infusion
Total hemoglobin (Fig. 6a) and clinical markers of hemolysis (Figure S7) stabilized by Week 26 post- BIVV003 infusion in all six participants with ≥ 26 weeks of follow-up. In Group 1, percent HbF level (1–11% at screening) increased to 14–39% by Week 26 in all four participants. Notably, one participant had an HbF level which dropped below 15% from Week 26 onwards (Fig. 6a). In Group 2, percent HbF level increased to 45–54% at Week 26.
Preliminary safety and efficacy results from PRECIZN-1: an ongoing Phase 1/2 study of BIVV003, a ZFN-modified autologous CD34 + HSC therapy for SCD. (a) Total Hb and Hb fractionation in all participants (n = 7) following BIVV003 infusion. *Indicates the Hb value from local lab, since the central lab value was not collected. (b) F cells over time for treated participants. (c) HbF/F cell levels above the threshold for preventing HbS polymerization. (d) Number of VOCs (severe pain crises, acute chest syndrome) reported pre- and post-BIVV003 infusion. Note, some participants had back-to-back VOC events. The total number of events is reported in Table S2. Hb, hemoglobin; HbA, adult hemoglobin; HbA2, variant adult hemoglobin; HbF, fetal hemoglobin; HbS, sickle hemoglobin; VOC, vaso-occlusive crisis
Percent F cells increased up to 99% and reached a stable level by 26 weeks of follow-up in six infused participants (one subject having reached only Day 29 post-infusion by the cutoff date). F cell levels persisted at 77% or greater in five participants for up to 104 weeks of follow-up (Fig. 6b). At Week 26, five of six participants had stably achieved the average level of ≥ 10 pg of HbF/F cells that inhibits sickle hemoglobin (HbS) polymerization25 and sustained this level for up to 104 weeks of follow-up (Fig. 6c). Group 2 participants generally attained higher HbF/F cell levels than Group 1 participants. (Fig. 6c).
Safety and tolerability
Overall, the AEs reported in the study were generally consistent with mobilization, apheresis, and conditioning treatment (Table S5). Two SAEs of sickle cell anemia with crisis in two patients were reported by the investigator as related to plerixafor. One SAE of nausea was reported by the investigator as related to busulfan. There were no related SAEs and only one Grade 2 AE of anxiety assessed by the investigator as related to BIVV003. As of the cutoff, three vaso-occlusive crises had been reported ~ 9 ,17 and 22 months after BIVV003 infusion in one participant in Group 1 (Fig. 6D). No other SCD-related events were reported.
Discussion
BIVV003 is a novel gene-edited autologous cell therapy in development for SCD (NCT03653247), in which HSPC are genetically modified with mRNA encoding ZFNs that disrupt a specific regulatory GATAA motif in the BCL11A ESE to reactivate HbF. ZFN-mediated gene editing does not require viral transduction or the use of DNA-annealing oligonucleotides, such as guide RNAs. In our preclinical studies, ZFN editing was highly efficient, as evidenced by high indel frequency, and led to HbF induction ex vivo, in plerixafor-mobilized HSC from both healthy donors and donors with SCD. ZFN-mediated gene editing did not impact terminal erythroid differentiation, consistent with previously published approaches targeting the ESE of BCL11A30,31,38,39,40,41. Erythroid progenies from ZFN gene-edited HSC from an SCD donor showed attenuated in vitro sickling, supporting the therapeutic impact of ZFN editing and HbF induction, and expanding on our previous studies in β-thalassemia31.
ZFN-mediated gene editing did not compromise the function and potency of hematopoietic progenitors, consistent with our previous studies30,31. Following engraftment of ZFN-edited HSPC into immunodeficient mice, similar human chimerism in circulation and bone marrow was observed compared to non-edited HSPC. Furthermore, the frequency of engrafted, gene-edited cells in total and sorted bone marrow populations persisted. Erythroid cells differentiated ex vivo from the chimeric bone marrow of primary recipients showed that ZFN-mediated gene editing resulted in effective HbF reactivation with no impact on enucleation, consistent with our in vitro findings. Our studies show a long-term and sustainable effect in an in vivo mouse model.
Long-term HSC were shown to preferentially undergo NHEJ-mediated repair following ZFN-mediated gene editing. Sequencing of input cells showed the most common edits were 15- and 13-bp deletions, due to MMEJ repair. After in vivo engraftment, we observed a reduced frequency of MMEJ repair alleles in engrafted versus input cells, suggesting that long-term HSC favor repair by NHEJ. Consistently, preclinical data from CRISPR-mediated gene editing in HSC suggest that long-term HSC favor NHEJ-repair and that MMEJ may be less common in these cell types40,41.
