Abstract
Retinal ganglion cells (RGCs) are among the first neurons to undergo apoptosis in diabetic retinopathy (DR), with their relationship to endoplasmic reticulum stress (ERS)-induced apoptosis still unclear. While glucocorticoid-induced transcript 1 (GLCCI1) has been shown to inhibit apoptosis, its role in ERS-induced apoptosis and its mechanisms in DR remain unclarified. Our findings indicated that GLCCI1 is predominantly localized in the ganglion cell layer and is downregulated in DR. GLCCI1 overexpression mitigated the apoptosis of RGCs and the swelling of endoplasmic reticulum and mitochondria under hyperglycemia, and downregulated ERS-induced apoptosis related markers (GRP78, CHOP and cleaved CASP3), whereas GLCCI1 knockdown has the opposite effect. In vivo, GLCCI1 overexpression not only prevents structural lesions but also protects against microvascular dysfunctions in the retinas of DR mice. We found that GLCCI1 directly interacts with HSP90AB1, which in turn interacts with GRP78. Additionally, GLCCI1 is an upstream regulator of HSP90AB1, which regulates GRP78. Thus, the impact of GLCCI1 on the ERS-induced apoptosis is mainly through the regulation of HSP90AB1, and subsequently inhibiting GRP78-initiated ERS-induced apoptosis. These findings offer a promising avenue for further treatment of DR.
Similar content being viewed by others
Introduction
According to the International Diabetes Federation (IDF), the global number of patients with diabetes mellitus (DM) was approximately 537 million in 2021, with a projected surge to 783 million by 2045 1. DR, a commonly occurring complication of DM affecting 22.27% of DM patients2, ranks as the principal cause of adult blindness3,4. Given the high incidence of DR (80%) occurring after 10 years of diabetes5, it is imperative and time-sensitive to investigate the early pathogenesis and preventive measures for DR.
Accumulated evidence suggests that neurodegeneration occurs prior to microvascular abnormalities and clinical signs in DR, making retinal neurodegeneration widely recognized as the primary cause of DR6,7,8,9. According to reports, ERS contributes to neurodegeneration in DM and DR patients10,11,12,13,14. The loss of neurons as well as apoptosis are hallmarks of neurodegeneration, and various stimuli can trigger neurodegeneration in mature neurons via the apoptosis pathway15,16,17. Thus, we speculate that the ERS-induced apoptotic pathway may be involved in the development of DR, but it has not been well elucidated in documents. Among all types of retinal neurocytes, RGCs is one of the first apoptotic cells in DR18,19. Therefore, investigating the endoplasmic reticulum stress (ERS)-induced apoptosis of RGCs is crucial for understanding the mechanism and prevention of DR.
Glucocorticoid-induced transcript 1 (GLCCI1), also known as FAM117, GCTR, GIG18, and TSSN1, is located on the short arm of chromosome 7 (7p21.3) and is responsible for the production of a 58 kDa protein (gene annotation from NCBI). GLCCI1 has been shown to be highly expressed in CD4(+) CD8(+) cells of the mouse thymus and during spermatogenesis in the testis20. It was reported that the inhibition of GLCCI1 caused T cells to undergo apoptosis, while overexpressing GLCCI1 promoted T-cell proliferation. These observations suggest that GLCCI1 may protect against apoptosis21. However, the expression of GLCCI1 and its relationship with DR and ERS-induced apoptosis, as well as its potential molecular mechanisms, are still largely unknown and urgently need to be explored.
In this study, based on streptozotocin (STZ)-induced type 1 diabetic mice and in vitro study, we demonstrated that GLCCI1 was downregulated in DR, resulting in HSP90AB1 downregulation, and was associated with ERS-induced RGCs apoptosis, possibly through the GLCCI1-HSP90AB1-GRP78 signaling pathway, suggesting that GLCCI1 could serve as a novel avenue for the treatment of DR.
Results
GLCCI1 expression was downregulated while ERS-induced apoptosis related markers were upregulated in DR mice
20 male C57BL/6J mice were randomly divided into two groups (DM: 150 mg/kg ,1% STZ intraperitoneal injection; Sham: equal volume of 0.1 mM citrate-citric acid buffer; n = 10/group). Before STZ injection, the average blood glucose level of the mice was 4.52 ± 0.71 mmol/L. Following the STZ injection, blood glucose levels were measured three times over a 72-hour period, revealing that 9 out of 10 mice had blood glucose levels exceeding 16.7 mmol/L, indicating that 90% of the mice successfully developed diabetes. The DM group gradually exhibited typical symptoms associated with diabetes, characterized by polydipsia, polyphagia, polyuria, and weight loss, while the blood glucose levels in the Sham group remained within the normal range. The average blood glucose levels in the DM group were significantly higher than those in the Sham group at 72 h, 2 weeks, 6 weeks, and 12 weeks (p < 0.05, Fig. 1A; Table 1). There was no difference in body weight between the two groups of mice prior to the intraperitoneal injection. By the time of execution, the body weight of the DM group was significantly lower than that of the Sham group (p < 0.05, Fig. 1B; Table 2). Changes in body weight and blood glucose levels in both groups confirmed the successful establishment of the DM model (p < 0.05) (Fig. 1A, B; Tables 1 and 2).
GLCCI1 is downregulated in diabetic retinopathy. Sham: injected with 0.1 mM citrate-citric acid buffer; DM: diabetes mellitus; After detecting changes in the retinal structure of DM, we subsequently refer to it as the DR group. DR: diabetic retinopathy group; C-CASP3: cleaved Caspase3. The data is described as mean ± standard error of the mean (SEM), and the results are analyzed using independent sample t-test. (A) Random blood glucose levels of mice after intraperitoneal injection at 72 h, 2 weeks, 6 weeks, and 12 weeks. (B) Body weight of mice in each group before STZ injection or before execution. (C) Representative IHC figures of GLCCI1 expression in the retinas of Sham and DR mice. GCL: ganglion cell layer; IPL: inner plexus layer; INL: inner nuclear layer; ONL: outer nuclear layer. scale bar = 100 μm. (D) Average intensity of GLCCI1 in IHC. (E) Average GLCCI1 intensity in GCL. GLCCI1 was predominantly located in ganglion cell layer (GCL) and was significantly lower in the DR group (DR vs. Sham: 0.438 ± 0.0481 vs. 1.000 ± 0.002, t = 116.7, p < 0.001). (F-H) The average thickness of IPL, INL and ONL in DR group were decreased (DR vs. Sham: IPL:20.83 ± 0.375 vs. 49.05 ± 0.608, t = 39.52, p < 0.001; INL: DR vs. Sham: 12.90 ± 0.297 vs. 26.64 ± 0.752, t = 16.99, p < 0.001; ONL: DR vs. Sham: 12.86 ± 0.270 vs. 38.69 ± 0.848, t = 29.01, p < 0.001). (I) Average number of RGCs per 100 μm in ganglion cell layer (GCL) in DR group was reduced (DR vs. Sham: 19.67 ± 1.20 vs. 48 ± 1.053, t = 26.88, p < 0.001). (J) qPCR results showed the relative Glcci1 mRNA expression level was decreased in DR (DR vs. Sham: 0.762 ± 0.0383 vs. 1.001 ± 0.0204, t = 5.51, p = 0.001, p < 0.05). (K) and (L) WB analysis of the protein expression level of GLCCI1 and ERS-induced apoptosis related markers in the mouse retina. GLCCI1 was downregulated in DR group (DR vs. Sham: 0.672 ± 0.0044 vs. 0.876 ± 0.0014, t = 44.11, p < 0.001), while the expression of ERS-induced apoptosis related markers was upregulated in the DR group (DR vs. Sham: GRP78:1.001 ± 0.0022 vs. 0.481 ± 0.0018, t=-185.06.51, p < 0.001; CHOP: 0.919 ± 0.0086 vs. 0.467 ± 0.0130, t=-28.99, p < 0.001; and cleaved Caspase3: 0.833 ± 0.0017 vs. 0.495 ± 0.0082, t=-40.13, p < 0.001). *p < 0.05, **p < 0.01. ns: not significant.
IHC results revealed that, compared to the Sham group, the average intensity of GLCCI1 in the DR group was significantly reduced (DR vs. Sham: 25.41 ± 0.176 vs. 3.51 ± 0.062, p < 0.001) (Fig. 1C, D). Our findings demonstrated that GLCCI1 was primarily localized in the ganglion cell layer (GCL), with a noticeable decrease in average intensity in the DR group (DR vs. Sham: 3.21 ± 0.065 vs. 60.01 ± 0.362, p < 0.001) (Fig. 1E). Additionally, structural analysis indicated that the thickness of the inner plexiform layer (IPL), inner nuclear layer (INL), and outer nuclear layer (ONL) in the DR group was significantly reduced compared to the Sham group (p < 0.001) (Fig. 1F-H). Furthermore, IHC results showed that the number of retinal ganglion cells (RGCs) in the DR group was decreased (p < 0.001) (Fig. 1I). The changes in the aforementioned structures indicate that pathological changes associated with DR have already developed.
Both qPCR and Western blot (WB) results indicated that the expression level of GLCCI1 mRNA and protein in the DR group were all downregulated (p < 0.001) (Fig. 1J-L). Western blot (WB) analysis also revealed that the upregulation of ERS-induced apoptosis related markers (GRP78, CHOP and cleaved Caspase3) in the DR group (p < 0.001) (Fig. 1K, L). These results suggest that the decreased expression of GLCCI1 and the ERS-induced apoptosis pathway may be involved in the occurrence and development of DR.
