Abstract
Podophyllotoxin (PTOX), produced by Linum album, is a monolignol that participates in plant defense strategies. Our previous study established that methyl jasmonate (MeJA) significantly stimulates PTOX production in L. album cells. However; the mechanisms by which MeJA regulates PTOX biosynthesis are uncovered. In the present study, we demonstrated that MeJA induces a time-dependent hydrogen peroxide (H2O2) and salicylic acid (SA) accumulation but reduces nitric oxide (NO) generation in L. album cells. PTOX biosynthetic genes such as PAL, CCR, CAD, and PLR were upregulated in response to MeJA exposure. Furthermore, the results of RT-qPCR revealed a positive correlation between the expression of PTOX biosynthetic genes and MeJA-induced upregulation of four miRNAs such as miR156, miR159, miR172, and miR396 at 12 h. Generally, this study revealed that MeJA mediates PTOX biosynthesis in L. album cells by inducing H2O2 and SA formation, which can probably upregulate the expression level of some miRNAs and biosynthetic genes in a redox balance-dependent manner.
Similar content being viewed by others
Introduction
Podophyllotoxin (PTOX) and its derivatives are a subcategory of polyphenolic metabolites and are synthesized by coupling two phenylpropane units1. Several reports have confirmed that Linum album Kotschy ex Boiss. is an important source of lignans2,3. PTOX is a monolignol derived from the phenylpropanoid pathway, serving as a biochemical barrier to intensify innate immunity under adverse conditions4. It has been proved that the biosynthesis of PTOX is a complex process, which can be regulated by multiple factors5.
In the previous study, we found that different concentrations of methyl jasmonate (MeJA) can direct primary metabolites such as carbohydrates and amino acids toward PTOX production by changing oxidant balance in L. album cells6. MeJA, a methylated derivative of jasmonic acid (JA), is a phytohormone in a dynamic regulatory network that can modulate various plant physiological functions7. This phytohormone is sensed via a variety of signatures that can reprogram cellular metabolic and genetic pathways by transferring signaling information8,9.
For instance, reactive oxygen species (ROS), particularly hydrogen peroxide (H2O2), are pivotal mediators that stimulate or amplify plant signal transduction in response to MeJA10,11. Similarly, nitric oxide (NO), a gaseous signaling molecule, transmits information within the plant cells alone or in combination with H2O2, which can also imbalance oxidative status. The imbalanced oxidative status can provoke various defense responses by affecting gene expression, enzyme activities, protein post-translational modification, and metabolite accumulation12,13.
Moreover, MeJA can alter the level of salicylic acid (SA), a phytohormone involved in plant innate immunity, an efficient component of different signaling cascades14. Numerous investigations have revealed that the JA and SA can interact with each other through complex regulatory networks in the form of priming, synergistic effects, and mutual antagonism15. Upon pathogen infection, it has been suggested that SA level can negatively regulate the JA signaling pathway via WRKY transcription factors like WRKY70, the W-box motif, and PLANT DEFENSIN 1.2 (PDF1.2) as the JA marker gene16. It has been reported that the SA molecule is perceived by two groups of receptors, the non-expression of pathogenesis-related gene 1 (NPR1) and NPR3/NPR4, which can trigger the immediate survival responses in interaction with ROS and reactive nitrogen species (RNS)17,18.
However, plants tightly restore cellular oxidative homeostasis through several enzymatic and non-enzymatic ROS scavengers including antioxidant enzymes and secondary metabolites19,20. These antioxidant agents have crucial roles in decreasing oxidative-induced damage to membranes, proteins, and nucleic acids21.
The induction of secondary metabolites production by MeJA-triggered redox imbalance appears to be mediated by the upregulation of specific biosynthetic genes22,23. Previously, we performed a transcriptome analysis for genes involved in PTOX biosynthesis like phenylalanine ammonia-lyase (PAL), cinnamoyl-CoA reductase (CCR), cinnamyl-alcohol dehydrogenase (CAD), and pinoresinol-lariciresinol reductase (PLR), under various elicitors in L. album5,24. PAL gene encodes the first enzyme of monolignols biosynthesis, catalyzing the deamination of phenylalanine to provide trans-cinnamic acid25. Also, a recent study on RNA-mediated silencing of the PLR gene in L. album hairy roots showed that this gene translates to a key enzyme that is directly involved in PTOX biosynthesis5.
A large body of investigations have indicated that gene expression can be effectively regulated by two main mechanisms such as transcription factors (TFs) and non-coding RNAs (ncRNAs)26,27. MicroRNAs (miRNAs) are a group of non-coding transcripts with small length (18–22 nucleotides)28. They are related to the network of genetic modification through transcriptional and translational regulatory actions under adverse conditions29. It has been observed that the level of miRNAs has a significant relationship with the production of polyphenols. The overexpression of miR156 has been shown to increase the transcript level of the dihydroflavonoil-4-reductase (DFR) gene, which is directly linked to the synthesized anthocyanin precursors30. It has been suggested that several miRNAs, especially miR396, contribute to carotenoids and flavonoid biosynthesis in Chlorophytum borivilianum31. Moreover, miR172 modulates the PTOX biosynthesis through MeJA in Podophyllum hexandrum cells in a ROS-non-responsive manner8. However, the mechanisms by which MeJA affects genes involved in PTOX biosynthesis in L. album cells have not been fully investigated. Therefore, this study aims to provide insight into the biochemical and molecular mechanisms regulating MeJA-induced PTOX production with a particular emphasis on the role of genes and miRNAs involved in its biosynthetic pathway.