A major consideration for the clinical development of gene therapies is high efficiency of therapeutic editing and the reduction of off-target editing. We demonstrate that ZFN-mediated gene editing in HSC from healthy donors and donors with SCD results in highly efficient and precise disruption of the BCL11A ESE. Genotype distributions of edited HSC were highly skewed towards biallelic editing, suggesting that ZFN-expressing cells are characterized by complete allelic disruption. Colonies with biallelic modifications also exhibited increased HbF levels in an edited allele- and GATAA site disruptive-dependent fashion. The high precision of ZFN-mediated gene editing was confirmed by sequencing a 12 kb region around the target site. High rates of unintended on-target large deletion of genes, including BCL11A, in CRISPR/Cas9 gene-edited SCD HSC were recently reported42,43. We observed that indels resulting from ZFN editing of healthy donor and SCD donor HSC were generally small and located well within the intronic region of BCL11A. The largest deletion was contained within the BCL11A ESE and all large insertions appeared to be random integrations. Our data also suggest that large deletions are less likely to arise in the long-term engrafting HSC as large deletion rates positively correlate with MMEJ repair usage44. However, a limitation of our study is that long-range sequencing was not conducted, which would have provided a more accurate characterization of potential large deletions, insertions, and complex events.
Interim safety and efficacy results confirm the potential therapeutic value of ZFN-mediated modification of the BCL11A ESE region and BIVV003 infusion to address current unmet needs of individuals with SCD. Five of six infused participants with more than 3 months of follow-up showed sustained increases in total Hb, HbF, and percent F cells. Participant 3, the one who displayed a non-sustained level of HbF expression, likely did not have adequate myeloablation and/or engraftment of ex vivo LT-HSC despite receiving a cell product with a high percentage of edited cells. This is supported by the large difference between the indel percentage in the drug product and after infusion (Table S4). BIVV003 was well tolerated in all seven participants infused to date, with all three post-infusion severe VOC events reported in one participant (participant 3) after treatment, which were associated with HbF levels below what is inhibitory of HbS polymerization. The manufacturing process used in group 1 has been shown to result in a reduction in the LT-HSPC population in the drug product (data not shown). This manufacturing process was also used in our study of ZFN-edited autologous HSPC transplant in patients with transfusion-dependent beta-thalassemia (NCT03432364), and, in addition to the different mobilization procedure and patient population, may have factored into the short-lived increases in HbF in RBC observed in that study. It may also have factored in the non-sustained HbF expression in participant 3, although unique aspects of the treatment of this patient or their biology may have contributed to this outcome. In group 2, a refined manufacturing process with improved culture conditions during CD34 + HSPC isolation and culture to leading to an increased number of LT-HSC in the drug product has been used. Remaining participants to be dosed in the Phase 1/2 study will receive drug product manufactured using the improved process. Despite this manufacturing challenge, there was natural enrichment of modified erythroid lineage cells in the blood with elevated HbF, sufficient for a clinical benefit in all participants, in terms of prevention of severe VOC and not requiring transfusion therapy with the exception of participant 3.
Our results show that ZFN-mediated disruption of the BCL11A ESE results in highly efficient editing and produces HSC that are capable of long-term multilineage engraftment in vivo, independent of mobilization strategy or disease state, and express increased levels of HbF as erythroid progeny. The HbF and Hb levels achieved in group 2 to date indicate an improvement compared to group 1 and appear comparable to those observed with exa-cel, a CRISPR-based, BCL11A ESE editing approach that targets the same site and that recently received marketing approval. Through clonal analysis of edited HSPC from healthy donors and donors with SCD, we have also shown enriched biallelic editing and a high frequency of small indels, supporting the high efficiency of ZFN gene editing. While highlighting the challenges of consistently manufacturing clinical grade, autologous, gene-edited HSPC therapy for patients with SCD, our preliminary data support further clinical development of this approach and suggest that BIVV003 represents a novel cell therapy for the potential treatment of SCD.
Online methods
Experimental approaches used in this study have been described previously30. All experiments were performed in accordance with relevant guidelines and regulations. All animal procedures were approved by the Sanofi Institutional Animal Care and Use Committee (IACUC) and performed in accordance. The study is reported in accordance with the ARRIVE guidelines (https://arriveguidelines.org). The PRECIZN-1 study was performed in accordance with the Declaration of Helsinki and local regulations and was approved by independent ethics committees or institutional review boards as described in the PRECIZN-1 section.
Cell culture
Human CD34 + cells were mobilized from de-identified healthy or sickle cell disease volunteers using granulocyte colony stimulating factor (G-CSF) and plerixafor or plerixafor as previously described35. Mobilized CD34 + HSPC and/or chimeric bone marrow cells were cultured for 3 days in a maintenance media consisting of X-VIVO 10 (VWR), 100 U/mL penicillin–streptomycin (ThermoFisher), 2 mM L-glutamine (Fisher Scientific), 100 ng/mL Recombinant Human Stem Cell Factor (SCF; ThermoFisher), 100 ng/mL Recombinant Human Thrombopoietin (TPO; ThermoFisher), and 100 ng/mL Recombinant Human Flt-3 Ligand (Flt-3L; ThermoFisher). Cells were then differentiated into erythroid cells using a three-step differentiation protocol developed by Giarratana et al. (2005)45. In brief, CD34 + cells were cultured for 7 days in Step1 media consisting of Iscove’s modified Dulbecco’s medium (IMDM) (ThermoFisher) supplemented with 1X GlutaMAX, 100 U/mL penicillin–streptomycin (ThermoFisher), 5% human AB + plasma, 330 ug/mL human holo-transferrin, 10 ug/mL human insulin, 2 U/mL heparin, 1 uM hydrocortisone (Sigma-Aldrich), 3 U/mL recombinant human erythropoietin (EPO) (ThermoFisher), 100 ng/mL SCF (ThermoFisher), and 5 ng/mL interleukin 3 (IL3; Sigma-Aldrich). On Day 7, cells were transferred to Step2 media i.e., Step1 media without hydrocortisone and IL3, and cultured for 3–4 days. Lastly, cells were cultured for 8–9 days in Step3 media i.e., Step2 media without SCF.