GLCCI1 overexpression mitigated ERS-induced RGCs apoptosis and promoted the vitality of RGCs under high glucose conditions in vitro
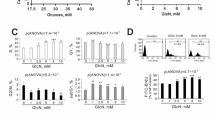
Immortalized mouse retinal ganglion cells (RGCs) were first identified by fluorescence staining using the RGC-specific marker BRN3A. At the same time, we also examined whether GLCCI1 was expressed in cells. The results of immunofluorescence showed that both BRN3A and GLCCI1 were expressed in cells (Fig. 2A), indicating that it is an RGCs cell line and can be used for following intervention experiments targeting GLCCI1. In vitro high glucose screening experiments, the experiments were divided into a normal glucose concentration group (Normal control), a mannitol group as an osmotic control (mannitol 50 mmol/L), and four different high glucose concentration groups (HG: including 25 mmol/L, 50 mmol/L, 100 mmol/L, and 150 mmol/L). Four time points were set (12 h, 24 h, 48 h, and 72 h). we assessed changes in cell vitality at different high glucose concentrations using the CCK8 assay, which indirectly and roughly reflects the level of cell apoptosis. Based on the results of the CCK8 assay, we plotted a high glucose concentration-time curve (Fig. 2B). Upon the overall curve, a significant decline in proliferation was observed only at 48 h in the HG 50 mmol/L group, where the proliferation ability dropped to 52.67 ± 0.882%, resulting in an inhibition rate of approximately 43.73%. The results from the one-way ANOVA analysis of data at 48 h indicated statistically significant differences among the groups (F = 719.76, p < 0.001). Therefore, we selected HG 50 mmol/L at 48 h as the conditions for subsequent high glucose models in vitro.
GLCCI1 overexpression mitigates ERS-induced RGC apoptosis under high glucose conditions in vitro. RGCs: the normal group; HG: the cell control group under a high glucose; HG-Vector: the empty plasmid control group; HG-OE: GLCCI1 overexpression plasmid group. (A) GLCCI1 and RGC-specific marker BRN3A were all expressed in RGCs. (B) The CCK8 assay results showed the changes in cell vitality at different time points and high glucose concentrations. The results showed that HG 50 mmol/L at 48 h was the suitable condition for the subsequent in vitro high glucose model (p < 0.001). (C) Glcci1 mRNA relative expression level in RGCs. Glcci1 mRNA was downregulated in HG group compared with RGCs group (HG vs. RGCs: 0.497 ± 0.0176 vs. 1.000 ± 0.0173, t = 20.36, p < 0.001) and was successfully overexpressed in HG-OE group compared with HG-Vector group (1.230 ± 0.0404 vs. 0.607 ± 0.0524, t=-9.421, p = 0.001, p < 0.01). (D and E) WB analysis and bar graph showing the relative expression levels of GLCCI1, ERS-induced apoptotic markers and three UPR sensors in RGCs. GLCCI1 expression was downregulated in HG (HG vs. RGCs: 0.185 ± 0.0011 vs. 1.124 ± 0.0020, t = 844.58, p < 0.001) and was successfully overexpressed in HG-OE group (HG-OE vs. HG-Vector: 1.069 ± 0.0004 vs. 0.117 ± 0.0003, t = 1965.12, p < 0.001); the expression of ERS-induced apoptotic related markers and three UPR sensors were increased in RGCs (HG vs. RGCs: GRP78: 0.508 ± 0.0012 vs. 0.317 ± 0.0021, t = 90.33, p < 0.001; CHOP: 0.575 ± 0.0023 vs. 0.332 ± 0.0007, t = 7.64, p = 0.017, p < 0.05; cleaved Caspase3: 0.902 ± 0.0002 vs. 0.423 ± 0.0003, t = 1446.32, p < 0.001); the expression of ERS-induced apoptotic related markers in HG-OE group (HG-OE vs. HG-Vector: GRP78: 0.192 ± 0.0035 vs. 0.451 ± 0.0052, t = 75.11, p < 0.001; CHOP: 0.161 ± 0.0002 vs. 0.567 ± 0.0013, t = 299.70, p < 0.001; cleaved Caspase3: 0.051 ± 0.0009 vs. 0.854 ± 0.0045, t = 177.97, p < 0.001). (F and E) Flow cytometry analysis and bar graph showing the percentage of apoptotic RGCs. GLCCI1 overexpression alleviated ERS-induced apoptosis rate (p < 0.001) and promoted the vitality of RGCs (p < 0.001). (H) The vitality of RGCs. ns: not significant; *P < 0.05, **P < 0.01.
The GLCCI1 overexpression experiment was categorized into four groups: the normal group (RGCs), the cell control group under a high glucose (HG), the empty plasmid control group (HG-Vector), and the GLCCI1 overexpression plasmid group (HG-OE). qPCR and WB Results showed that the relative expression level of GLCCI1 mRNA and protein in HG group were all downregulated compared with RGCs group (p < 0.001) (Fig. 2C-E); The relative expression level of GLCCI1 mRNA and protein in HG-OE group were all successfully overexpressed compared with HG-Vector group (p < 0.01) (Fig. 2C-E). The results of flow cytometry showed that high glucose increased cell apoptosis rate (HG vs. RGCs: 18.99% vs. 8.40%, p < 0.001), while overexpression of GLCCI1 inhibited cell apoptosis (HG-OE vs. HG-Vector: 11.46% vs. 17.60%, p < 0.001) (Fig. 2F and G). The CCK-8 results also revealed that high glucose suppressed the vitality of RGCs (HG vs. RGCs: 58.47 ± 4.69% vs. 100 ± 1.38%, p < 0.001) (Fig. 2H), while GLCCI1 overexpression enhanced the vitality of RGCs under high glucose (HG-OE vs. HG-Vector: 80.84 ± 4.95% vs. 53.54 ± 7.96%, p < 0.001) (Fig. 2H). WB results indicated that ERS-induced apoptosis related markers and UPR sensors in HG group were all upregulated compared with RGCs group (GRP78, CHOP, cleaved Caspase3,) (p < 0.05) (Fig. 2D and E). However, GLCCI1 overexpression (HG-OE group) downregulated the expression of ERS-induced apoptosis-related markers compared with HG-Vector group (p < 0.05) (Fig. 2D and E). These data suggest that GLCCI1 may protect RGCs by inhibiting GRP78-mediated ERS-induced apoptosis.
GLCCI1 knockdown exacerbated ERS-induced apoptosis and inhibited the vitality of RGCs, and this effect can be reversed by the ERS inhibitor 4-phenylbutyric acid (4-PBA) in vitro
To better understand the function of GLCCI1, we knocked down GLCCI1 and try to rescue its effect by 4-PBA, the experiment groups included RGCs, Vector, GLCCI1 knockdown (KD) and 4-PBA. Both qPCR and WB results indicated that the relative expression of GLCCI1 mRNA (p < 0.01) and protein (p < 0.01) in KD group were successfully knocked down compared with Vector group (Fig. 3A-C). However, the addition of 4-PBA (4-PBA) group had no effect on the expression levels of GLCCI1 mRNA (p = 0.146, p > 0.05) or protein (p = 0.173, p > 0.05). Our findings indicated that, compared with the Vector group, GLCCI1 knockdown (KD) enhanced the apoptosis of RGCs (p < 0.01) and compromised the vitality (p < 0.01) (Fig. 3D-F), and upregulated the relative expression of ERS-induced apoptosis-related markers (GRP78, CHOP, cleaved Caspase3) (p < 0.05). Additionally, these above upregulated proteins were rescued by the ERS inhibitor 4-PBA (p < 0.001) (Fig. 3D-F). These findings further support the hypothesis that GLCCI1 may primarily exert its anti-apoptotic function through suppressing the GRP78-mediated ERS-induced apoptosis in vitro.
GLCCI1 knockdown exacerbates ERS-induced RGC apoptosis. CASP12: Caspase12, C-CASP3: cleaved Caspase3. (A) Glcci1 mRNA relative expression level in RGC. Glcci1 mRNA was successfully knocked down (KD vs. Vector : 0.078 ± 0.0008 vs. 0.703 ± 0.0491, t = 12.725, p < 0.001); 4-PBA had no effect on the expression levels of GLCCI1 mRNA (4-PBA vs. KD: 0.083 ± 0.0023 vs. 0.078 ± 0.0008, t = 1.801, p = 0.146, p > 0.05). (B and C) WB images and bar graph of the relative protein expression levels. GLCCI1 protein expression level was successfully knocked down (KD vs. Vector: 0.268 ± 0.0062 vs. 0.837 ± 0.0042, t = 76.155, p < 0.001); 4-PBA had no effect on the expression levels of GLCCI1 protein (4-PBA vs. KD: 0.279 ± 0.0030 vs. 0.268 ± 0.0062, t = 1.656, p = 0.173, p > 0.05). GLCCI1 knockdown led to the upregulation of ERS-induced apoptosis pathway related markers (KD vs. Vector: GRP78: 0.923 ± 0.0038 vs. 0.397 ± 0.0027, t = 112.52,p < 0.001; CHOP: 1.101 ± 0.322 vs0.917 ± 0.0029, t = 5.687, p = 0.029, P < 0.05; cleaved Caspase3: 0.989 ± 0.0017 vs. 0.745 ± 0.0029, t = 73.55, p < 0.001). 4-PBA reverted those above upregulated proteins in KD group (4-PBA vs. KD: GRP78: 0.567 ± 0.0042 vs. 0.923 ± 0.0038, t = 62.78, p < 0.001; CHOP: 0.515 ± 0.0092 vs. 1.101 ± 0.0322, t = 17.487, p < 0.001; cleaved Caspase3: 0.584 ± 0.0019 vs. 0.989 ± 0.0017, t = 158.29, p < 0.001). (D and E) Flow cytometry results and bar graph showing the proportion of apoptotic RGCs. The apoptosis rate (early and late apoptosis) of RGCs was elevated following the knockdown of GLCCI1(KD vs. Vector: 22.18 ± 0.58% vs. 8.41 ± 0.51%, t = 84.91, p < 0.001), and 4-PBA rescued the results of GLCCI1 knockdown (4-PBA vs. KD: 13.27 ± 0.15% vs. 22.18 ± 0.58%, p < 0.001). (F) Cell vitality is illustrated as a bar graph. The vitality of RGCs was reduced following the knockdown of GLCCI1(KD vs. Vector: 40.64 ± 1.14% vs. 99.93 ± 2.37%, t = 22.51, p < 0.001), and which was alleviated by 4-PBA(4-PBA vs. KD: 72.39 ± 1.03% vs. 40.64 ± 1.14%, t = 20.71, p < 0.001). ns: not significant. *p < 0.05, **p < 0.01.
