Abstract
Preeclampsia (PE) is an important research subject in obstetrics. Nevertheless, the underlying mechanisms of PE remain elusive. PE-related expression datasets (GSE96983, GSE96984 and GSE24129) were downloaded from the Gene Expression Omnibus (GEO) database. Firstly, the differentially expressed messenger RNAs (DE-mRNAs), DE-microRNA (DE-miRNAs) and DE-long non-coding RNA (DE-lncRNAs) between PE and control cohorts were identified, and the ceRNA network was constructed. Then candidate hub genes were obtained through five algorithms by the protein-protein intersection (PPI) network of the mRNAs. Further, five hub genes were identified by receiver operating characteristic (ROC) curve and gene expression profiles: DAXX, EFNB1, NCOR2, RBBP4 and SOCS1. The function of 5 hub genes was analyzed and the interaction between drugs and hub genes was predicted. A total of 5 small molecule drugs were predicted, namely benzbromarone, 9,10-phenanthrenequinone, chembl312032, insulin and aldesleukin. AGAP2-AS1 was mainly located in exosome and cytoplasm. Agap2-as1-related regulatory subnetworks were extracted from ceRNA networks which included 41 mRNAs, 2 miRNAs and 1 lncRNA, including the regulated relationship pairs AGAP2-AS1-hsa-miR-497-5p-SRPRB, and AGAP2-AS1-hsa-miR-195-5p-RPL36. In summary, we constructed a competitive endogenous RNA (ceRNA) network to identify five potential biomarkers (DAXX, EFNB1, NCOR2, SOCS1 and RBBP4) of PE. The in-depth analysis of the AGAP2-AS1 regulatory network will help to uncover more important molecules closely related to PE and provide a scientific Reference.
Similar content being viewed by others
Introduction
Preeclampsia (PE) is a prevalent acute and severe obstetric disorder, characterized by elevated blood pressure and the presence of excess protein in the urine. It can cause severe multiorgan functional damage and adversely affect the health of both mother and child. If left undetected or untreated, it can result in maternal and neonatal mortality, particularly in severe instances, thus emerging as a significant contributor to global maternal and neonatal death rates1. The diagnosis of PE is made by a systolic blood pressure of 140mmHg and/or a diastolic blood pressure of 90mmHg after 20 weeks of gestation, combined with urinary protein quantity and urine protein/creatinine ratio of 0.3 or random urinalysis results. In the absence of proteinuria, the diagnostic criteria for PE can also include the presence of organ or system involvement, such as cardiovascular, respiratory, hepatic, renal, hematological, gastrointestinal, neurological, or placental-fetal abnormalities2,3. PE impacts 5-8% of pregnancies, correlating with a higher likelihood of negative pregnancy outcomes like placental detachment, preterm delivery, and restricted fetal growth, leading to stillbirth4,5. In recent years, the occurrence rate of PE has been rising annually, but the study of its related pathological mechanism is relatively limited. It is currently believed that factors such as the infiltration capacity of placental trophoblast cells and uterine spiral artery remodeling are closely related to the pathogenesis of PE, but the exact mechanism remains unclear6,7. Therefore, it is critical to explore the pathologic mechanisms of PE occurrence in order to find effective and accessible clinical biomarkers for PE screening in the field of obstetrics, and then to develop effective PE treatment and prevention measures in the clinic.
Research indicates that less than 2% of the human genome is made up of genes coding for proteins, with the bulk comprising non-coding genes, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs)8. The significance of non-coding RNAs in regulating genes has gained increasing recognition in recent years. miRNAs, a category of small, naturally occurring single-stranded noncoding RNAs, are crucial in the progression of diseases. LncRNAs, an RNA molecule with a gene length of more than 200 nucleotide units, lack an open-reading coding frame and have no ability to encode proteins, but they possess a broad array of biological functions, capable of regulating gene expression both at the transcriptional and post-transcriptional stages9. LncRNAs play a role in numerous cellular activities, including cell differentiation, immune responses, metastasis in cancer cells, proliferation, and resistance to drugs10,11,12. In addition, lncRNAs can be involved in several biological processes including growth and development, and material metabolism at the epigenetic level13. It has been established that lncRNAs and miRNAs are critical in the onset and progression of PE14,15,16. There is a variety of miRNAs expressed differently in the placentas of patients with PE compared to normal pregnancies, suggesting that these miRNAs may be associated with PE17. Furthermore, some studies have indicated that lncRNAs can be involved in PE progression by affecting the function of trophoblast cells18,19.