BCL11A Editing of human CD34 + Cells Using ZFNs
Zinc-finger nucleases (ZFN)
ZFN were designed and assembled as previously described28. The ZFN are composed of a triple flag domain, a nuclear localization signal, an engineered DNA-binding domain and a mutated obligate heterodimer FokI nuclease domain. The pGEM-based expression vectors contain a T7 promoter 5′ and 3′ UTRs and a synthetic 64-bp poly(A) stretch for optimized mRNA production. Compared to a ZFN pair described earlier (Psatha, Ref. 31) ZFN activity and specificity was further optimized. On the one hand, this was achieved by fine tuning DNA binding affinity through extensive screening of zinc finger domain variants containing mutations which remove nonspecific phosphate contacts to the DNA backbone. The ZFNs described in this publication both contain three variant amino acids in the zinc finger domain compared to the original left and right BCL11A targeting ZFNs. On the other hand, ZFN performance was further improved via optimization of the DNA cleavage kinetics by mutating one residue in the FokI domain of the right ZFN. Both approaches are described in Reference 29 (Miller et al.).
The amino acid sequence in the zinc finger DNA binding domains is as follows:
F1 | F2 | F3 | F4 | F% | F6 |
Left ZFN DQSNLRA | RNFSTM | STGNTN | TSGSLTR | DQSNLRA | AQCCLFH |
Right ZFN DQSNLRA | QKAHLIR | QKGTLGE | RGRDLSR | RRDNLHS |
Electroporation
To edit CD34 + cells, cells were thawed and cultured in the CD34 maintenance media described above for 1 or 2 days at 37C, 5% CO2 in a humidified incubator. CD34 + cells were electroporated using a MaxCyte GT transfection instrument (MacCyte) or Lonza Nucleofector 4D in the presence of the ZFN mRNAs. After transfection, cells were incubated in the CD34 maintenance media at 30C for 18–24 h and then at 37C for 24 h. Two days after electroporation, cells were harvested, resuspended in CryoStor CS10 (10% dimethyl sulfoxide [DMSO]), and transferred to cryovials. The cells were then cryopreserved and stored in liquid nitrogen.
Transplantation of human CD34 + HSPC in NSG mice
Female NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratories) at the age of 6–7 weeks, pre-treated with 10 mg/kg/day Baytril water for two days were sub-lethally irradiated at 300 rad 1 day before transplant. For primary recipients, human CD34 + cells were injected into the mice via the tail vein at 1 million viable cells per mouse.
Flow cytometry
Enucleation
To determine the enucleation rate of the erythroid differentiation culture at Day 20, cells were stained with NucRed (used for living cells; ThermoFisher) per the manufacturer’s instructions (2 drops/mL) and fluorescein isothocyanate (FITC)-conjugated anti-CD235 (Glycophorin A) antibody (Clone JC159-Dako-Agilent), washed with PBS (ThermoFisher) supplemented with 0.5% BSA (Sigma-Aldrich).
HbF staining
To determine the percentage of HbF + cells, cells were fixed and permeabilized according to ThermoFisher protocol. Cells were also stained with Phycoerythrin (PE)-conjugated anti CD235 antibody (ThermoFisher) and NucBlue (Hoechst 33,342, used for fixed cells; ThermoFisher). HbF levels were detected using Allophycocyanin (APC)-conjugated anti-HbF antibody (ThermoFisher) after gating on the enucleated (Hoechst low) CD235 positive cell fraction.
For enucleation and HBF staining methods, the acquisition of stained cells was performed on FACSCanto and the analysis was done using FlowJo software.
Ex vivo staining and sorting
To assess the degree of human chimerism, fractions of cells in peripheral blood (at 8-, 12-, 16-, and 19-weeks post-engraftment) and bone marrow (at 12 and 19 weeks post-engraftment) were stained with mCD45 (Biolegend) and hCD45 (BD Biosciences) antibodies respectively and FACS analysis was performed. In addition, hematopoietic lineage analysis was performed by staining bone marrow cells with anti-hCD3 (clone UCHT1), anti-hCD19 (clone SJ25C1), anti-hCD45, anti-hCD33, anti-hCD38, anti-hCD34, anti-hCD71, and anti-hCD56 antibodies (BD Biosciences) and lineage cocktail (Biolegend) (Table S6).