GLCCI1 knockdown resulted in DR-like retinas in normal mice, and GLCCI1 overexpression attenuated retinal damage in DR mice
60 male C57BL/6J mice were randomly divided into two groups: the DM group (150 mg/kg, 1% STZ peritoneal injection) and the Sham group (equal volume of 0.1 mM citrate-citric acid buffer), with 30 mice in each group. Before STZ injection, the average blood glucose level of the mice was 4.48 ± 0.68 mmol/L. Following the STZ injection, blood glucose levels were measured three times over a 72-hour period, revealing that 29 out of 30 mice (96.7%) had blood glucose levels exceeding 16.7 mmol/L. Furtherly, Sham group mice were randomly divided into three groups (n = 10/group): Sham, AAV2/9 vehicle (Veh(-)) and AAV2/9 shGLCCI1(KD) group; and DM mice were randomly divided into DR, AAV2/9 vehicle (Veh(+)) and AAV2/9 GLCCI1 overexpression group (OE) (n = 10/group). One week after hyperglycemia, the corresponding reagents were injected into the vitreous cavities. However, the detection of weight and blood glucose is still conducted according to the grouping of Sham and DM. Typical diabetes symptoms including polydipsia, polyphagia, polyuria, weight loss and hyperglycemia were displayed in the DM group, while not in the Sham group (p < 0.05, Tables 3 and 4, supplementary Fig. 1A and B). To observe the viral transfection efficiency of AAVs in live mice, we first using fluorescein fundus angiography (FFA) and OCT, indicating successful transfection and functional expression of the AAVs overexpression or knockdown (Fig. 4A-E). Subsequently, after euthanizing the mice, we assessed the expression levels of GLCCI1 through IHC and Western blotting, further confirming the success of the AAVs intervention (Fig. 4F-K).
GLCCI1 expression was associated with the structure of the retina. Sham: normal control group; Veh (-): AAV2/9 vehicle; KD: AAV2/9 sh-GLCCI1 knockdown; DR: diabetic retinopathy; OE: GLCCI1 overexpression. Veh (+): AAV2/9 vehicle in DR group; RNFL/GCL/IPL: retinal nerve fiber layer/ganglion cell layer/inner plexiform layer, INL: inner nuclear layer, OPL: outer plexiform layer, ONL: outer nuclear layer, OLM/RPE: outer limiting membrane/retinal pigment epithelium. (A) Representative FFA images of mouse retinas. The black arrow indicates the location of the lesion in the fundus, while the white arrow shows the microvascular changes in the retina (microaneurysms). (B) Representative OCT images of mouse retinas. (C) Local magnification of the images in panels A (1–6) are shown in panels B (1–6). (D) Bar chart of the average thickness of the retina. The whole thickness of retina decreased in DR group compared with Sham group (DR vs. Sham: 200.20 ± 0.369 μm vs. 226.58 ± 0.441 μm, t = 45.88, p < 0.001); the whole thickness of retina decreased in KD group compared with Veh (-) group (KD vs. Veh (-): 195.86 ± 0.507 μm vs. 205.90 ± 0.343 μm, t = 16.38, p < 0.001); and the whole thickness of retina increased in OE group compared with Veh (+) group (OE vs. Veh(+): 220.88 ± 0.399 μm vs. 195.93 ± 0.518 μm, t = 38.18, p < 0.001) (E) Bar chart of the average thickness of the RNFL/GCL/IPL. The thickness of the RNFL/GCL/IPL decreased in DR vs. Sham: 56.48 ± 0.375 μm vs. 64.81 ± 0.460 μm, t = 14.05, p < 0.001; the thickness of the RNFL/GCL/IPL in KD vs. Veh (-): 51.90 ± 0.501 μm vs. 57.63 ± 0.375 μm, t = 9.152, p < 0.001; and the whole thickness of retina in OE vs. Veh (+): 62.54 ± 0.325 μm vs. 52.37 ± 0.355 μm, t = 21.16, p < 0.001. (F) Representative IHC images of GLCCI1 expression in the groups. (G) Bar chart of GLCCI1 average intensity in groups. (H) Bar chart of GLCCI1 average intensity in ganglion cell layer (GCL). (I) Bar chart of RGCs number. RGCs number in KD vs. Veh (-): 20.125 ± 0.6665 vs. 34.125 ± 0.7892, t = 13.55, p < 0.001; RGCs number in OE vs. Veh (+):36.875 ± 1.3421 vs. 17.625 ± 0.4605, t = 13.57, p < 0.001. (J) and (K) WB results and bar chart of the expression of ERS-induced apoptosis-related markers and three UPR sensors in the mouse retina. GLCCI1 knockdown resulted in the upregulation of ERS-induced apoptosis related markers(KD vs. Veh(-)): GLCCI1: 0.411 ± 0.0001 vs. 0.623 ± 0.0002, t = 826.86, p < 0.001; GRP78: 0.782 ± 0.0002 vs0.483 ± 0.0002, t = 1273.24, p < 0.001; CHOP: 1.206 ± 0.0018 vs. 0.653 ± 0.0025, t = 179.42, p < 0.001; C-CASP3: 0.910 ± 0.0003 vs. 0.392 ± 0.0047, t = 110.79, p < 0.001. GLCCI1 overexpression resulted in the downregulation of ERS-induced apoptosis related markers in vivo (OE vs. Veh(+)):GLCCI1:1.031 ± 0.0001 vs. 0.588 ± 0.004, t = 1273.24, p < 0.001; GRP78: 0.365 ± 0.0006 vs. 1.117 ± 0.0015, t = 1142.27, p < 0.001; CHOP: 0.909 ± 0.002 vs. 1.172 ± 0.0039, t = 59.71, p < 0.001; C-CASP3: 0.211 ± 0.0007 vs. 0.840 ± 0.0020, t = 292.64, p < 0.001. ns: not significant; * P < 0.05; ** P < 0.01. Scale bar = 100 μm.
To detect the retinal microvascular dysfunctions, fluorescein fundus angiography (FFA) was performed on the mouse fundus. Results showed that, compared with Vehicle (-) group, GLCCI1 knockdown (KD) resulted in an increased formation of microaneurysms in the retinas of normal blood glucose mice (Fig. 4A). In the fundus images of the DR group and Vehicle (+) group mice, exudates can be observed (indicated by black arrows), and the FFA shows vascular dilation and /or neovascularization in the corresponding areas (indicated by white arrows). This indicates that these mice experience microvascular dysfunctions. However, in the overexpression of GLCCI1 group (OE), no significant microvascular dysfunctions, such as microaneurysms, vascular dilation, or neovascularization, were observed in the fundus and FFA examinations (Fig. 4A). This indicates that GLCCI1 can effectively prevent retinal microvascular dysfunctions in DR mice, demonstrating a protective effect on the microvasculature in the retinas of these mice.
In order to detect changes in retinal structure, OCT was performed on the eyes of mice in live. OCT results revealed that: the whole thickness of retina decreased in DR group compared with Sham group (p < 0.001), indicating successful DR modeling; the whole thickness of retina decreased in KD group compared with Veh (-) group (p < 0.001); and the whole thickness of retina increased in OE group compared with Veh (+) group (p < 0.001) (Fig. 4B-E). The OCT results also showed a similar trend in the changes of retinal nerve fiber layer/ganglion cell layer/inner reticular layer (RNFL/GCL/IPL) in each group (p < 0.001) (Fig. 4B-E).
IHC results showed that, compared to Veh (-) group, the average intensity of GLCCI1 in the KD group was noticeable reduced (KD vs. Veh (-): 4.97 ± 0.022 vs. 13.54 ± 0.048, p < 0.001) (Fig. 4F, G), while compared to Veh (+) group, the average intensity of GLCCI1 in the OE group was increased successfully (OE vs. Veh (+):12.32 ± 0.024 vs. 6.64 ± 0.017, p < 0.001) (Fig. 4F, G). The results also suggested that the average intensity of GLCCI1 in the GCL of KD group was knocked down compared with Veh (-) group (KD vs. Veh (-): 2.92 ± 0.016 vs. 7.40 ± 0.034, p < 0.001); the expression level of GLCCI1 in OE group was overexpressed compared with Veh (+) group (OE vs. Veh (+): 9.73 ± 0.018 vs. 2.13 ± 0.015, p < 0.001); the expression level of GLCCI1 in DR group was decreased compared with Sham group (p < 0.001) (Fig. 4H). Additionally, The IHC results demonstrated that a reduction in GLCCI1 expression led to a decrease in the thickness of the IPL, INL and ONL, conversely, the overexpression of GLCCI1 improved the thickness of these layers in the DR group (p < 0.001) (supplementary Fig. 1C-E). Furthermore, the level of GLCCI1 expression was correlated with the number of RGCs. Knockdown of GLCCI1 decreased the number of RGCs, whereas overexpression of GLCCI1 protected against RGC loss (p < 0.001) (Fig. 4I).
WB analysis revealed a negative correlation between GLCCI1 expression and the expression of ERS-induced apoptosis-related markers (GRP78, CHOP and cleaved Caspase3) (p < 0.001), and the increase in the expression of these proteins was subsequently attenuated upon GLCCI1 overexpression (p < 0.001) (Fig. 4J-K). These results indicate that GLCCI1 not only protects the structural integrity of the mouse retina but also preserves the functional integrity of the retinal microvasculature.