Results
MeJA accelerates H2O2, NO, and SA formation
To reveal the mechanisms underlying MeJA-induced oxidative responses, we monitored the changes in the H2O2 production in L. album-elicited cells. In this analysis, we observed a time-dependent H2O2 formation, which significantly accelerated after 6 and 12 h (a 9 and 15% increase over the controls, respectively), albeit its level balanced at other times following MeJA treatment (Fig. 1a). The content of NO was also detected and we found that MeJA treatment diminished the cellular level of NO differentially over some time after compared to the control cells, reaching a relatively stable level after 72 h (Fig. 1b). The data clearly showed that the highest decrease in the amount of NO occurred at 48 h after MeJA treatment (63% less that control samples). On the other hand, our results showed that MeJA led to a change in endogenous level of SA, so that it increased during the first 12 h of MeJA elicitation and then reached a constant level until 72 h (Fig. 1c).
MeJA induces the activity of antioxidant enzymes
Considering the observed induction of H2O2 and SA levels under MeJA elicitation, it was hypothesized that this phytohormone might perturb the oxidative balance, leading to the activation of enzymatic antioxidant reactions. Therefore, we estimated the activities of three crucial antioxidant enzymes including, SOD, POD, and CAT subjected to a time course of MeJA elicitation. We recognized the activity of SOD showed a rise at 24 h, which was 1.38 times more than the control samples (Fig. 2a). Our results also indicated that CAT activity increased substantially throughout all time points of MeJA treatment (Fig. 2b). Moreover, POD activity initially provoked at 6 h and 12 h of MeJA elicitation and then peaked after 48 h (a 54% increase over the untreated condition) (Fig. 2c).
Analysis of the activity of (a) SOD, (b) CAT, and (c) POD antioxidant enzymes after 6 h to 72 h of exposure to exogenous MeJA in L. album cells. Values are the mean of three biological replicates ± standard deviation (SD). Different letters imply significant differences obtained using Duncan test at α = 5%.
MeJA provokes PAL and TAL enzyme activities
Other than the enzymatic antioxidants, the activity of PAL and TAL enzymes, which are responsible for the biosynthesis of a broad spectrum of antioxidant metabolites such as lignans were assayed under MeJA elicitation. MeJA resulted in a significant induction of PAL activity within 24 h of treatment in comparison with the control cells (Fig. 3a). Interestingly, the treated cells represented a clear oscillation in TAL activity in response to MeJA elicitation over a time course, so after an early increase at 6 h and 12 h, its activity increased again by 48 h of treatment (Fig. 3b).
MeJA induces the expression of specific genes involved in lignans production
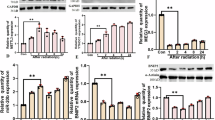
Under MeJA treatment, L. album cells experienced transient upregulation in the genes related to lignans production. Comparative analyses of PAL, CCR, CAD, and PLR genes expression revealed considerable correlations between their transcript levels (Fig. 4). In the case of PAL gene, its expression in the treated cells showed an increase at 12 h and 48 h after elicitation. Afterward, a decrease in the expression level of this gene was found in the treated cells (Fig. 4a). Considering changes in CCR expression in response to MeJA, its transcript level was strongly upregulated at 6 h and 12 h after elicitation. Also, the transcripts of CCR gene diminished at 24 h of MeJA treatment (Fig. 4b). The relative expression of CAD gene increased significantly after 12 h and 72 h of treatment, achieved up to 3.7 and 2.5-fold increase, respectively (Fig. 4c). In the present study, the expression level of PLR gene, which is directly involved in the biosynthesis of lignans, represented an increase at 12 h and 48 h, which was 1.74 and 1.66 times over the control group, respectively (Fig. 4d).
Analysis of the relative expression of (a) PAL, (b) CCR, (c) CAD, and (d) PLR genes after 6 h to 72 h of exposure to exogenous MeJA in L. album cells. Values are the mean of three biological replicates ± standard deviation (SD). Different letters imply significant differences obtained using Duncan test at α = 5%.
MeJA alters the expression levels of miRNAs
Due to the lack of information about miRNAs associated with PTOX biosynthesis in L. album, we checked MeJA-induced changes in the expression pattern of four conserved miRNAs that are probably involved in this process (Fig. 5). MeJA treatment led to upregulation of all investigated miRNAs with a fold-change between 1.5 and 3.5 in L. album cells. Our data revealed that the transcript levels of miR156 and miR159 upregulated remarkably at 12 h after treatment. However, the expression of miR156 decreased after 72 h of exposure, while miR159 transcripts increased at this time (Fig. 5a,b). On the other hand, the initial response time of miR172 and miR396 to MeJA was at 12 h, and then the downregulation occurred after 24 h of treatment. The lowest amount of miR172 expression was observed at 72 h after MeJA treatment, while the level of miR396 transcripts drastically downregulated after 48 h (Fig. 5c,d).
Analysis of the relative expression of (a) miR156, (b) miR159, (c) miR172, and (d) miR396 genes after 6 h to 72 h of exposure to exogenous MeJA in L. album cells. Values are the mean of three biological replicates ± standard deviation (SD). Different letters imply significant differences obtained using Duncan test at α = 5%.
Time-dependent PTOX production in MeJA-elicited cells
HPLC analysis was performed to detect the PTOX content of L. album cells exposed to MeJA treatment at different time courses (24, 48, and 72 h). As shown in Fig. 6, we found that a significant increase in PTOX content occurred at 24 h after MeJA treatment, which reached its highest level after 72 h.