To purify and sort HSC populations, an enrichment/depletion strategy using magnetic cell separation (MACS) was used (Table S7). Bone marrow cells were first stained with hCD19-biotin (clone HIB19), hCD3-biotin (clone HIT3a), mB220-biotin, mTER119biotin and -m-ckitbiotin (BD Biosciences) and then incubated with -antibiotin- beads (Miltenyi Biotec). The positive fraction and depleted fraction were separated using LS columns (Miltenyi Biotec) placed in the magnetic field of a MACS. After separation, the positive fraction was stained with StreptavidinAPC-, hCD3-FITC, hCD19-PE, hCD45-BV510 (BD Biosciences) and the depleted fraction was stained with hCD34-FITC (BD Biosciences), hCD19APC (BD, clone SJ25C1), -LinAPC- (Biolegend), StreptavidinAPC-, hCD45-BV510, hCD33-PE-CF594 (BD Biosciences), and hCD38-PECy-7 (Biolegend). Cell populations were sorted and processed for MiSeq analysis.
In vitro HSPC flow staining
Purified CD34 + cells were stained with anti-CD34, anti-CD38, anti-CD45rA, anti CD49f. (BD), anti CD123, anti CD90 (ebioscience) and CD34hCD38-CD90 + CD45RA-CD49f. + (LT-HSC), CD34hCD38-CD90-CD45RA-CD49f.- (MPP), CD34hCD38-CD90-CD45RA-CD49f. + (ST-HSC), CD34hCD38-CD90 + CD45RA- (HSC),CD34hCD38-CD90-CD45RA + (LMPP), CD34hCD38 + CD123 + CD45RA- (CMP), CD34hCD38 + CD123-CD45RA- (MEP), CD34hCD38 + CD123 + CD45RA + (GMP), CD34 low and CD34- were sorted and processed for MiSeq.
Assessment of RBC sickling
Sickling of CD34 + HSPC-derived RBCs was assessed under hypoxic and normoxic conditions. Approximately 2–10 × 106 RBCs were pelleted by centrifugation and resuspended in 800 µL phosphate buffered saline (PBS) supplemented with 10 mmol/L-1 glucose and 0.2% w/v (bovine serum albumin (BSA). Two duplicate 24-well plates were prepared using 400 µL of each sample. The hypoxia plate was incubated at 1% oxygen for 2 h in a hypoxia chamber and the control plate incubated in normoxic conditions. Following incubation, all cell samples were fixed with 40 µL of fresh 25% glutaraldehyde (EM grade), before pelleting by centrifugation and resuspension in 50 µL. Sickling was measured by imaging flow cytometry (ImageStream). The number of sickled or normal cells is reported as a percentage of all cells.
Reverse phase UPLC
Reverse phase high-performance liquid chromatography (RP UPLC or RPLC) of human globin chains was used to determine any changes in the ratios of γ-globin, β-globin, and α-globin protein. Under conditions of varying pH (such as used in the UPLC gradient), the globin chains (β, δ, α, G γ and A γ) were denatured and separated from the heme groups. Hemolysates from 1.0 × 106 RBCs obtained from Day 18 or 21 cultures were washed twice with PBS and resuspended in 50 µL of UPLC grade water for lysis. The sample was then vortexed and the hemolysate clarified by centrifugation at 10,000 × g for 5 min at 4 °C. The hemolysate supernatant was then transferred to a new tube or plate and stored at -80 °C until analysis. For the UPLC runs, 20 µL of hemolysate was injected directly into the column for each assay. UPLC analyses were performed on a Waters Acquity BEH300 C4 column (C4 Reversed Phase, 300 A Pore Size 2.1 × 100 mm, 1.7 μm particle size; Waters Acquity; Product No. 186004496) on an Agilent 1290 Infinity II Ultra High-Performance Liquid Chromatographer (Agilent). Globin chains were eluted with a two solvent system (100% UPLC H2O + 0.1% trifluoroacetic acid and solvent B, 100% acetonitrile + 0.1% trifluoroacetic acid) and a multi-step RPLC elution program starting at 35% Solvent B and gradually increasing to 38% Solvent B over a time period of 4 min, increasing again to 42.5% Solvent B over a time period of 18 min, an increase to 70% Solvent B in 4 min followed by a 4 min flush of the column at 70% Solvent B and re-equilibration down to 35% solvent B within 5 min. The flow rate was 0.2 mL/min. Quantitation of the amounts of the various globins was obtained by integration of the peak areas monitored by absorbance at 220 nm with reference wavelength of 360 nm using software provided with the UPLC unit (Agilent OpenLab Chemstation Rev. C.01.07).
Colony forming unit CFU) assay
Human CD34 + enriched were washed with PBS (1X) / 0.5% BSA and added to 300 mL media (IMDM with GlutaMAX; Invitrogen; Cat No. 31980030). The cells were then mixed in semi solid methycellulose media supplemented with cytokines (MethoCult H4435 Classic; StemCell Technologies; Cat No. 04445) per the manufacturer’s instructions. In brief, 500 cells per well were plated in duplicates (1.1 mL cell culture per well) in six well meniscus free cell culture plates (SmartDish; StemCell Technologies; Cat No. 27301). The plates were incubated for 14 days in a humidified incubator at 37 °C with 5% CO2 and then scored for total progenitors and type of colony using an automated colony counter (STEMvision; StemCell Technologies; Cat No. 22000). The automated colony counter recorded the total number of erythroid colony forming units (CFU E), erythroid burst forming units (BFU E), granulocyte colony forming unit / macrophage colony forming unit / granulocyte, macrophage colony forming units (CFU G/M/GM) and granulocyte, erythrocyte, macrophage, megakaryocyte forming units (CFU GEMM) in each well.