GLCCI1 overexpression alleviated ER swelling and ERS-induced apoptosis
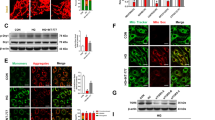
We explored the function of GLCCI1 in vitro and in vivo and further evaluated the function of GLCCI1 via subcellular structure analysis. TEM was used to examine ER stress, which was evidenced by direct morphological signs of swollen/enlarged ER lumens resulting from substantial and persistent ERS leading to extensive ER swelling22,23. To further determine the function of GLCCI1 in modulating subcellular structures, TEM was used to assess RGCs subjected to various treatments. The findings revealed significant enlargement of the endoplasmic reticulum (ER) lumen in the high glucose (HG) group, with particularly pronounced swelling observed in the negative control group (vector), an effect that is indicative of severe ballooning. Conversely, the overexpression of GLCCI1 mitigated the swelling of the ER lumen in the presence of high glucose (Fig. 5A). Additionally, swollen mitochondria were observed in both the HG and Vector groups; however, this swelling was attenuated by the overexpression of GLCCI1 (Fig. 5B). The above data provide convincing evidence supporting the inhibitory effect of GLCCI1 on ERS and ERS-induced apoptosis at the subcellular structural level.
HSP90AB1 interacts with both GLCCI1 and GRP78
We previously identified the function of GLCCI1 at the subcellular level, but the underlying molecular mechanism remains to be explored. The results of Co-IP/mass spectrometry (MS) showed that 51 proteins were detected in the IgG group, and 313 were detected in the Flag-GLCCI1 group. By removing non-specific binding proteins from the IgG group and arranging them from high to low according to their coverage %, a total of 276 relatively specific proteins were obtained (as shown in Fig. 6A). Among them, we screened and analyzed proteins with coverage ≥ 20% (a total of 39, as shown in Supplementary Table 1). Subsequent GO and Pathway enrichment, as well as protein interaction network analysis (Fig. 6B-D) showed that the 5 proteins of the endoplasmic reticulum protein processing pathway (HSPA8/HSPA5/HSP90AB1/HSP90AA1/P4HB) were closely related to our objectives. Therefore, we performed protein-protein docking with GLCCI1 to search for proteins that directly act on them.
Analysis of co-IP/MS and protein‒protein docking results. (A) Venn diagram of MS detection results. (B) GO analysis on specific proteins with coverage ≥ 20% (39 proteins). (C) String results showed the protein-protein interactions between 39 proteins. (D) KEGG pathway analysis of the 39 proteins. (E) Protein-protein docking between GLCCI1 and HSP90AB1. (F) Protein-protein docking between HSP90AB1 and GRP78.
The protein-protein docking results showed that the only protein capable of binding to GLCCI1 is HSP90AB1. Specifically, in the molecular interaction model with the highest HDOCK and HADDOCK scores, the Lys82 residue of GLCCI1 formed a hydrogen bond with Glu542 of HSP90AB1, with a bond length of 2.0 Å (GLCCI1 Lys82/HSP90AB1Glu542, 2.0 Å). Additionally, two other interaction sites were identified, namely, GLCCI1 Ala97/HSP90AB1 Lys551 [H]2.3 Å and GLCCI1 Phe253/HSP90AB1 Asp529 [H]2.3 Å (Fig. 6E and Supplementary Vedio1). Furthermore, the protein‒protein docking results revealed that HSP90AB1 is capable of interacting with GRP78 via specific interaction sites, namely, (HSP90AB1 Glu10/GRP78 Arg510, [H]2.1 Å and [H]2.3 Å; HSP90AB1 His6/GRP78 Glu536, [H]2.5 Å; and HSP90AB1 His6/GRP78 Glu539, [H]1.7 Å) (Fig. 6B and Supplementary Vedio2).
The co-IP results further suggested that Flag-GLCC1 can bind to HSP90AB1 but does not coprecipitate with GRP78 (Fig. 7A). Conversely, Flag-HSP90AB1 could bind to both GLCCI1 and GRP78 (Fig. 7.B). Additionally, GST-GLCCI1 and 6His-HSP90AB1 pulldown experiments confirmed the interaction between GLCCI1 and HSP90AB1 (Fig. 7C-D). In vitro, double fluorescence labeling was performed, and colocalization of GLCCI1 (green) with HSP90AB1 (red) and of HSP90AB1 (red) with GRP78 (green) was successfully observed (Fig. 7E and F). These results indicated that GLCCI1 interacts with HSP90AB1, and HSP90AB1 can interact with GRP78, which may be associated with the inhibition of ERS-induced apoptosis.
Co-IP, pull-down and double fluorescence labeling results. (A) Results of the co-IP of GLCCI1-FLAG in RGC-KO cells. (B) Results of co-IP of HSP90AB1-flag in RGCs. (C) Purified GST-GLCCI1 was used to pull down purified 6His-HSP90AB1 from E. coli. (D) Purified 6His-HSP90AB1 was used to pull down purified GST-GLCCI1 from E. coli. (E) Double fluorescence labeling of GLCCI1 (green) and HSP90AB1 (red) in RGCs. Arrows indicate areas of merging. (F) Double fluorescence labeling of GRP78 (green) and HSP90AB1 (red) in RGCs. Arrows indicate areas of merging.
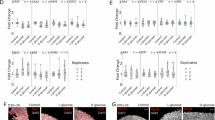
The regulation of GLCCI1 on the ERS-induced apoptosis pathway (initiated by GRP78) depends on the intermediary of HSP90AB1
GLCCI1 is upstream of GRP78 has been proved by previous experiments. To further investigate the regulatory relationship among GLCCI1, HSP90AB1and GRP78, we conducted the rescue experiments and assessed the changes of RGCs vitality. The qPCR and Western blot results indicated that, compared with the vector (+/-) group, the expression levels of GLCCI1 mRNA and protein were significantly reduced (p < 0.001) (Fig. 8A-C). This knockdown led to a downregulation of HSP90AB1 (mRNA: 0.300 ± 0.0035 vs. 0.629 ± 0.0089, p < 0.001; Protein: 0.327 ± 0.0003 vs. 0.516 ± 0.0001, p < 0.001) (Fig. 8A, B and D), upregulation of GRP78 (0.971 ± 0.001 vs. 0.629 ± 0.0003, p < 0.001) (Fig. 8B and E) and decrease of RGC vitality (0.559 ± 0.0064 vs. 1.000 ± 0.0002, p < 0.001) (Fig. 8F). Conversely, as a rescue, concurrent overexpression of HSP90AB1 an knockdown of GLCCI1 (GLCCI1(-) + HSP90AB1(+) vs. GLCCI1(-)) (p < 0.001) reduced the expression of GRP78 (0.736 ± 0.0003 vs. 0.971 ± 0.001, p < 0.001) (Fig. 8B and E) and enhanced the vitality of RGCs (1.112 ± 0.0008 vs. 0.559 ± 0.0064, p < 0.001) (Fig. 8F). This means HSP90AB1 overexpression reverted the effect of GLCCI1 knockdown.
Relationship between GLCCI1 and HSP90AB1. GLCCI1(-): GLCCI1 knockdown group; HSP90AB1(+): HSP90AB1 overexpression group; GLCCI1(-) + HSP90AB1(+): GLCCI1 knockdown and HSP90AB1 overexpression group. GLCCI1(+): GLCCI1 overexpression group; HSP90AB1(-): HSP90AB1 knockdown group; GLCCI1(+) + HSP90AB1(-): GLCCI1 overexpression and HSP90AB1 knockdown group. M: marker. (A) The relative mRNA expression level of GLCCI1 and HSP90AB1. The mRNA expression of GLCCI1(-) vs. Vector (+/-): 0.267 ± 0.0032 vs. 0.761 ± 0.0048, t = 85.24, p < 0.001; The mRNA expression of HSP90AB1(+) vs. Vector (+/-): 1.011 ± 0.0029 vs. 0.629 ± 0.0089, t = 12.54, p < 0.001. (B-E) WB and bar graph results of GLCCI1 knockdown and HSP90AB1 overexpression. GLCCI1 was successfully knocked down (GLCCI1(-) vs. Vector (+/-): 0.210 ± 0.0002 vs. 0.856 ± 0.0005, t = 1259.44, p < 0.001); GLCCI1 knockdown led to HSP90AB1 downregulation (0.327 ± 0.0003 vs. 0.516 ± 0.0001, t = 577.67, p < 0.001) and GRP78 upregulation (GLCCI1(-) vs. Vector (+/-)):0.971 ± 0.0001 vs. 0.629 ± 0.0003, t = 577.67, p < 0.001). HSP90AB1 overexpression cannot change the expression of GLCCI1 (HSP90AB1(+) + GLCCI1(-) vs. GLCCI1(-): 0.209 ± 0.0001 vs. 0.210 ± 0.0002, t = 2.152, p = 0.098, p > 0.05), but can reverse the effect of GLCCI1 knockdown and led to the downregulation of GRP78(HSP90AB1(+) + GLCCI1(-) vs. GLCCI1(-):0.736 ± 0.003 vs. 0.971 ± 0.0001, t = 745.27, p < 0.001). (F) Cell vitality of RGCs. The reduction of cell vitality caused by GLCCI1 knockdown was reversed by the overexpression of HSP90AB1 (GLCCI1(-) vs. GLCCI1(-) + HSP90AB1(+): 0.559 ± 0.0064 vs. 1.112 ± 0.0008, t = 86.08, p < 0.001). (G) The relative mRNA expression level of GLCCI1 and HSP90AB1. GLCCI1 mRNA overexpressed: GLCCI1(+) vs. Vector (+/-): 1.019 ± 0.0139 vs. 0.656 ± 0.0057, t = 24.12, p < 0.001; The mRNA expression of HSP90AB1 knocked down: HSP90AB1 (-) vs. Vector (+/-): 0.420 ± 0.0006 vs. 0.772 ± 0.0002, t = 541.94, p < 0.001. (H-K) WB and bar graph results of GLCCI1 overexpression and HSP90AB1 knockdown. GLCCI1 was overexpressed compared with Vector group (vs. Vector:1.133 ± 0.0002 vs. 0.566 ± 0.0001, t = 2558.50, p < 0.001). GLCCI1 overexpression resulted in the upregulation of HSP90AB1(vs. Vector: 0.916 ± 0.0002 vs. 0.772 ± 0.0002, t = 591.42, p < 0.001) and the inhibition of GRP78 (vs. Vector: 0.621 ± 0.0001 vs. 0.980 ± 0.0003, t = 1212.92, p < 0.001). Simultaneous knockdown of HSP90AB1 and overexpression of GLCCI1 (GLCCI1(+) + HSP90AB1(-) vs. GLCCI1(+)) reverted the effect of GLCCI1 overexpression and led to the upregulation of GRP78 (vs. GLCCI1(+): 0.852 ± 0.0008 vs. 0.621 ± 0.0001, t = 279.92, p < 0.001). (L) Cell vitality of RGCs. The increasement of cell vitality induced by GLCCI1 overexpression was reversed by the knockdown of HSP90AB1 (GLCCI1(+) + HSP90AB1(-) vs. GLCCI1(+): 1.163 ± 0.0049 vs. 1.554 ± 0.0109, t = 32.60, p < 0.001). * P < 0.05; ** P < 0.01, ns: not significant.