Overview of the responses of L. album cells to MeJA
In this study, PLS-DA analysis was first performed to obtain clear information on the correlation shares of sampling times and MeJA treatment in PTOX accumulation. The subset of results revealed PLS components 1 and 2 (PLS1, and PLS2) accounted for 16.9 and 50.7% of the total variance within the data set, respectively (Fig. 7a). Score plot of PLS-DA results also indicated that the separation of sample replicates from each other was completed. Moreover, the variability from sampling time is classified by Component 1, while the variability caused by MeJA treatment is classified by Component 2. It is obvious that among the respective components, time-dependent responses can explain a larger percentage of data set variability than responses to MeJA treatment (50.7 and 16.9%, respectively). Also, MeJA-treated cells at time series 6, 12, and 24 h are not very different from each other, because there is an overlap of the 95% confidence interval (CI) between these groups.
Summary of dimension reduction and classification of biochemical and molecular responses in L. album cells exposed to MeJA at the 6, 12, 24, 48, and 72 h time points. (a) Partial Least Squares Discriminate Analysis (PLS-DA) scores plot is used to classify data obtained from the untreated and treated cells. MeJA treatment are shown as M6, M12, M24, M48, and M72. Controls are shown with signs C6, C12, C24, C48, and C72. (b) Hierarchical Cluster Analysis (HCA) is applied to cluster data based on Pearson correlations coefficient. Data are shown for three replications for each variable during MeJA treatment. Colours in the HCA map show the magnitude and direction of correlations. The blue box means a strong negative correlation, and the red box signifies a strong positive correlation. The primary clusters are numbered 1 to 5 in the picture. (c) A summary of this research in which SA molecule can play a crucial role in MeJA responses in L. album cells. An increase in SA can affect H2O2 as a crucial second messenger for inducing phenylpropanoid pathway through changes in the expression levels of miR156, miR159, miR172, miR396, and CCR.
On the other hand, HCA analysis showed similarities between different measured factors in response to MeJA according to Pearson correlation coefficient (Red and blue colors illustrate a positive and negative correlation, respectively) (Fig. 7b). Based on the results, we found five main clusters (1–5) with different response patterns, which are indicated by the red dashed lines in Fig. 7b. This pattern revealed a strong correlation among the samples treated with MeJA and a clear distinction between MeJA-treated cells compared with the untreated cells as a time-dependent manner. Each cluster represents different patterns of response to MeJA; cluster 1: CCR gene expression to SOD activity; cluster 2: miR396 gene expression to miR156 gene expression; cluster 3: POD activity to TAL activity; cluster 4: PLR gene expression to CAD gene expression; cluster 5: PTOX content to NO content.
Moreover, Fig. 7c indicated compounds correlated with the SA in response to MeJA in L. album. Accordingly, there was a positive correlation amongst the level of SA and H2O2 content, SOD activity, and CCR, miR158, miR172, miR396 and miR159 genes expression.
Discussion
In our previous work, it has been well approved that different concentrations of MeJA can act as an effective inducer of primary and secondary metabolite accumulation in L. album cells using H2O2 and NO molecules as second messengers. This study was undertaken to determine the underlying biochemical and molecular mechanisms of PTOX accumulation in response to MeJA (50 µM) at different time courses in L. album cells. In the present study, an increase in the level of ROS, determined by H2O2 content, was found at 6 h and 12 h of exposure to MeJA treatment, which either can act as a signal clue or as a harmful agent. It leads to the disturbance of the cell’s redox homeostasis, which can induce enzymatic and non-enzymatic defense reactions32,33. Also, NO content as a reductive substance was substantially decreased during MeJA treatment, providing evidence for NO/MeJA antagonism in L. album cells. This hypothesis is contrary to various studies, which suggest that MeJA might elicit NO burst as a synergistic signal, regulating a variety of plants’ physiological responses8. Probably, NO action in different plants is closely related to the quantitative nature of treatment, which can differently trigger signal transduction cascades13,34.
On the other hand, the experimental results showed a transient increase in SA content that can be suggestive of the involvement of this molecule in a combinatory effect of MeJA signaling in L. album cells. These results were consistent with our previous studies which reported that SA serves as an important mediator for lignans production in L. album cells12,19. It seems that SA has a synergistic relationship with the MeJA signaling pathway at the early hours of treatment in L. album cells, while based on Fig. 7c data, it probably acts as a negative intermediator of MeJA-induced PTOX biosynthesis as the treatment continues. Numerous findings have represented that SA actions in plant resistance might be ascribed to trigger signaling pathways reprogramming defense-related gene expression and enzyme activity35. Accordingly, our results showed that the increase in SA level probably provokes the generation of H2O2 as a second messenger, which can subsequently lead to the regulation of expression and activity of enzymes involved in PTOX biosynthesis in L. album cells exposed to MeJA (Fig. 7b and c). Samari et al.12 also indicated that SA through inducing H2O2 production can regulate lignan biosynthesis in L. album cells.
A large body of evidence suggests that plants have a set of antioxidant equipment including enzymatic and non-enzymatic agents, which can combat with ROS overproduction23,36. In the present study, we found that MeJA treatment distinctively activates CAT, POD, and SOD antioxidant enzymes in a time-dependent manner. The changes in the activities of these antioxidant enzymes may correspond with differences in their catabolism process and reaction sites37. These findings are consistent with the previous observations of Samari et al.38 and Tashackori et al.39, who reported that these enzymes have a central role in scavenging excess amounts of ROS molecules in L. album in response to chitosan and digested cell wall of Piriformospora indica, respectively.