Miseq indel analysis (or genotyping the targeted loci by deep sequencing)
Disruption of targeted BCL11A ESE loci was quantitated by MiSeq deep sequencing of a 140-bp amplicon using procedures described previously30. The primer sequences bearing sample-specific barcodes and the Illumina flow cell-specific sequences (P5 and P7) are provided in Methods Table 1. We used a MiSeq Reagent Kit v2 (300 cycles) for deep sequencing of the amplicon.
N: mixed base containing 25% of each nucleotide of ACGT (to increase sequence diversity). We merged paired-end reads using SeqPrep (https://github.com/jstjohn/SeqPrep), then trimmed reads using a custom script to remove any remaining adapter sequence as well as the 4 random nucleotides from the left primer. The script also filtered out sequences where the first and last 20 bp do not match the reference amplicon sequence. We then removed reads with lower quality bases using prinseq46(-min_qual_score 15) and aligned remaining sequences to the human genome (hg38) using bowtie247.
We filtered out sequences that aligned outside the target site but kept reads that failed to align. As a last filtering step, we removed reads ≤ 70 bp long to filter out potential PCR artifacts.
Finally, we aligned sequences to the expected amplicon using the Needleman-Wunsch algorithm as implemented in seq-align (https://github.com/noporpoise/seq-align). To count indels, a window of 21 bp was defined around the central GATAA motif (GATAA ± 8 bp each side), and contiguous gaps in either the query or reference sequence that overlapped this window were considered indels.
Single-cell clonal expansion analysis
Single-cell clonal expansion analysis was performed as described previously30. Briefly, ZFN-edited CD34 + cells were seeded at 2 cells/well into 96-well non-TC treated plates (Corning) using FACSAria III. The cells were subjected to 3-step expansion in erythroid differentiation media for 20 days; genomic DNA and protein lysate were collected at Day 14 and 20, for deep sequencing and RP-UPLC analysis, respectively. Additionally, at Day 20, cells were stained with GlyA-FITC antibody and Nuc Red for enucleation rate.
We called genotypes of single clones as above, with the following differences: For a subset of clones from sample 1 and all clones from samples 3 and 4, we used a larger amplicon (471 bp) (MethodsTable 2) that covered common SNPs to potentially distinguish homozygous clones from clones where one allele failed to amplify due to a deletion covering the primer sites. For the individuals with informative heterozygous SNP, this method allows confirmation of whether the originally assigned homozygous clones (biallelic edits with same editing pattern) are true homozygotes or false homozygous where one allele failed to amplify due to a deletion covering the primer sites. We used a MiSeq Reagent Kit v2 (500 cycles) for deep sequencing of the larger amplicon. We also included reads that failed merge with SeqPrep due to the overlap being too small. We called SNPs using bcftools mpileup48 on all clones.
We only considered clones with read counts > 50 reads and allele frequencies consistent with a clonal population (i.e. 2 major alleles with read counts of 50% each or one major allele at ~ 100% frequency). Specifically, we used the following criteria: For heterozygotes, the top 2 indels needed to each comprise 40% to 60% of reads and together more than 85%, and the third most common indel constitute less than 5% of read sequences. For homozygotes, the most common sequence needed to comprise more than 85% of reads, with the second indel less than 5%.
For clones with uncharacterized alleles (including homozygotes and clones that failed to amplify), we amplified a ~ 12 kb region centered on the target site (Methods Table 3) with PrimeSTAR® GXL DNA Polymerase (R050A, Takara). After purification of the ~ 12 kb PCR products, we fragmented and tagged the DNA with adapter sequences using Nextera XT DNA library prep kit (FC-131–1096, Illumina), following the instruction from the manufacturer. We amplified the library using Nextera XT Index Kit v2 Set A (FC-131–2001, Illumina) and pooled the library for deep sequencing with MiSeq Reagent Kit v2 (300 cycles).
We trimmed adapters from reads using Trimmomatic49, then aligned reads to the genome (hg38) using SpeedSeq align50. We used LUMPY51to call large indels and bcftools mpileup48 to call small indels and SNPs. Moreover, each alignment was visually inspected in Integrated Genomics Viewer (IGV) to assure that editing events were not missed by the variant callers, and to filter out events likely reflecting PCR artifacts (e.g. structural variants at the ends of the amplicon).
Association of HbF with editing in single clones
We used linear regression to test the association of indels with HbF, adjusting for the differences between donors by incorporating a donor-specific covariate, specifically:
where Y represents the HbF levels, \({\beta }_{0}\) is the intercept, \({\beta }_{indels}\) is the slope (beta) coefficient for the editing level variable and \({\beta }_{donor}\) is the slope coefficient for the donor effect variable\({X}_{donor}\) . We represented HbF levels as the ratio of gamma-globin over all beta-like globins.