Recently, the significance of non-coding RNAs (ncRNAs), including lncRNAs and miRNAs, in gene regulation has increasingly been acknowledged. LncRNAs, a type of RNA molecule exceeding 200 nucleotides in length, don’t encode proteins but are crucial in regulating gene expression20. LncRNAs can regulate the transcriptional, post-transcriptional and epigenetic levels of gene expression by interacting with DNA, RNA and proteins21. Conversely, miRNAs are small RNA molecules, approximately 22 nucleotides long, primarily regulating gene expression by binding to the mRNAs of target genes, leading to their degradation or inhibiting their translation22.
Abnormal expression and dysfunction of lncRNAs and miRNAs have been widely observed during disease development, and they can influence cell proliferation, apoptosis, differentiation, and cell cycle regulation, thus affecting disease onset and progression23,24. In the context of PE, numerous research investigations have proposed that lncRNAs and miRNAs might play crucial roles in the onset and advancement of this condition25,26. For instance, a previous discovered found that the lncRNA AGAP2-AS1 was aberrantly expressed in tumor cells and was involved in tumorigenesis and progression27. In addition, down-regulated lncRNA AGAP2-AS1 causes pre-eclampsia by impairing the trophoblast phenotype as a competing endogenous RNA for jun dimerization protein 2 (JDP2)28. However, the specific regulatory mechanism and its specific role manages to be investigated for other downstream of AGAP2-AS1 in PE occurrence.
In this investigation, the transcriptomic data and pertinent clinical data of placental tissues (GSE96983, GSE96984, GSE24129) were acquired from the Gene Expression Omnibus (GEO) repository, specifically focusing on individuals diagnosed with PE. The lncRNA-miRNA-mRNA regulatory network of PE was constructed. Furthermore, PE-related biomarkers were identified by bioinformatics analysis. By deeply studying the function and regulatory mechanism of AGAP2-AS1, our objective is to elucidate the precise involvement of these factors in the onset and progression of PE, thus establishing a significant theoretical foundation for the timely detection and management of PE. In addition, by analyzing the regulatory network of AGAP2-AS1, we anticipate discovering other important molecules related to PE and further resolve the pathogenesis of PE.
Results
Creation of ceRNA network
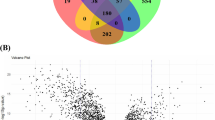
There were 4388 differentially expressed messenger RNAs (DE-mRNAs) between PE and control cohorts (Fig. 1A,B). In total, 4129 differentially expressed long non-coding RNAs (DE-lncRNAs) (PE vs. control) were identified, comprising 1597 up-regulated lncRNAs and 2532 down-regulated lncRNAs (Fig. 1C,D). There were 46 differentially expressed microRNAs (DE-miRNAs) (PE vs. control), consisting 17 up-regulated miRNAs and 29 down-regulated miRNAs (Fig. 1E,F). There were 1318 miRNAs predicted, and 40 miRNAs were obtained finally (Fig. 2A,B). In total, 7599 mRNAs were predicted, and 1254 mRNAs finally were got (Fig. 2C,D). Afterwards, the ceRNA network was created, including 1203 mRNAs, 39 miRNAs, and 138 lncRNAs (Fig. 2E). For instance, the regulated relationship pairs included AC092168.2-hsa-miR-324-5p-RBBP4, and ADAMTSL4-AS1-hsa-miR-331-3p- SOCS1 (Fig. 2E). The results of enrichment analysis for the mRNAs suggested that, 1517 Gene Ontology- biological process (GO BP) items, 87 Gene Ontology- cellular component (GO CC) items, 113 Gene Ontology-molecular function (GO MF) items, and 85 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were enriched (Supplementary Tables 1–2), including kidney epithelium development, focal adhesion, DNA-binding transcription activator activity, human papillomavirus infection, etc. (Fig. 2F,G).
Construction of ceRNA network. (A) The Venn map of candidate miRNA between mirtarbase and StarBase database. (B) The Venn map between candidate miRNA and DE-miRNA. (C) The Venn map of candidate mRNA between mirtarbase and StarBase database. (D) The Venn map between candidate mRNA and DE-mRNA. (E) Construction of ceRNA network. Blue circles indicate lncRNA, green circles indicate miRNA, red circles indicate mRNA. (F) GO enrichment analysis of mRNAs. (G) KEGG enrichment analysis of mRNAs.