Furthermore, GLCCI1 overexpression resulted in the upregulation of HSP90AB1(vs. Vector: mRNA: 0.963 ± 0.0013 vs. 0.756 ± 0.0032, p < 0.001; protein: 0.916 ± 0.0002 vs. 0.772 vs. 0.0002, p < 0.001) (Fig. 8G, H and J) and the inhibition of GRP78 (protein: 0.621 ± 0.0001 vs. 0.980 ± 0.0003, p < 0.001) (Fig. 8H and K) and elevation of RGCs vitality (1.554 ± 0.0109 vs. 1.000 ± 0.0044, p < 0.01) (Fig. 8L); Conversely, simultaneous knockdown of HSP90AB1 and overexpression of GLCCI1 (GLCCI1(+) + HSP90AB1(-) vs. GLCCI1(+)) reverted the effect of GLCCI1 overexpression and led to the upregulation of GRP78 (0.852 ± 0.0008 vs. 0.621 ± 0.0001, p < 0.001) (Fig. 8H and K) and the inhibition of cell vitality (1.163 ± 0.0049 vs. 1.554 ± 0.0109, p < 0.001) (Fig. 8L).
These data indicated that the expression level of GLCCI1 affects the expression level of HSP90AB1, while changes in HSP90AB1 expression cannot influence the expression level of GLCCI1. However, HSP90AB1 can reverse the effects of altered GLCCI1 expression, specifically by restoring GRP78 expression levels and cell vitality. This demonstrated that GLCCI may function as an upstream regulator of HSP90AB1, which in turn acts as an upstream regulator of GRP78, thereby influencing cellular survival.
Materials and methods
Animals
C57BL/6J male mice (aged 6 weeks; weighting between 19 and 22 g) were obtained from BEIJING HFK BIOSCIENCE Co., Ltd. In addition to providing adequate food and water, the mice were kept in a controlled environment that included a temperature of 22 ± 2 °C, a relative humidity of 60–65%, and a 12-hour light/dark cycle. During the process of developing the diabetes model, the mice underwent an initial adaptive feeding period lasting one week. Subsequently, the mice were subjected to a 12-hour period of fasting and water deprivation, after which a 150 mg/kg dose of STZ was administered. STZ was dissolved in a solution of 0.1 mM citric acid sodium citrate buffer (pH 4.5) prior to injection. If the concentration of glycemia was ≥ 16.7 mM one week after the administration of STZ, the model was considered successful and intravitreal injection was administered.
Before optical coherence tomography (OCT), intravitreal injection, or decapitation, the anesthetic (1% pentobarbital sodium dissolved in double distilled water, 80 mg/kg) was intraperitoneally injected into the mice, and the depth of anesthesia was evaluated through the pinch-paw reflex. After the mice were anesthetized, tropicamide eye drops were administered to dilate pupae, and gatifloxacin gel was subsequently applied to safeguard the cornea. The retinal structure was examined using OCT, after which the intravitreal injection was performed under a microscope using a Hamilton microliter syringe with a 32-gauge needle (Hamilton) under a microscope. Mice in the normal control group (sham) and DM groups were injected with 2 µL of PBS, and mice in the empty adenovirus groups (vehicle (-): pAAV-U6-shRNA(NC)-CMV-MCS-WPRE, 5.98E + 12v.g./ml, or vehicle (+): pcAAV-CMV-MCS-WPRE,1.35E + 13v.g./ml), or in the Glcci1-shRNA-AAV2/9 group (KD: pAAV-U6-shRNA(Glcci1)-CMV-MCS-WPRE, 5.38E + 12v.g./ml ) and Glcci1-AAV2/9 group (OE: pcAAV-CMV-Glcci1-HA-WPRE, 1.24E + 13v.g./ml) were all injected with 2 µL of related adenoviruses. At 12 weeks after injection, the mice were subjected to OCT again. After the measurements, mice were euthanized via a lethal intraperitoneal injection of pentobarbital (250 mg/kg), then perfused, and fixed with 4% paraformaldehyde (if utilized for immunohistochemistry), after which the eyes were enucleated.
Immunohistochemistry (IHC) for retinal GLCCI1
After being washed with phosphate-buffered saline (PBS) 3 times (5 min/each), the sections were handled with 3% H2O2 for 10 min in the dark to abolish the activity of endogenous peroxidase. Next, the sections were blocked with normal goat serum for 10 min, incubated with rabbit primary antibodies overnight at 4 °C (normal rabbit serum was used as the negative control), and incubated with biotin labeled secondary antibody at 37 °C for 2 h. Antibodies including rabbit anti-rat GLCCI1(1:200; Thermo Fisher, MA, USA), were used. To amplify the positive signal, horseradish peroxidase-conjugated streptavidin (HRP-SA) was added to the sections, which were subsequently stained with fresh diaminobenzidine (DAB) and observed under a microscope. After being washed with double distilled water, nuclear counterstaining was performed with hematoxylin. Finally, the sections were dehydrated with an ascending gradient of ethanol, cleared in xylene, sealed with neutral gum and observed under a microscope. The IHC score was quantified with ImageJ software (Maryland, USA).
Enucleated eyeballs of the mice were immediately fixed with formaldehyde-75% alcohol-acetic acid-normal saline (FAS) for longer than 24 h. Next, the tissues were dried in increasing concentrations of ethanol, clarified in xylene (twice), and finally encased in paraffin. Paraffin Sect. (4 μm) were prepared, dewaxed in xylene and hydrated in a descending gradient of ethanol. Afterward, the antigens were extracted from the sections using 200 ml of citrate buffer (composed of 36 ml of 0.1 mM citric acid and 164 ml of 0.1 mM trisodium citrate) through a high-pressure and high-temperature process for 2 min, followed by natural cooling to room temperature (RT). The sections were incubated with 3% H2O2 for 12 min in the absence of light to inhibit the activity of endogenous peroxidase after being rinsed with PBS four times (5 min/wash). Afterward, the sections were blocked using regular goat serum for 10 min, subsequently incubated overnight (≥ 16 h) at 4 °C with primary antibodies (normal goat serum was used as the negative control). Afterward, the sections were incubated with a biotin-labeled secondary antibody at 37 °C for 2 h.
The antibodies used included rabbit anti-rat GLCCI1 (1:200; Thermo Fisher, MA, USA). To amplify positive signals, horseradish peroxidase-conjugated streptavidin (HRP-SA) was added to the sections, which were subsequently stained with fresh diaminobenzidine (DAB) and observed under a microscope. After washing with double distilled water, nuclear counterstaining was performed with hematoxylin. Increasing concentrations of ethanol were used to dry the sections, which were then cleared with xylene and sealed with neutral gum, and observed under a microscope observing. The IHC score was quantified with ImageJ software (Maryland, USA).
Real-time quantitative PCR (qPCR)
An RNA extraction reagent (TRIzol, Invitrogen, USA) was used to extract RNA from cells and tissues, and 250 ng of RNA was used for cDNA synthesis (Thermo Scientific, Thermo Fisher Scientific, Inc.). Applied Biosystems’ SYBR Green PCR Master Mix was used for qPCR, with an initial denaturation step of 95 °C for 150 s followed by 40 cycles of 95 °C for 30 s and extension at 60 °C for 30 s. The qPCR procedure involved melting curve analysis, during which the qPCR product was gradually heated from 55 to 95 °C. The primers (5′-3′) utilized in this analysis were as follows: β-actin (forward: AGCAAGCAGGAGTACGATGA, reverse: GGTGTAAAACGCAGCTCAGTA); Glcci1 (forward: ACTTAGGCAACAGCTACAACG, reverse: AGGGTACACGGGAAACAGAT); Hsp90ab1 (forward: AAGTGCACCATGGAGAGGAA, reverse: ACTTGGAAGGGTCAGTCAGG). The expression of each gene of interest was standardized against that of β-actin, and the levels of gene expression (fold change) were reported as 2−ΔΔCt values.
Western blotting
The collection and lysis of RGCs and tissues were performed using radioimmunoprecipitation assay (RIPA, enhanced type, Beyotime) buffer, which included the protease inhibitor PMSF (Solarbio, Beijing, China). We separated the protein samples using SDS-PAGE gels and then transferred them onto polyvinylidene fluoride (PVDF) membranes manufactured by MILLIPORE (Bedford, MA, USA). Afterward, the membrane was subjected to blocking with 1% BSA (Bioss, China) for 2 h. Subsequently, the membranes were incubated at 4 °C (≥ 16 h) with a primary antibody, followed by 2 h of incubation with a secondary antibody (1:3000; Bioss, China). The visualization of the bands was achieved using an enhanced chemiluminescence (ECL) kit (Boster, Wuhan, China) and a Bio-Rad ChemiDoc XRS + system manufactured by Bio-Rad (1708265). The primary antibodies used in this study including: those manufactured by Thermo Fisher (MA, USA), were anti-ACTB (1:3000) and anti-GLCCI1 (1:2000); the antibodies manufactured by Proteintech (USA), were anti-GRP78 (1:2000), and anti-CHOP (1:1000). Other antibodies used included anti-HSP90AB1 (1:2000; Zenbio, Chengdu, China), and anti-cleaved CASP3 (1:1500; Affinity, USA). The blots were quantified with ImageJ software (Maryland, USA).