Biallelic enrichment analysis
To calculate the enrichment of biallelic clones, we first calculated the expected number of clones using an equation analogous to the Hardy–Weinberg equilibrium52:
where \({f}_{unedited}\) and \({f}_{edited}\) are the frequencies of unedited and edited alleles respectively. \({f}_{unedited}^{2}\) represents the expected frequency of unedited clones,\({2f}_{unedited}{f}_{edited}\) the expected frequency of monoallelic edited clones and \({f}_{edited}^{2}\) the expected frequency of biallelic edited clones. We multiplied the frequencies by the total number of clones to obtain the expected number of wild-type, monoallelic edited, and biallelic edited clones. Finally, we constructed a contingency table with the observed and expected number of clones and calculated significant deviations using a chi-squared test. This analysis was done separately for each donor.
Homozygote enrichment analysis
To calculate the enrichment of homozygote clones, we first permuted each allele 100,000 times. Then, we derived a P value by counting the number of permutations that lead to a count of homozygotes higher than observed and dividing that number by 100,000.
Microhomology indels identification
We used the CRISPR RGEN Microhomology online tool53 to predict MMEJ indels, using the following sequence as input:
CGCCCCCA CCCTAATCA GAGGCCAA ACCCTTC CTGGAGCC TGTGATAA AAGCAAC TGTTAG CTTGCAC TAGACTA GCTTCA AAGTT GTATT. We used Pearson correlation to relate pattern scores with MMEJ-based indel frequencies. In addition, we inferred microhomology sequences around small (< 50 bp) and large (> 50 bp) deletions using mhscanR44.
PRECIZN-1: Bivv003 phase 1/2 study design and methodology
PRECIZN-1 (NCT03653247) is an ongoing, first-in-human, open-label, single arm, multi-site, US-based study to evaluate safety, tolerability, and efficacy of BIVV003 in adult participants age 18–40 years with severe SCD . Eligibility was based on having HbSS or HbSβ0 genotype, and at least one of the following: (1) history of clinically significant neurologic event (stroke) or any neurological deficit lasting > 24 h; (2) history of 2 or more episodes of ACS in the 2-year period preceding informed consent; (3) six or more pain crises (requiring intravenous [IV] pain management) in the prior 2 years; (4) 2 or more episodes of priapism with subject seeking medical care or following a pre-approved management plan in the prior 2 years; (5) regular (≥ 8) RBC transfusions in the preceding year to prevent vaso-occlusive clinical complications; (6) tricuspid valve regurgitant jet velocity ≥ 2.5 m/s by echocardiography. Hydroxyurea (HU) use was permitted but one had to be willing to discontinue HU at least 30 days prior to stem cell mobilization through Day 100 post-transplantation.
There was an initial 12-week screening period to assess participants’ suitability for the study. At the screening visit, participant 2 reported cerebral infarction, participant 4 reported stroke, and participant 7 reported cerebral microembolism. No stroke was reported after the screening visit in the study. Transfusion therapy was used within 2 months of mobilization and prior to myeloablative conditioning to target levels of HbS < 30% and total Hb at least 9–10 g/dL. Enrolled participants underwent plerixafor mobilization (240 μg/kg/day for up to 3 days) and apheresis to collect autologous CD34 + HSC with a target of 10 × 106 CD34 + HSC/kg for manufacturing BIVV003 using a LOVO device. Additional apheresis cycles were allowed to collect the minimum cell dose and unmodified rescue aliquots. Selected CD34 + cells were cultured in the presence of cytokines (SCF, FLT-3L, TPO). Two days after CD34 + cell isolation autologous HSC were transfected ex vivo with ZFN messenger ribonucleic acids (mRNAs) targeting the ESE region of the BCL11A locus using a MaxCyte device. Transfected cells were cultured in the presence of cytokines for another two days and then harvested, formulated in cryopreservation medium, aliquoted, frozen using a controlled-rate freezer and stored in the vapor phase of liquid nitrogen at ≤ −150 °C. For CD34 + cell culture in Group 1 Lonza X-Vivo 10 Medium and ThermoFisher cytokines were used, in Group 2 Cell Genix media and cytokines. In addition, for Group 2 manufacturing the LOVO instrument was upgraded with software version 3.0 which improved CD34 + cell recovery. Myeloablation of the participant’s bone marrow was performed using busulfan 3.2 mg/kg/day IV × 4 days, with pharmacokinetically adjusted dosing. At least 72 h after the final busulfan dose, a single IV infusion of 3–20 × 106 CD34 + HSC/kg was administered.
Participants were monitored for hematopoietic engraftment and recovery, adverse events (AE), clinical and laboratory hemolysis markers, total Hb and HbF, percentage of F cells, and sickle-cell-related events post-BIVV003 infusion.
Key study endpoints
Primary endpoints (to evaluate safety and tolerability of BIVV003) were survival at post transplantation Day 100, Week 52, and Week 104 (last study visit), successful engraftment, and the occurrence of adverse events and serious adverse events. Secondary endpoints (to assess the success and kinetics of stem-cell collection, manufacturing, and engraftment) were CD34 + HSC yield from stem-cell mobilization, yield of ZFN-edited CD34 + HSC, time to initial neutrophil recovery following infusion (first of three consecutive days with absolute neutrophil count ≥ 500/μL), time to platelet recovery post infusion (first of three consecutive measurements with a platelet count ≥ 50,000/μL at least 7 days after last platelet transfusion) and clinical assessments after BIVV003 infusion. Data cutoff for study endpoints was September 2022.
Statistical analyses
No formal statistics were performed. All statistics are presented as descriptive.