The increase in PAL and TAL enzyme activities in the early hours of MeJA treatment can reflect the occurrence of a defensive reaction at this stage. PAL and TAL are essential enzymes for the conversion of the aromatic amino acids phenylalanine and tyrosine to cinnamic and p-coumaric acids, which can be employed as simple precursors to produce various non-enzymatic antioxidant compounds such as PTOX19,25. These results are in agreement with our previous study, where we found an increase in PAL and TAL activity in L. album cells treated with the MeJA6. Furthermore, our findings showed that PAL activity dynamics are closely associated with PAL gene expression alterations in response to MeJA treatment. Also, we pursued the expression of other genes related to PTOX biosynthesis including CCR, CAD, and PLR. As expected, it was ascertained that these specific genes were significantly upregulated during MeJA treatment. There was also a slight positive correlation between PAL, PLR, and CAD genes expression and PTOX content (Fig. 7b).
A lag time was observed between the highest level of PLR gene expression at 48 h and the highest content of PTOX at 72 h of treatment. These findings agreed with our earlier studies, which have reported that PTOX accumulation in response to light24 and fungal elicitors39 correlated significantly with PLR gene expression level. Additionally, it has been showen that down-regulation of PLR gene expression by using RNA interference (RNAi) strategy caused the depletion of PTOX in L. album hairy roots due to a reduction in PTOX precursors5. These observations emphasized that the PLR gene has functional roles in PTOX biosynthesis in L. album.
Little is known about the expression of miRNAs in L. album cells. However, we selected four conserved miRNAs such as miR156, miR159, miR172, and miR396, to determine their expression levels in L. album cells exposed to a time course of MeJA treatment. According to a recent study, the prediction of miRNAs in L. usitatissimum revealed that 15 miRNAs such as miR156, miR157, miR159, miR160, miR164, miR166, miR167, miR171, miR172, miR319, miR395, miR396, miR399, miR5219, and miR5523 are involved in the biosynthetic pathway of lignans40. The RT-qPCR results showed that the transcript levels of miR156, miR159, miR172, and miR396 were upregulated after 12 h under MeJA treatment as an early response, and then, their expression was downregulated.
The HCA heatmap revealed that there are positive correlations between SA content and miR156, miR159, miR172, and miR396 gene expression. Moreover, it represented positive correlations between H2O2 content and the transcript level of these miRNAs. Therefore, it can be assumed that MeJA affects the expression level of the mentioned miRNAs in L. album cells by perturbing cellular oxidative homeostasis. Recent studies have demonstrated that miRNAs directly respond to ROS overproduction by targeting genes that encode proteins with oxidant and antioxidant effects in an indirect manner, as well as by targeting genes that regulate downstream redox signaling pathways41. In cereals, miR172 was identified to regulate salt tolerance positively by targeting the INDETERMINATE SPIKELET 1 (IDS1), which can bind to the promoter of enzymatic ROS scavengers including CAT, ascorbate peroxidase (APX), glutathione peroxidase (GPX) genes to suppress their transcription42.
Furthermore, Fig. 7b depicted a positive correlation between the expression level of the PLR gene with miR172 and miR156 as upstream gene regulators. A recent investigation has predicted that these miRNAs likely target PLR in L. usitatissimum40. DSPC network also represented that miR159 can regulate PTOX accumulation in a positive manner in L. album cells treated by MeJA (Fig. 8). Also, our observation showed that there was a negative correlation amongst miR156, miR172, and PTXO accumulation (Fig. 8). Likewise, a study has represented that miR172 is a crucial checkpoint in the phenylpropanoid pathway by regulation of fucosyltransferase (FUT) gene expression43.
On the contrary, Hazra et al.8 reported that MeJA in P. hexandrum can down-regulate eight selected miRNAs (e.g., miR172i, miR035, miR1438, miR2275, miR8291), which play significant roles in modulating the expression of phenylpropanoid pathway genes. In Prunus persica, small RNA deep sequencing ascertained that miR159 is linked to MYBs as its targets, thus regulating the phenylpropanoid pathway and lignin deposition44. Also, Wu et al.45 reported that monolignols production is regulated by several miRNAs such as miR396b, miR395, miR397, miR408, miR2936, miR502, miR1061-3p, miR1318, and miR3627 in pear fruit. This evidence suggested that miR156, miR159, miR172, and miR396 probably contribute to the expression of target genes, which may directly and/or indirectly act as regulatory checkpoints for PTOX biosynthesis.
Materials and methods
Cell culture and treatment
Seeds of L. album used in this study were harvested according to the relevant legislation and international guidelines. All permissions for plant collection were provided by the Research Ethics Committee of Tarbiat Modares University. Each plant was taxonomically identified by Dr. Shahrokh Kazempour-Osaloo, a professor of plant systematic. A voucher specimen of L. album was deposited in the Herbarium of the Department of Plant Biology of Tarbiat Modares University, Iran. As previously described by Sagharyan and Sharifi3, callus was initiated by culturing sterile leaves for 30 days on the solid MS medium46 with 2 mg L− 1 of NAA and 0.4 mg L− 1 of Kinetin. Subculture of callus was performed after 14 days in the same medium composition. In brief, cell suspension culture was provided by the desegregation of friable Calli (2 g) in 30 mL of liquid MS medium fortified with 2 mg L− 1 of NAA and 0.4 mg L− 1 of Kinetin in 100 mL Erlenmeyer flasks. The cultures were incubated on an orbital shaker at 22 °C at 110 rpm before treatment. According to a representative growth curve (data not shown), seven-day-old cell cultures were treated with a final concentration of MeJA (0 and 50 µM) using the previous experiment based on growth yield and PTOX accumulation in L. album cells6. A set of time points (6, 12, 24, 48, and 72 h) was considered for harvesting the cells after treatment.