RGCs culture in vitro
Immortalized mouse-retinal ganglion cells (RGCs) obtained from Qingqi Biotechnology Development Co., Ltd. (Shanghai, China). RGCs were cultured in a 37 ℃ and 5% CO2 humidified incubator using DMEM/F12 medium from HyClone in the USA supplemented with 9% fetal bovine serum and 1% penicillin and streptomycin from HyClone. The RGCs (approximately 90% confluent) were treated with 0.25% trypsin, centrifuged at 1000 revolutions per minute (rpm) for 5 min, and subsequently passaged every 3 to 4 days. In the present study, the low-glucose medium used contained 5.5 mM D-glucose (standard glucose medium), while the ERS induced high glucose medium used was DMEM/F12 medium supplemented with 50mM D-glucose; These medium are consistent with our previous RGCs high glucose model and reports24.
Plasmid synthesis and transfection
Shanghai Justscience Company successfully created and produced RNA oligos. The Oligo sequence of the knockdown group Vector group is 5’- UCUCCGAACGUGUCACGUTT-3’, and the sequence of si-Glcci1 is 5’- GAAAGAGUGCCAACUUTT-3’; The sequence of si-Hsp90ab1 is 5’- GGAAGAGAAGGUGAAA-3’. Diethyl pyrocarbonate (DEPC) water (Beyotime, China) was used to dissolve the RNA oligos to 20µM. To transfect the plasmids into RGCs, Lipofectamine 2000 was used (Invitrogen, USA), and the cells were incubated for 8 h. Then, it was replaced with complete culture without antibiotics. After 48 h, the expression levels of GLCCI1mRNA and protein were detected using qPCR and WB.
Cell vitality detection
Vitality tests were conducted using a Cell Counting Kit-8 (CCK-8) (Beyotime, China). A total of 5 × 103 RGCs were incubated for 24 h in 96-well plates and subsequently subjected to different treatments for 48 h. Next, 10 µl of CCK-8 solution was added to the cells, which were incubated for 2 h, after which the absorbance at 450 nm was measured to determine the vitality of the RGCs.
Flow cytometry for apoptosis
Flow cytometry was used to evaluate apoptosis using Annexin V-FITC/PE or Annexin V-PE/7AAD kit (Elabscience, Wuhan, China). Using trypsin, the cells were harvested and washed in PBS before being stained with 5 µl of Annexin V-PE or Annexin V-FITC, in addition to 5 µl of either 7AAD or PE, for 15 min. Subsequently, RGCs were examined using flow cytometry (ACEA NovoCyteTM). The procedures were conducted in strict accordance with the manufacturer’s instructions, and the FlowJo software package (TreeStar, Ashland, OR) was used to analyze the results.
Transmission electron microscopy (TEM)
The cellular ultrastructure (endoplasmic reticulum and mitochondrial morphology) was observed via TEM (HT7700, Hitachi, Japan). RGCs were fixed in 2.5% glutaraldehyde for two hours at RT and then incubated overnight at 4 °C. Next, the cells were subjected to four 15-minute rinses with PBS. The cells were subsequently postfixed with 1% osmium tetroxide (OsO4) for 2 h, followed by two 5-minute washes with PBS. RGCs were then stained with 2% uranyl acetate for 2 h, dehydrated using acetone, and finally embedded in epoxy resin. TEM was conducted by Kang Chen Biotech, Inc. (Beijing, China).
Optical coherence tomography (OCT)
The mice retinas were observed via OCT (Leica Microsystems, Germany). Testing was conducted at specified time points based on experimental requirements. The mice were thoroughly anesthetized with 1% pentobarbital sodium, administered at a dosage of 80 mg/kg, until there was no response to tail or toe pinching. Mydriatic eye drops were used for pupil dilation, and gatifloxacin gel was applied to moisten and protect the cornea. After dilation was fully achieved, the mice were stabilized for imaging. The positioning and focus were adjusted on the experimental platform prior to conducting the scans. The testing procedures were carried out by experienced professionals from the Liaoning Provincial Key Laboratory for the Prevention and Treatment of Age-Related Macular Degeneration at Jinzhou Medical University.
Fluorescein fundus angiography (FFA)
The mice fundi were observed via fundus camera equipped with fluorescein image acquisition system (Leica Microsystems, Germany). The pupils of mice were dilated after anesthesia as OCT, and 1% fluorescein sodium (Zeye Biotechnology Co., Ltd, Shanghai, China) was injected intraperitoneally. 1 min later, the testing procedures were also carried out by experienced professionals as mentioned in OCT.
Screening of stable cell lines and coimmunoprecipitation/mass spectrometry (co-IP/MS)
A recombinant plasmid (Flag-Glcci1 or Hsp90ab1-puromycin resistant) was constructed and introduced into RGCs. After transfection, puromycin (2 µg/mL) or neomycin (50 µg/mL) was added to screen the transformants. During a span of fourteen days, stable cell lines were screened and subsequently maintained with either 1 µg/mL puromycin or 25 µg/mL neomycin. According to the manufacturer’s instructions, we used the ‘3×FlagTag Protein IP Assay Kit with Magnetic Beads’ (Beyotime, China) to perform protein concentration. A mixture of protease inhibitors and a phosphatase inhibitor (Thermo Fisher Scientific) was added to IP lysis buffer to prepare cell lysates, which were subsequently incubated for 30 min on ice. Afterward, using a centrifuge at 14,000 rpm and 4 °C, the lysate was centrifuged for 15 min. The concentration of proteins in the supernatant was measured and standardized for all samples. The Flag-GLCCI1or Flag-HSP90AB1 protein was isolated using Flag-Trap magnetic beads (Beyotime, China). Blots of Flag-GLCCI1 were probed with a rabbit anti-GLCCI1 antibody (1:2000; Thermo Fisher, MA, USA) and analyzed via mass spectrometry (MS) at Beijing Qinglian Baiao Biotechnology Co., Ltd. (Beijing, China).
Recombinant protein purification and pull-down assay
Justscience Company (Shanghai, China) synthesized the recombinant plasmids containing GST-Glcci1 and 6His-Hsp90ab1, which were subsequently introduced into the Escherichia coli BL21 strain to express the recombinant proteins. The purification of GST-pulldown and His-pulldown products was conducted using a GST-tag or His-tag protein purification kit, which was obtained from Beyotime (China). Briefly, GST-Glcci1 and 6His-Hsp90ab1 were produced in Escherichia coli BL21 (DE3) cells at 22 °C overnight and then purified using BeyoGold™ GST-tag or His-tag Purification Resin. Pulldown experiments were performed with purified GST-GLCCI1 and 6His-HSP90AB1 proteins. Following 1 h of interaction at a low temperature with GST-GLCCI1 and 6His-Hsp90Ab1, the mixture was divided into two groups, to which 50 µl of BeyoGold™ GST-tag or His-tag Purification Resin was added; the mixture was allowed to interact overnight on a shaker at 4 °C. For GST-GLCCI1 pulldown, after washing with chilled lysis buffer 5 times, the GST-tagged protein was eluted 7 times using 0.5 ml of elution buffer (containing glutathione). Next, the pulldown samples were subjected to SDS-PAGE and subsequently analyzed by WB. After performing the 6His-HSP90AB1 pulldown, the column was washed 5 times with 0.5 ml of nondenatured detergent. Subsequently, the column was washed 6 times with 0.5 ml of nondenatured eluent. Additionally, the pulldown samples were subjected to SDS‒PAGE and immunoblotting.
Protein‒protein docking
The AlphaFold Protein Structure Database was used to obtain the 3D configurations of the proteins. The evaluation of protein‒protein interactions and interaction sites were conducted using HDOCK and HADDOCK25,26. The outcomes were additionally evaluated via PDBePISA, and ultimately, the sites of interaction were displayed using PyMOL.
Dual immunofluorescence staining for GLCCI1/HSP90AB1 and HSP90AB1/GRP78
A total of 2 mL of RGCs suspension was added to a 12-well plate (2.0 × 105 cells/mL), and aseptic slides were placed on the plates. After being cultured, the RGCs were washed three times in PBS, with each round lasting 5 minutes. After incubating for 20 min at RT, the RGCs were treated with 4% paraformaldehyde; subsequently, a total of three times, the RGCs were washed in PBS for 5 minutes each. Subsequently, the cells were rendered permeable for a duration of 30 min using 0.5% Triton-X100, followed by three additional washes with PBS lasting 5 minutes each. Following a 1-hour blockade with 5% BSA, the cells were subjected to an overnight incubation (> 14 h) at 4 °C, after which the primary antibodies were added to a humidified chamber (rabbit anti-GLCCI1 (1:100, Thermo Fisher, MA, USA), mouse anti-HSP90AB1 (1:100, Zenbio, Chengdu, China), and rabbit anti-GRP78 (1:100, Proteintech, USA)). After incubation with the secondary antibody against HSP90AB1, the sections were then incubated at RT with a concentration of 1:500 of Alexa Fluor 594 secondary antibody (Thermo Fisher). For immunofluorescence analysis, the sections were incubated with biotinylated secondary antibodies against GLCCI1 and GRP78 for 2 h at RT, followed by incubation with the avidin-biotin complex for 0.5 h at RT. Subsequently, the cells were subjected to three washes with PBS, mounted and stained with DAPI using Fluoroshield mounting medium. A Leica SP5 II laser scanning confocal microscope was used to obtain confocal images, which were utilized to demonstrate the colocalization of GLCCI1-HSP90AB1 and HSP90AB1-GRP78.
Statistical analysis
SPSS 26.0 statistical software was used for analyzing all the data. Samples were subjected to independent sample t-test. Multiple group comparisons were conducted using one-way ANOVA. The results are presented as means ± SEMs. For all tests, P < 0.05 was considered to indicate statistical significance. The abbreviation ‘ns’ denotes no statistical significance.