Data availability
Next-generation sequencing data generated during the current study are available in the Sequence Read Archive (BioProject PRJNA1053294, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1053294). Qualified researchers may request access to patient level data and related study documents. Patient level data will be anonymized, and study documents will be redacted to protect the privacy of our trial participants. Further details on Sanofi’s data sharing criteria, eligible studies, and process for requesting access can be found at: https://www.vivli.org/.Other datasets generated during the current study are available from the corresponding author on reasonable request.
References
Frenette, P. S. & Atweh, G. F. Sickle cell disease: old discoveries, new concepts, and future promise. J Clin Invest 117, 850–858 (2007).
Hebbel, R. P., Osarogiagbon, R. & Kaul, D. The endothelial biology of sickle cell disease: inflammation and a chronic vasculopathy. Microcirculation 11, 129–151 (2004).
Kato, G. J. et al. Sickle cell disease. Nat Rev Dis Primers 4, 18010 (2018).
Khemani, K., Katoch, D. & Krishnamurti, L. Curative Therapies for Sickle Cell Disease. Ochsner J 19, 131–137 (2019).
Lettre, G. & Bauer, D. E. Fetal haemoglobin in sickle-cell disease: from genetic epidemiology to new therapeutic strategies. Lancet 387, 2554–2564 (2016).
Perrine, R. P., Brown, M. J., Clegg, J. B., Weatherall, D. J. & May, A. Benign sickle-cell anaemia. Lancet 2, 1163–1167 (1972).
Kar, B. C. et al. Sickle cell disease in Orissa State. India. Lancet 2, 1198–1201 (1986).
Pincez, T. et al. Variation and impact of polygenic hematologic traits in monogenic sickle cell disease. Haematologica 108, 870–881 (2023).
Drysdale, C. M. et al. Hematopoietic-Stem-Cell-Targeted Gene-Addition and Gene-Editing Strategies for beta-hemoglobinopathies. Cell Stem Cell 28, 191–208 (2021).
Steinberg, M.H. et al. The risks and benefits of long-term use of hydroxyurea in sickle cell anemia: A 17.5 year follow-up. Am J Hematol 85, 403–408 (2010).
Venkatesan, V., Srinivasan, S., Babu, P. & Thangavel, S. Manipulation of Developmental Gamma-Globin Gene Expression: an Approach for Healing Hemoglobinopathies. Mol Cell Biol 41 (2020).
Wienert, B., Martyn, G. E., Funnell, A. P. W., Quinlan, K. G. R. & Crossley, M. Wake-up Sleepy Gene: Reactivating Fetal Globin for beta-Hemoglobinopathies. Trends Genet 34, 927–940 (2018).
Hsieh, M. M. et al. Nonmyeloablative HLA-matched sibling allogeneic hematopoietic stem cell transplantation for severe sickle cell phenotype. JAMA 312, 48–56 (2014).
Hsieh, M. M. et al. Allogeneic hematopoietic stem-cell transplantation for sickle cell disease. N Engl J Med 361, 2309–2317 (2009).
Mentzer, W. C., Heller, S., Pearle, P. R., Hackney, E. & Vichinsky, E. Availability of related donors for bone marrow transplantation in sickle cell anemia. Am J Pediatr Hematol Oncol 16, 27–29 (1994).
Robinson, T. M. & Fuchs, E. J. Allogeneic stem cell transplantation for sickle cell disease. Curr Opin Hematol 23, 524–529 (2016).
Esrick, E. B. et al. Post-Transcriptional Genetic Silencing of BCL11A to Treat Sickle Cell Disease. N Engl J Med 384, 205–215 (2021).
Frangoul, H. et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and beta-Thalassemia. N Engl J Med 384, 252–260 (2021).
Basak, A. et al. BCL11A deletions result in fetal hemoglobin persistence and neurodevelopmental alterations. J Clin Invest 125, 2363–2368 (2015).
Sankaran, V. G. et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322, 1839–1842 (2008).
Sankaran, V. G. et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature 460, 1093–1097 (2009).
Bauer, D. E. et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–257 (2013).
Weber, L. et al. Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci Adv 6 (2020).
Metais, J. Y. et al. Genome editing of HBG1 and HBG2 to induce fetal hemoglobin. Blood Adv 3, 3379–3392 (2019).
Li, C. et al. In vivo HSPC gene therapy with base editors allows for efficient reactivation of fetal gamma-globin in beta-YAC mice. Blood Adv 5, 1122–1135 (2021).
Humbert, O. et al. Therapeutically relevant engraftment of a CRISPR-Cas9-edited HSC-enriched population with HbF reactivation in nonhuman primates. Sci Transl Med 11 (2019).
Canver, M. C. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015).
Vierstra, J. et al. Functional footprinting of regulatory DNA. Nat Methods 12, 927–930 (2015).
Miller, J. C. et al. Enhancing gene editing specificity by attenuating DNA cleavage kinetics. Nat Biotechnol 37, 945–952 (2019).
Chang, K. H. et al. Long-Term Engraftment and Fetal Globin Induction upon BCL11A Gene Editing in Bone-Marrow-Derived CD34(+) Hematopoietic Stem and Progenitor Cells. Mol Ther Methods Clin Dev 4, 137–148 (2017).