Analysis of H2O2 content
The content of H2O2 was photometrically analyzed using the protocol of Velikova et al.47 with a slight modification. To determine H2O2 content, 0.2 g of fresh cells were lysed in 2 mL of pre-cooled TCA (0.1%) on the ice bath. After centrifugation (12,000×g, 4 °C, 20 min), the supernatant (500 µL) was mixed with 500 µL of PO43- (100 mM; pH ~ 7) and 1 mL of KI (1 M) in the dark condition for 30 min. After that, the absorption of the sample was assayed at 390 nm, and the concentration of H2O2 in µmol H2O2 g-1 FW was evaluated using the standard curve.
Measurement of NO content
NO generation was monitored through the measurement of intercellular nitrite concentration by means of Griess reagent48. For this experiment, 0.2 g of frozen cells were extracted by 1.5 mL of PO43- (100 mM; pH ~ 7). Following centrifugation (12,000×g for 15 min at 4 °C), 500 µL of Griess reagent and PO43- buffer (100 mM; pH ~ 7) were mixed to an adequate volume of the resulting supernatant at room temperature for 30 min. The content of NO was assessed using a spectrophotometer at 540 nm. The concentration of NO was determined from a calibration curve of sodium nitrite as µmol g-1 FW.
Quantification of SA by HPLC
To evaluate the content of SA in treated and untreated cells, the extraction was performed as described previously by Owen et al.49. In brief, 200 mg of the frozen samples were powdered and extracted with methanol. For HPLC analysis, the supernatant obtained from the methanolic extraction was evaporated. The residue was dissolved in 4 mL of acetonitrile, washed thrice with n-hexane, and then dried under air condition again. The final residue was resuspended in 0.5 mL of methanol for SA quantification by HPLC system (Agilent Technologies; 1260 Infinity, USA) via a C-18 column (250 × 4.6 mm; ID; 5 µM; Alltech). The eluent phase consisted of solvent A (aqueous acetic acid (2%)) and Solvent B (methanol). Column and instrumentation for analysis were carried out as standardized by Zafari et al.50. The wavelength for the SA detection was set at 278–300 nm using a UV dual-array detector. The determination of the SA peak was conducted by comparing retention time with the analytical standard.
Assessment the activity of antioxidant enzymes
The total soluble proteins were assayed using the protocol of Bradford51, with bovine serum as the calibration standard. Superoxide dismutase (SOD) activity was estimated according to the prevention of nitroblue tetrazolium (NBT) photoreduction52. Catalase (CAT) activity was measured using the procedure of Cakmak and Marschner53. Peroxidase (POD) activity was assessed according to the capacity cell extraction for guaiacol oxidation in 1 min54.
Extraction and assay of PAL and TAL activity
The activity of PAL and TAL enzymes was assayed based on Beaudoin-Eagan and Thorpe55 method through measuring the contents of cinnamic and p-coumaric acids, respectively. In brief, 100 µL of crude enzyme extract was added to 200 µL of phenylalanine (PAL) and tyrosine (TAL) (0.1 M) dissolved in PO43- buffer (0.1 M; pH 8.8). Then, the reaction mixtures were incubated in a water bath (PAL ~ 37 °C, TAL ~ 30 °C) for 1 h. After that, the activity of PAL and TAL enzymes was stopped by adding 50 µL of HCl (0.05 M). The reaction mixtures were extracted with 2 mL of ethyl acetate three times. The ethyl acetate phase was collected and dried at room temperature. The residue was resuspended by 1 mL of sodium hydroxide (0.05 M). The absorbance of the sample was measured at 290 nm (PAL) and 330 nm (TAL) via spectrophotometer (Cary 100-UV-vis, Agilent, USA). The activity of PAL and TAL enzymes was calculated based on cinnamic and p-coumaric acids standard curves and expressed as µg cinnamic/p-coumaric acid mg-1 protein h-1.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from each sample using the One-Step RNA reagent based on the manufacturer’s procedure. The RNA quantity and purity parameters were characterized using Nanodrop ND-1000 (ThermoFisher Scientific) and agarose gel electrophoresis. Only RNA extracts with an A260/A280 ratio between 1.8 and 2.1 and an A260/A230 ratio superior to 1.7 were selected for cDNA synthesis. To synthesize cDNA, 1 µg of total RNA was used for reverse transcription reaction by using a first-strand cDNA synthesis kit (RevertAid, Thermo Fisher, USA), following the manufacturer’s instructions. The expression of PAL, CCR, CAD, and PLR genes was analyzed by quantitative real-time chain reaction (RT-qPCR) using SYBR PCR Master mix (4x) ROX reference dye (Amplicon). Appropriate gene primers were listed in Table 1 as previously reported by Yousefzadi et al.24. The qPCR program performed an initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s, and a final extension at 72 °C for 10 s (The Applied Biosystems® StepOne™, Thermo Fisher, USA). Data were visualized using the StepOne software, v2.3 (Applied Biosystems). The relative expression level of the mentioned genes was reported after normalization by the expression level of β-Actin as described by Livak and Schmittgen59.
miRNAs primers design
For identification of candidate miRNAs, a similar procedure as used earlier was employed56. Briefly, we downloaded L. usitatissimum miRNA sequences from the Sanger Institute miRBase Sequence Database (https://www.mirbase.org) (release; 2021)57. Pre-miRNA sequences of L. usitatissimum with a range of 0–4 mismatches in the mature miRNAs were identified and served as candidate miRNAs. The homology search was performed using the Blast algorithm from the publicly available Flax Expressed Sequence Tag (ESTs) database at http://blast.ncbi.nlm.nih.gov/Blast.cgi. Subsequently, the primers were designed by Oligo 7 software (version 7.0) as described by Samavatian et al.58 with some modifications. Several principles of designing primers for miRNAs were considered as follows: (i) optimum annealing temperature was 60 °C, ranging from 55 °C to 66 °C, and (ii) optimum primer length was 22 bp, ranging from 18 bp to 25 bp. All primers were purchased from Pishgam company (Iran).