Discussion
ERS has been reported to be closely related to DM and DR27,28,29, however, whether ERS-induced apoptosis is associated with DR remains obscure. Our results demonstrated that GLCCI1 is a vital protective factor that helps to inhibit ERS-induced RGC apoptosis. GLCCI1 knockdown resulted in RGC apoptosis or retinal impairment similar to that observed in DR mice; however, this damage was alleviated by the ERS inhibitor 4-PBA or GLCCI1 overexpression, indicating that GLCCI1 may function by inhibiting ERS-induced apoptosis. Mechanistically, these effects occurred mainly through the GLCCI1-HSP90AB1-GRP78 pathway.
GLCCI1 reportedly impedes cell death and promotes cell proliferation31,31. However, the specific involvement of GLCCI1 in the retina of DR mice has not been documented, particularly in relation to ERS-induced apoptosis, and the underlying mechanism involved remains unexplored. To assess the function of GLCCI1 in ERS-induced apoptosis, we examined ERS-induced apoptosis related markers (GRP78, CHOP and cleaved Caspase3) expression level32,33,34,35. The IHC, qPCR and WB results in this study demonstrated that GLCCI1 was primarily localized in the GCL and exhibited decreased expression in the retinas of DR mice. Additionally, the expression of markers of ERS-induced apoptosis was found to be increased in the retinas of DR mice (Fig. 1). These findings suggest a potential correlation between GLCCI1 and ERS-induced apoptosis as well as between GLCCI1 and DR. Therefore, we postulated that GLCCI1 may protect RGCs from ERS-induced apoptosis.
The unfolded protein response (UPR) encompasses three distinct pathways that are regulated by the sensors PERK, ATF6, and IRE1. These sensors are activated when the upstream modulator and chaperone GRP78 dissociate in response to harmful stimuli. As a result, PERK, ATF6, and IRE1 activate the common downstream transcriptional target CHOP36,37,38,39. As a consequence of ERS, the UPR is initiated, leading to the activation of three UPR pathways40,41. The findings revealed that the overexpression of GLCCI1 mitigated ERS-induced apoptosis in RGCs exposed to high glucose conditions (Fig. 2). In turn, the knockdown of GLCCI1 resulted in the upregulation of apoptosis markers induced by ERS (GRP78, CHOP and cleaved Caspase3), an effect that was reversed by the ERS inhibitor 4-PBA (Fig. 3). These data indicate that GLCCI1 indeed alleviates ERS-induced apoptosis and may exert its effect by inhibiting GRP78 expression and associated pathway.
In vivo, AAV2/9-shGlcci1 and AAV2/9-Glcci1 were administered to knock down and overexpress GLCCI1, respectively, in the retinas of C57BL/6J normal and DR mice. As observed in the DR group, the retinal alterations observed in the AAV2/9-shGlcci1 group were similar. Conversely, the overexpression of GLCCI1 mitigated retinal damage in DR mice, as depicted in Fig. 4. To investigate the subcellular effects of GLCCI1 knockdown and overexpression, TEM was used to evaluate the morphology of the ER in RGCs. The TEM results (Fig. 5) provided additional confirmation that GLCCI1 overexpression alleviated the excessive glucose-induced enlargement of the ER and mitochondria, consistent with the in vivo and in vitro outcomes.
To further elucidate the molecular mechanism of GLCCI1, we performed coimmunoprecipitation/mass spectrometry (co-IP/MS) and protein‒protein docking, and identified the protein of interest: HSP90AB1. As a molecular chaperone, HSP90AB1 can bind to a variety of proteins, including ubiquitin ligases, kinases, and transcription factors. It also inhibits ERS, helps protein folding, misfolded protein degradation and maintains protein stability under various stress conditions42,43,44,45. According to previous reports, HSP90AB1 is linked to the growth and spread of certain cancer cells, and increased HSP90AB1 expression has also been documented in the management of neurodegenerative disorders such as Huntington’s disease 46–50.
The direct interactions between GLCCI1 and HSP90AB1 as well as HSP90AB1 and GRP78 were verified by protein‒protein docking, co-IP (and pull-down assays for GLCCI1-HSP90AB1) and double-label immunofluorescence in RGCs (Figs. 6, 7 and 8, supplementary video 1–2). These findings suggested that HSP90AB1 potentially acts as an intermediary in the interaction between GLCCI1 and GRP78, and hinders ERS and ERS-induced apoptosis.
To determine the regulatory relationship between GLCCI1 and HSP90AB1, we carried out rescue experiments (Fig. 8). And the results showed that the expression level variation of GLCCI1 led to the variation of HSP90AB1, while the expression level of HSP90AB1 could not influence the expression level of GLCCI1, but it can reverse the effects of GLCCI1, including the impact on the expression of GRP78 and cell vitality. Therefore, we confirmed GLCCI1 is the upstream of HSP90AB1, which in turn is the upstream of GRP78. The relationship between HSP90AB1 and ERS has been previously reported3. Our findings further elucidated the underlying mechanisms of this interaction.
In summary, GLCCI1 can regulate the expression of HSP90AB1 positively; while HSP90AB1 can regulate the expression of GRP78 negatively; The regulation of GRP78 by GLCCI1 depends on the expression level of HSP90AB1. And ultimately, the impact on ERS-induced apoptosis was achieved through GLCCI1-HSP90AB1-GRP78 pathway (Fig. 9). The interaction of HSP90AB1-GRP78 inhibits the binding of GRP78 to unfolded/misfolded proteins, thereby inhibiting the occurrence of ERS; Whether the GLCCI-HSP90AB1-GRP78 complex exists, as well as its function, is currently unclear and requires further research. At the same time, there are some other limitations in this study: Firstly, we only studied the function and mechanism of GLCCI1 in the model, and the mechanism of GLCCI1 in the human body still need to be investigated; Secondly, we mainly conduct research in RGCs, and further research is needed to determine whether this pathway is also involved in other retinal cells and interactions.
Partial schematic diagram of the mechanism by which GLCCI1 inhibits ERS-induced apoptosis in RGCs. RER: rough endoplasmic reticulum; HG: high glucose; DR: diabetic retinopathy; ERS: endoplasmic reticulum stress. UPR sensors: PERK, ATF6 and IRE1. (1) GLCCI1 overexpression leads to the upregulation of HSP90AB1; (2) HSP90AB1 overexpression suppresses the expression of GRP78; (3) HSP90AB1 overexpression promotes protein folding, thus alleviating ERS caused by the accumulation of misfolded proteins; (4) HSP90AB1 promotes the degradation of misfolded proteins; and (5) HSP90AB1 binds to misfolded proteins, thereby inhibiting ERS.
However, GLCCI1 demonstrates crucial importance in maintaining the normal structure of the retina. RGCs are among the first neurons to undergo apoptosis in DR. Overexpression of GLCCI1 can inhibit the apoptosis of RGCs, which means that GLCCI1 provides significant protective effects against the early neurodegenerative symptoms of DR. By researching and utilizing GLCCI1, we can detect early neurodegenerative signs of DR, implement appropriate protective measures, and effectively reduce both the incidence of the disease and the rate of blindness associated with it. Additionally, the unique characteristics of GLCCI1 suggest promising applications in the field of neurodegenerative diseases in the future.
Conclusion
Based on the results of this study, we can draw the following conclusions:
-
(1)
The downregulation of GLCCI1 is related to DR induced by ERS-induced apoptosis. Overexpression of GLCCI1 inhibits the apoptosis of RGCs, improves the structure of various layers and vessels of DR retina, and alleviates the swelling of endoplasmic reticulum and mitochondria under stress;
-
(2)
GLCCI1 regulates the GRP78-mediated ERS-induced apoptosis pathway by regulating the expression of HSP90AB1;
-
(3)
GLCCI1 interacts with HSP90AB1, HSP90AB1 interacts with GRP78, and the direct interaction of HSP90AB1 to GRP78 inhibits the ERS-induced apoptosis pathway.
Data availability
Data is provided within the manuscript or supplementary information files.
Change history
16 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41598-024-83001-6
Abbreviations
- DR:
-
Diabetic retinopathy
- DM:
-
Diabetes mellitus
- ERS:
-
Endoplasmic reticulum stress
- GLCCI1:
-
Glucocorticoid induced transcript 1
- IDF:
-
International Diabetes Federation
- PBS:
-
Phosphate-buffered saline
- STZ:
-
Streptozotocin
- OCT:
-
Optical coherence tomography
- TEM:
-
Transmission electron microscopy
- GCL:
-
Ganglion cell layer
- IPL:
-
Inner plexiform layer
- INL:
-
Inner nuclear layer
- ONL:
-
Outer nuclear layer
References
Saeedi, P. et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes. Res. Clin. Pract. 157, 107843 (2019).
Teo, Z. L. et al. Global prevalence of diabetic retinopathy and projection of burden through 2045: Systematic review and meta-analysis. Ophthalmology 128, 1580–1591 (2021).
Kang, Q. & Yang, C. Oxidative stress and diabetic retinopathy: Molecular mechanisms, pathogenetic role and therapeutic implications. Redox Biol. 37, 101799 (2020).
Sun, L. et al. Single cell RNA sequencing (ScRNA-Seq) deciphering pathological alterations in streptozotocin-induced diabetic retinas. Exp. Eye Res. 210, 108718 (2021).
Chang, W., Lajko, M. & Fawzi, A. A. Endothelin-1 is associated with fibrosis in proliferative diabetic retinopathy membranes. PLoS One 13, e0191285 (2018).
Antonetti, D. A. The neuroscience of diabetic retinopathy. Vis. Neurosci. 38, E001 (2021).
Wang, W. & Lo, A. Diabetic retinopathy: Pathophysiology and treatments. Int. J. Mol. Sci. 19 (2018).
Catalani, E. & Cervia, D. Diabetic retinopathy: A matter of retinal ganglion cell homeostasis. Neural Regen. Res. 15, 1253–1254 (2020).