Psatha, N. et al. Disruption of the BCL11A Erythroid Enhancer Reactivates Fetal Hemoglobin in Erythroid Cells of Patients with beta-Thalassemia Major. Mol Ther Methods Clin Dev 10, 313–326 (2018).
Alavi, A. et al. Interim Safety and Efficacy Results from a Phase 1/2 Study of Zinc Finger Nuclease-Modified Autologous Hematopoietic Stem Cells for Sickle Cell Disease (PRECIZN-1). Blood 140, 4907–4909 (2022).
Alavi, A. et al. Preliminary Safety and Efficacy Results from Precizn-1: An Ongoing Phase 1/2 Study on Zinc Finger Nuclease-Modified Autologous CD34+ HSPCs for Sickle Cell Disease (SCD). Blood 138, 2930–2930 (2021).
Yannaki, E. et al. Hematopoietic stem cell mobilization for gene therapy: superior mobilization by the combination of granulocyte-colony stimulating factor plus plerixafor in patients with beta-thalassemia major. Hum Gene Ther 24, 852–860 (2013).
Yannaki, E. et al. Hematopoietic stem cell mobilization for gene therapy of adult patients with severe beta-thalassemia: results of clinical trials using G-CSF or plerixafor in splenectomized and nonsplenectomized subjects. Mol Ther 20, 230–238 (2012).
Hsieh, M. M. & Tisdale, J. F. Hematopoietic stem cell mobilization with plerixafor in sickle cell disease. Haematologica 103, 749–750 (2018).
Lessard, S. et al. Zinc Finger Nuclease-Mediated Disruption of the BCL11A Erythroid Enhancer Results in Enriched Biallelic Editing, Increased Fetal Hemoglobin, and Reduced Sickling in Erythroid Cells Derived from Sickle Cell Disease Patients. Blood 134, 974–974 (2019).
Brendel, C. et al. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J Clin Invest 126, 3868–3878 (2016).
Brendel, C. et al. Preclinical Evaluation of a Novel Lentiviral Vector Driving Lineage-Specific BCL11A Knockdown for Sickle Cell Gene Therapy. Mol Ther Methods Clin Dev 17, 589–600 (2020).
Demirci, S. et al. BCL11A enhancer-edited hematopoietic stem cells persist in rhesus monkeys without toxicity. J Clin Invest 130, 6677–6687 (2020).
Wu, Y. et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med 25, 776–783 (2019).
Leibowitz, M. L. et al. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat Genet 53, 895–905 (2021).
Park, S.H. et al. Comprehensive analysis and accurate quantification of unintended large gene modifications induced by CRISPR-Cas9 gene editing. Sci Adv 8, eabo7676 (2022).
Owens, D. D. G. et al. Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res 47, 7402–7417 (2019).
Giarratana, M. C. et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol 23, 69–74 (2005).
Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10 (2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Chiang, C. et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods 12, 966–968 (2015).
Layer, R. M., Chiang, C., Quinlan, A. R. & Hall, I. M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15, R84 (2014).
Canver, M. C. et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem 292, 2556 (2017).
Bae, S., Kweon, J., Kim, H. S. & Kim, J. S. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods 11, 705–706 (2014).
Acknowledgements
This study was funded by Biogen, Sangamo, and Sanofi. We would like to thank the investigators, research staff, donors, and trial volunteers who participated in these studies. Medical writing and editing support were provided by Neil M. Thomas and Rebecca S. Jones of Fishawack Communications Ltd., part of Fishawack Health. This support was funded by Sanofi.
Funding
Sanofi, Biogen, Sangamo Therapeutics.
Author information
Authors and Affiliations
Contributions
All authors had access to study data, had full editorial control of the manuscript, and provided their final approval of all content.
Corresponding author
Ethics declarations
Competing interests
AH and SL are employees of Sanofi and may hold shares and/or stock options in the company. HL, KM, BV, DL, DR, PR, RP, TH, YL, GMR, IG and VH were employees of Sanofi at the time of the study. AR, MC, and BH are employees of Sangamo and may hold shares and/or stock options in the company. RB and BC were employees of Sangamo at the time of study. NU and JT have no conflict of interest. MA receives honorarium from Bristol Myers Squibb, hold stock from Cytodine Inc, and participates in speaker bureaus for Bristol Myers Squibb, Gilead, and Abbvie. MCW has served as consultant for Ensoma, Inc., Vertex pharmaceuticals, AllCells, Inc, and BioLabs, Inc.
Informed consent
The study (NCT03653247) was performed in accordance with the Declaration of Helsinki and local regulations. The study was approved by an independent ethics committee or institutional review board at each trial site (UCSF Benioff Children’s Hospital Oakland 2018–041; National Institutes of Health 526,145; Western Institutional Review Board 1–1,134,061-1/20,181,878; UC Davis Clinical Committee 1,363,599–4). All participants provided written informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lessard, S., Rimmelé, P., Ling, H. et al. Zinc finger nuclease-mediated gene editing in hematopoietic stem cells results in reactivation of fetal hemoglobin in sickle cell disease. Sci Rep 14, 24298 (2024). https://doi.org/10.1038/s41598-024-74716-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74716-7