RT-qPCR of miRNAs
To determine the expression levels of miRNAs, 1.5 µg of total RNA sample was polyadenylated with poly (A) polymerase (PAP) at 38 °C for 45 min using the Poly (A) Tailing Kit. Then, PAP-treated total RNA was reverse transcribed by a poly (T) adapter universal reverse transcription (RT)-primer (5′ GCGTCGACTAGTACAACTCAAGGTTCTTCCAGTCACGACGTTTTTTTTTTTTTTTTTT-3′) using SuperScript III Reverse Transcriptase kit (Invitrogen) according to the manufacture’s instruction. Quantitative real-time PCR was performed on Applied Biosystems® 48 Plates (The Applied Biosystems® StepOne™, Thermo Fisher). The reaction mixture was 4 µL of SYBR PCR Master mix (4x) ROX reference dye, 1 µL of cDNA, 0.5 µL of forward primer, 0.5 µL of Universal-R as a reverse primer, and ddH2O in a final volume of 20 µL. All primers utilized for this experiment are listed in Table 2. Amplifications was set based on the program 15 min pre-denaturation at 95 °C, and 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 70 °C for 30 s. The melting curve analysis condition was 95 °C for 15 s, 65 °C increased to 95 °C with temperature enhancement of 0.5 °C every 1 s). Negative control was composed of a No-template reaction. The relative expression of the miRNAs was determined by the 2−∆∆Ct method employing the U6 gene as an internal control59.
Measurement of PTOX content
Lignan accumulation was quantified by HPLC (Agilent 1260 HPLC system using a UV detector, CA, USA) as described by Yousefzadi et al.2 method. So, 0.2 g of dried cells were subject to extract in methanol (80%, v/v) by sonication, as previously reported by Chashmi et al.60. The supernatant was collected and evaporated to dryness. Finally, the residue was re-suspended in 250 µL of methanol before HPLC operation. A C18-ODS3 column (5 µM; 250 × 4.6 mm) was utilized as a stationary phase. The mobile phase was composed of acetonitrile and water with a standardized gradient system following Yousefzadi et al.2 method.
Statistical analysis
In the present study, the normality of data was checked by the Shapiro-Wilk test. All analyses were conducted at least three times, each with three independent biological and technical replications. To evaluate differences between treated and untreated cells, the analysis of variance (ANOVA) test and Duncan’s multiple range test (α = 0.05) were applied by using SPSS software (ver. 25). The expression levels of genes and miRNAs were assayed by GenEx (ver. 6.1.). Next, we used Partial Least Squares Discriminate Analysis (PLS-DA), Hierarchical Cluster Analysis (HCA), and Debiased Sparse Partial Correlation (DSPC) algorithm to find interconnections between PTOX biosynthesis specific genes and miRNAs in relation to changes of oxidative status using web-based software MetaboAnalyst, which scores all the features according to Pearson correlation coefficient and obtained a detailed analysis report (https://www.metaboanalyst.ca).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 6MPTOX:
-
6-Methoxypodophyllotoxin
- CCR:
-
Cinnamoyl-CoA reductase
- CAD:
-
Cinnamyl-alcohol dehydrogenase
- H2O2 :
-
Hydrogen peroxide
- MeJA:
-
Methyl jasmonate
- miRNAs:
-
MicroRNAs
- ncRNAs:
-
Non-coding RNAs
- NO:
-
Nitric oxide
- PAL:
-
Phenylalanine ammonia-lyase
- PLR:
-
Pinoresinol-lariciresinol reductase
- PTOX:
-
Podophyllotoxin
- RNS:
-
Reactive nitrogen species
- ROS:
-
Reactive oxygen species
- SA:
-
Salicylic acid
References
Gordaliza, M., García, P. A., del Corral, J. M., Castro, M. A. & Gómez-Zurita, M. A. Podophyllotoxin: distribution, sources, applications and new cytotoxic derivatives. Toxicon. 44, 441–459 (2004).
Yousefzadi, M., Sharifi, M., Chashmi, N. A., Behmanesh, M. & Ghasempour, A. Optimization of podophyllotoxin extraction method from Linum album cell cultures. Pharm. Biol. 48, 14211425 (2010).
Sagharyan, M. & Sharifi, M. Metabolic and physiological changes induced by exogenous phenylalanine in Linum album cells. J. Plant. Growth Regul. 43, 2785–2801 (2024).
Fuss, E. Lignans in plant cell and organ cultures: an overview. Phytochem Rev. 2, 307–320 (2003).
Tashackori, H. et al. RNAi-mediated silencing of pinoresinol lariciresinol reductase in Linum album hairy roots alters the phenolic accumulation in response to fungal elicitor. J. Plant. Physiol. 232, 115–126 (2019).