Ola, M. S., Alhomida, A. S. & LaNoue, K. F. Gabapentin attenuates oxidative stress and apoptosis in the diabetic rat retina. Neurotox. Res. 36, 81–90 (2019).
Tian, J. et al. Ginkgo biloba leaf extract attenuates atherosclerosis in streptozotocin-induced diabetic ApoE-/- mice by inhibiting endoplasmic reticulum stress via restoration of autophagy through the MTOR signaling pathway. Oxid. Med. Cell. Longev. 2019, 8134678 (2019).
Kong, D. Q., Li, L., Liu, Y. & Zheng, G. Y. Association between endoplasmic reticulum stress and risk factors of diabetic retinopathy. Int. J. Ophthalmol. 11, 1704–1710 (2018).
McLaughlin, T. et al. Cellular stress signaling and the unfolded protein response in retinal degeneration: Mechanisms and therapeutic implications. Mol. Neurodegener. 17, 25 (2022).
Sanchez-Chavez, G., Hernandez-Ramirez, E., Osorio-Paz, I., Hernandez-Espinosa, C. & Salceda, R. Potential role of endoplasmic reticulum stress in pathogenesis of diabetic retinopathy. Neurochem. Res. 41, 1098–1106 (2016).
Yang, L. et al. Role of endoplasmic reticulum stress in the loss of retinal ganglion cells in diabetic retinopathy. Neural Regen. Res. 8, 3148–3158 (2013).
Nandakumar, S., Rozich, E. & Buttitta, L. Cell cycle re-entry in the nervous system: From polyploidy to neurodegeneration. Front. Cell. Dev. Biol. 9, 698661 (2021).
Sohn, E., Lim, H. S., Kim, Y. J., Kim, B. Y. & Jeong, S. J. Annona atemoya leaf extract improves scopolamine-induced memory impairment by preventing hippocampal cholinergic dysfunction and neuronal cell death. Int. J. Mol. Sci. 20 (2019).
Zheng, J., Min, S., Hu, B., Liu, Q. & Wan, Y. Nrdp1 is involved in hippocampus apoptosis in cardiopulmonary bypass-induced cognitive dysfunction via the regulation of ErbB3 protein levels. Int. J. Mol. Med. 43, 1747–1757 (2019).
Arroba, A. I. et al. Somatostatin protects photoreceptor cells against high glucose-induced apoptosis. Mol. Vis. 22, 1522–1531 (2016).
Simo, R., Stitt, A. W. & Gardner, T. W. Neurodegeneration in diabetic retinopathy: Does it really matter?. Diabetologia 61, 1902–1912 (2018).
Miazek, A. & Malissen, B. Two genes, three messengers: Hybrid transcript between a gene expressed at specific stages of T-cell and sperm maturation and an unrelated adjacent gene. Immunogenetics 54, 681–692 (2003).
Kiuchi, Z. et al. GLCCI1 is a novel protector against glucocorticoid-induced apoptosis in T cells. Faseb. J. 33, 7387–7402 (2019).
Ohoka, N. et al. SNIPER(TACC3) induces cytoplasmic vacuolization and sensitizes cancer cells to bortezomib. Cancer Sci. 108, 1032–1041 (2017).
Li, X., Xu, C. & Yang, P. C-Jun NH2-terminal kinase 1/2 and endoplasmic reticulum stress as interdependent and reciprocal causation in diabetic embryopathy. Diabetes 62, 599–608 (2013).
Wang, Q. et al. An in vitro model of diabetic retinal vascular endothelial dysfunction and neuroretinal degeneration. J. Diabetes Res. 2021, 9765119 (2021).
Honorato, R. V. et al. Structural biology in the clouds: The WeNMR-EOSC ecosystem. Front. Mol. Biosci. 8, 729513 (2021).
van Zundert, G. et al. The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016).
Cao, Z. H. et al. Endoplasmic Reticulum Stress and Destruction of Pancreatic Beta Cells in Type 1 Diabetes. Chin. Med. J. (Engl). 133, 68–73 (2020).
Elmasry, K. et al. Role of Endoplasmic Reticulum Stress in 12/15-Lipoxygenase-Induced Retinal Microvascular Dysfunction in a Mouse Model of Diabetic Retinopathy. Diabetologia. 61, 1220–1232 (2018).
De Franco, E. et al. YIPF5 Mutations Cause Neonatal Diabetes and Microcephaly through Endoplasmic Reticulum Stress. J. Clin. Invest. 130, 6338–6353 (2020).
Wu, C. et al. Baicalein attenuates pyroptosis and endoplasmic reticulum stress following spinal cord ischemia-reperfusion injury via autophagy enhancement. Front. Pharmacol. 11, 1076 (2020).
Xiong, Y. et al. Neuroprotective mechanism of L-cysteine after subarachnoid hemorrhage. Neural Regen. Res. 15, 1920–1930 (2020).
Pan, T. J. et al. Antimetastatic effect of fucoidan-sargassum against liver cancer cell invadopodia formation via targeting integrin AlphaVbeta3 and mediating AlphaVbeta3/Src/E2F1 signaling. J. Cancer 10, 4777–4792 (2019).
Xiao, F. et al. Intermedin facilitates hepatocellular carcinoma cell survival and invasion via ERK1/2-EGR1/DDIT3 signaling cascade. Sci. Rep. 11, 488 (2021).
Do, M. et al. PERK activation by SB202190 ameliorates amyloidogenesis via the TFEB-induced autophagy-lysosomal pathway. Aging (Albany NY) 14, 1233–1252 (2022).
Kara, M. et al. Ameliorative effects of the sesquiterpenoid valerenic acid on oxidative stress induced in HepG2 cells after exposure to the fungicide benomyl. Antioxidants 10 (2021).
Wu, Z. H., Chiu, C. H., Chen, C. C., Chyau, C. C. & Cheng, C. H. Amelioration of cyclosporine a-induced acute nephrotoxicity by cordyceps cicadae mycelia via Mg(+2) reabsorption and the inhibition of GRP78-IRE1-CHOP pathway: In vivo and in vitro. Int. J. Mol. Sci. 24 (2023).
Wu, Q., Zhang, H., Nie, H. & Zeng, Z. Anti-nogo-a antibody promotes brain function recovery after cardiopulmonary resuscitation in rats by reducing apoptosis. Mol. Med. Rep. 21, 77–88 (2020).
Kwon, J. H. et al. Hyperthermia disturbs and delays spontaneous differentiation of human embryoid bodies. Biomedicines 8 (2020).
Lin, X. et al. DSCC1 interacts with HSP90AB1 and promotes the progression of lung adenocarcinoma via regulating ER stress. Cancer Cell Int. 23, 208 (2023).
Cheng, H. M. et al. HSP90Β promotes osteoclastogenesis by dual-activation of cholesterol synthesis and NF-ΚB signaling. Cell Death Differ. 30, 673–686 (2023).
Sun, X. et al. Suppression of breast cancer-associated bone loss with osteoblast proteomes via Hsp90Ab1/moesin-mediated inhibition of TGFβ/FN1/CD44 signaling. Theranostics 12, 929–943 (2022).
Haase, M. & Fitze, G. HSP90AB1: Helping the good and the bad. Gene 575, 171–186 (2016).
Wang, D. et al. HSP90AB1 as the druggable target of maggot extract reverses cisplatin resistance in ovarian cancer. Oxidative Med. Cell. Longev. 2023, 9335440 (2023).
Zhang, H. et al. HSP90AB1 promotes the proliferation, migration, and glycolysis of head and neck squamous cell carcinoma. Technol. Cancer Res. Treat. 21, 2081107206 (2022).
Zhang, S. et al. Capecitabine regulates HSP90AB1 expression and induces apoptosis via Akt/SMARCC1/AP-1/ROS axis in T cells. Oxidative Med. Cell. Longev. 2022, 1012509 (2022).
Wang, D. et al. Maggot extract inhibits cell migration and tumor growth by targeting HSP90AB1 in ovarian cancer. J. Clin. Med. 11 (2022).
Mielcarek, M. & Isalan, M. A minimal region of the HSP90AB1 promoter is suitable for ubiquitous expression in different somatic tissues with applicability for gene therapy. Front. Mol. Biosci. 10, 1175407 (2023).
Funding
This work was supported by the Foundation of Education Department of Liaoning Province [LJKMZ20221241], the Natural Science Foundation of Liaoning Province of China [2023-MS-312].
Author information
Authors and Affiliations
Contributions
The study was designed by Xuezheng Liu and Zhongfu Zuo. Jiayou Liu, Hongdan Yu, Shengxue Yu, Mengren Liu, Xinyuan Chen and Wenqiang Liu conducted the experiments. Xuezheng Liu and Zhongfu Zuo provided supervision for the study. Protein-protein docking was performed by Mengren Liu and Xinyuan Chen. Wenqiang Liu interpreted the results. Funding was provided by Xuezheng Liu and Zhongfu Zuo. Figures were designed by Jiayou Liu and Mengren Liu and Xinyuan Chen. The draft was written by Jiayou Liu, Hongdan Yu, Shengxue Yu and Zhongfu Zuo. The manuscript was critically reviewed by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and informed consent
The study is reported in accordance with ARRIVE guidelines. No human subjects were recruited for this study. The animal experiments in this study were approved by the Animal Care Committee of Jinzhou Medical University, and all experiments involving animals were also conducted following the guidelines of the National Institutes of Health (NIH) for the Care and Use of Laboratory Animals.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: In the original version of this Article an affiliation was omitted for Xuezheng Liu. The correct affiliations are listed in the Correction Notice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary Material 2
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, J., Yu, H., Yu, S. et al. GLCCI1 alleviates GRP78-initiated endoplasmic reticulum stress-induced apoptosis of retinal ganglion cells in diabetic retinopathy by upregulating and interacting with HSP90AB1. Sci Rep 14, 26665 (2024). https://doi.org/10.1038/s41598-024-75874-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-75874-4