Sagharyan, M., Sharifi, M. & Samari, E. Methyl jasmonate redirects the dynamics of carbohydrates and amino acids toward the lignans accumulations in Linum album cells. Plant. Physiol. Biochem. 198, 107677 (2023).
Bhavanam, S. & Stout, M. Seed treatment with jasmonic acid and methyl jasmonate induces resistance to insects but reduces plant growth and yield in rice, Oryza sativa. Front. Plant. Sci. 12, 691768 (2021).
Hazra, S., Bhattacharyya, D. & Chattopadhyay, S. Methyl jasmonate regulates podophyllotoxin accumulation in Podophyllum hexandrum by altering the ROS-responsive podophyllotoxin pathway gene expression additionally through the down regulation of few interfering miRNAs. Front. Plant. Sci. 8, 164 (2017).
Wang, H., Kou, X., Wu, C., Fan, G. & Li, T. Nitric oxide and hydrogen peroxide are involved in methyl jasmonate-regulated response against Botrytis Cinerea in Postharvest blueberries. J. Agric. Food Chem. 68, 13632–13640 (2020b).
Ishikawa, T. et al. Metabolome analysis of response to oxidative stress in rice suspension cells overexpressing cell death suppressor Bax inhibitor-1. Plant. Cell. Physiol. 51, 9–20 (2010).
Sadeghnezhad, E., Sharifi, M. & Zare-Maivan, H. Profiling of acidic (amino and phenolic acids) and phenylpropanoids production in response to methyl jasmonate-induced oxidative stress in Scrophularia striata suspension cells. Planta. 244, 75–85 (2016).
Samari, E. et al. Interactions between second messengers, SA and MAPK6 signaling pathways lead to chitosan-induced lignan production in Linum album cell culture. Ind. Crop Pro. 177, 114525 (2022).
Abedi, M., Karimi, F., Saboora, A. & Razavi, K. Elicitation of flavonoid biosynthesis in cell suspension cultures of Haplophyllum virgatum by sodium nitroprusside. Plant. Biosyst. 157, 992–1002 (2023).
Du, L. et al. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature. 457, 1154–1158 (2009).
Schweiger, R., Heise, A. M., Persicke, M. & Müller, C. Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types. Plant. Cell. Environ. 37, 1574–1585 (2014).
Li, N., Han, X., Feng, D., Yuan, D. & Huang, L. J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20, 671 (2019).
Ding, Y. et al. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 173, 1454–1467 (2018).
Lv, Z. Y. et al. Phytohormones jasmonic acid, salicylic acid, gibberellins, and abscisic acid are key mediators of plant secondary metabolites. World J. Tradit. 7, 307–325 (2021).
Esmaeili, S. et al. Exogenous melatonin induces phenolic compounds production in Linum album cells by altering nitric oxide and salicylic acid. Sci. Rep. 13, 4158 (2023).
Soltani, M. et al. Putrescine induces lignans biosynthesis through changing the oxidative status and reprogramming amino acids and carbohydrates levels in Linum album hairy roots. Plant. Cell. Tissue Organ. Cult. 153, 387–402 (2023).
Blokhina, O., Virolainen, E. & Fagerstedt, K. V. Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 91, 179–194 (2003).
D’Onofrio, C., Cox, A., Davies, C. & Boss, P. K. Induction of secondary metabolism in grape cell cultures by jasmonates. Funct. Plant. Biol. 36, 323–338 (2009).
Khataee, E., Karimi, F. & Razavi, K. Alkaloids production and antioxidant properties in Catharanthus roseus (L.) G. Don. Shoots and study of alkaloid biosynthesis-related gene expression levels in response to methyl jasmonate and putrescine treatments as eco-friendly elicitors. Biol. Futura. 70, 38–46 (2019).
Yousefzadi, M. et al. The effect of light on gene expression and podophyllotoxin biosynthesis in Linum album cell culture. Plant. Physiol. Biochem. 56, 41–46 (2012).
Barros, J. et al. Role of biofunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants. 2, 16050 (2016).
Chiasson, D. M. et al. Soybean SAT1 (Symbiotic Ammonium Transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc. Natl. Acad. Sci. 111, 4814–4819 (2014).
Ebrahimi Khaksefidi, R. et al. Differential expression of seven conserved microRNAs in response to abiotic stress and their regulatory network in Helianthus annuus. Front. Plant. Sci. 6, 741 (2015).
Liu, W. W., Meng, J., Cui, J. & Luan, Y. S. Characterization and function of MicroRNAs in plants. Front. Plant. Sci. 22, 2200 (2017).
Shriram, V., Kumar, V., Devarumath, R. M., Khare, T. S. & Wani, S. H. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front. Plant. Sci. 14, 817 (2016).
Davies, K. M. et al. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 131, 259–268 (2003).
Kajal, M. & Singh, K. Small RNA profiling for identification of miRNAs involved in regulation of saponins biosynthesis in Chlorophytum borivilianum. BMC Plant. biol. 17, 1–14 (2017).
Amir, S. B., Rasheed, R., Ashraf, M. A., Hussain, I., & Iqbal, M. Hydrogen sulfide mediates defense response in safflower by regulating secondary metabolism, oxidative defense, and elemental uptake under drought. Physiol. Plant. 172, 795–808 (2021).
Wang, H., Kou, X., Wu, C., Fan, G. & Li, T. Methyl jasmonate induces the resistance of postharvest blueberry to gray mold caused by Botrytis Cinerea. J. Sci. Food Agric. 100, 4272–4281 (2020a).
Belchí-Navarro, S., Rubio, M. A., Pedreño, M. A. & Almagro, L. Production and localization of hydrogen peroxide and nitric oxide in grapevine cells elicited with cyclodextrins and methyl jasmonate. J. Plant. Physiol. 237, 80–86 (2019).
Chen, J. et al. Reprogramming and remodeling: transcriptional and epigenetic regulation of salicylic acid-mediated plant defense. J. Exp. Bot. 71, 5256–5268 (2020).
Zhao, H., Sun, X., Xue, M., Zhang, X. & Li, Q. Antioxidant enzyme responses induced by whiteflies in tobacco plants in defense against aphids: catalase may play a dominant role. PLoS One. 11, e0165454 (2016).
Wang, C., Xue, M., Bi, M., Li, Q. & Hu, H. Temporal effect of tobacco defense responses to Myzus perskae (Sulzer) (Homoptera: Aphididae) induced by Bemisia tabaci (Gennadius) (Homoptera: Aleyrodidae) B biotype. Acta Entomol. Sin. 53, 314–322 (2010).
Samari, E., Sharifi, M., Ghanati, F., Fuss, E. & Chashmi, N. A. Chitosaninduced phenolics production is mediated by nitrogenous regulatory molecules: NO and PAs in Linum album hairy roots. Plant. Cell. Tissue Organ. Cult. 140, 63–576 (2020).
Tashackori, H. et al. Physiological, biochemical, and molecular responses of Linum album to digested cell wall of Piriformospora Indica. Physiol. Mol. Biol. Plants. 2, 2695–2708 (2021).
Harenčár, Ľ. & Ražná, K. Prediction of microRNAs involved in lignan and cyanogenic glycosides biosynthesis of Flax (Linum usitatissimum L). Plant. Growth Regul. 1–19 (2023).
Cimini, S. et al. Redox balance-DDR-miRNA triangle: relevance in genome stability and stress responses in plants. Front. Plant. Sci. 10, 461601 (2019).
Cheng, X. et al. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New. Phytol. 230, 1017–1033 (2021).
Wong, M. M., Cannon, C. H. & Wickneswari, R. Identification of lignin genes and regulatory sequences involved in secondary cell wall formation in Acacia auriculiformis and Acacia mangium via de novo transcriptome sequencing. BMC Genom. 12, 342 (2011).
Zhu, H. et al. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant. Biol. 12, 149 (2012).
Wu, J. et al. Identification of miRNAs involved in pear fruit development and quality. BMC Genom. 15, 953 (2014).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 (1962).
Velikova, V., Yordanov, I. & Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant. Sci. 151, 59–66 (2000).
Kaur, G. et al. Exogenous nitric oxide (NO) interferes with lead (Pb)-induced toxicity by detoxifying reactive oxygen species in hydroponically grown wheat (Triticum aestivum) roots. PLoS One. 10, e0138713 (2015).
Owen, R. et al. Isolation, structure elucidation and antioxidant potential of the major phenolic and flavonoid compounds in brinedolive drupes. Food Chem. Toxicol. 41, 703–717 (2003).
Zafari, S., Sharifi, M., Mur, L. A. J. & Chashmi, N. A. Favouring NO over H2O2 production will increase Pb tolerance in Prosopis farcta via altered primary metabolism. Ecotox Environ. Safe. 142, 293–302 (2017).
Bradford, M. M. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein dye binding. Ann. Bot. 72, 248–254 (1976).
Giannopolitis, C. N. & Ries, S. K. Superoxide dismutases: I. occurrence in higher plants. Plant. Physiol. 59, 309–314 (1977).
Cakmak, I. & Marschner, H. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant. Physiol. 98, 1222–1227 (1992).
Pandolfini, T., Gabbrielli, R. & Comparini, C. Nickel toxicity and peroxidase activity in seedlings of Triticum aestivum L. Plant. Cell. Environ. 15, 719–725 (1992).
Beaudoin-Eagan, L. D. & Thorpe, T. A. Tyrosine and phenylalanine ammonia lyase activities during shoot initiation in tobacco callus cultures. Plant. Physiol. 78, 438–441 (1985).
Barozai, M. Y. K., Din, M. & Baloch, I. A. Identification of microRNAs in ecological model plant Mimulus. J. Biophys. chem. 2, 322–331 (2011).
Griffiths-Jones, S., Saini, H. K., Van Dongen, S. & Enright, A. J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, 154–158 (2007).
Samavatian, H., Soltani, B. M. & Yousefi, F. Susceptible and resistant wheat cultivars show different miRNAs expression patterns in response to Zymoseptoria Tritici. J. Plant. Pathol. 105, 43–447 (2023).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25, 402–408 (2001).
Chashmi, N. A. et al. Analysis of 6-methoxy podophyllotoxin and podophyllotoxin in hairy root cultures of Linum album Kotschy ex Boiss. Med. Chem. Res. 22, 745–752 (2013).
Acknowledgements
The authors would like to thank Tarbiat Modares University, Iran for providing this project’s laboratory facilities support. This work is based upon research funded by Iran National Science Foundation (INSF) under project NO. 4028433.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
MS performed the experiments, analyzed data and wrote the manuscript. MSH supervised the study. ES advised the study and edited the final manuscript. FK was the study advisor.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sagharyan, M., Sharifi, M., Samari, E. et al. Changes in MicroRNAs expression mediate molecular mechanism underlying the effect of MeJA on the biosynthesis of podophyllotoxin in Linum album cells. Sci Rep 14, 30738 (2024). https://doi.org/10.1038/s41598-024-78715-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-78715-6