Acquisition of biomarkers
The protein-protein interaction (PPI) network of the mRNAs contained 128 nodes and 123 edges, which displayed that RAC1 interacted with various proteins, including ROR2, PLD1, EFNB1, etc. (Fig. 3A). Then, 17 hub genes were identified, namely RAC1, PRKCE, TIAM1, JAK1, EFNB1, DAXX, PTPN13, MAP3K5, NCOR2, RBBP4, MYH14, ADCY1, SOCS1, FOXO1, WNT5A, NF2, and VEGFA (Fig. 3B). And 11 candidate genes were gained, namely DAXX, EFNB1, FOXO1, MAP3K5, NCOR2, NF2, PRKCE, RBBP4, SOCS1, TIAM1, and WNT5A (Fig. 3C,D). The expression of DAXX, EFNB1, NCOR2, SOCS1, and RBBP4 between PE and control cohorts were significantly different, and showed the same expressive trend in GSE96984 and GSE24129 (Fig. 3E,F). In PE, the expression of DAXX, EFNB1, NCOR2, and SOCS1 was found to be increased, whereas RBBP4 exhibited decreased expression. Therefore, these five genes were defined as the biomarkers. The results of functional similarity demonstrated that the similarity scores of DAXX and NCOR2 were greater than 0.6, indicating these two genes had a strong similarity (Fig. 3G,H). Finally, the relevant analysis showed all five biomarkers correlated with each other, and RBBP4 was negatively correlated with NCOR2, EFNB1, DAXX, and SOCS1 (Fig. 3I). However, NCOR2, EFNB1, DAXX, and SOCS1 were positively correlated with each other (Fig. 3I).
Screening of biomarkers. (A) Construction of PPI network. Red circles indicate up-regulated mRNAs, blue circles indicate down-regulated mRNAs. (B) The hub gene was obtained by BottleNeck, EcCentricity, Closeness, Betweenness and Stress algorithms. ROC curve of hub genes in GSE96984 (C) and GSE24129 dataset (D). The expression of hub genes between PE and control cohorts in GSE96984 (E) and GSE24129 dataset (F). (G) GO semantic similarity box plot of hub genes. (H) Raincloud plot of relatedness of the GO terms. (I) The diagram depicts Pearson correlations between hub genes. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Enrichment analysis
The enrichment analysis displayed the top 10 GO categories and KEGG pathways. Among the GO results, all biomarkers were involved in lymphocyte mediated immunity and adaptive immune response (Fig. 4A-E). The DAXX, EFNB1, NCOR2, and SOCS1 were involved in these items in high expressed cohort, yet RBBP4 enriched in these items in low expressed cohort (Fig. 4A-E). The DAXX and SOCS1 were involved in positive regulation of immune effector process, T cell differentiation, and regulation of leukocyte mediated immunity in high expressed cohort, while RBBP4 was involved in these items in low expressed cohort (Fig. 4A,D,E). Interestingly, DAXX and SOCS1 enriched in the same items, such as response to interferon gamma, positive regulation of leukocyte mediated immunity, etc. (Fig. 4A,D). Additionally, EFNB1 and NCOR2 also enriched in the same items, such as endoplasmic reticulum lumen, collagen containing extracellular matrix, regulation of peptidase activity, etc. (Fig. 4B,C). Of the KEGG results, all biomarkers were involved in allograft rejection, antigen processing and presentation, graft versus host disease, natural killer cell mediated cytotoxicity, cytokine-cytokine receptor interaction, autoimmune thyroid disease (Fig. 5A,B). The DAXX, EFNB1, NCOR2, and SOCS1 were involved in these items in high expressed cohort, yet RBBP4 enriched in these items in low expressed cohort (Fig. 5A-E). The DAXX, NCOR2, and SOCS1 were involved in Toll like receptor signaling pathway in high expressed cohort (Fig. 5A,C,D). EFNB1 and RBBP4 enriched in the same items, such as type I diabetes mellitus, systemic lupus erythematosus, hematopoietic cell lineage, etc. (Fig. 5B,E). And EFNB1 was involved in these pathways in high expressed cohort, while RBBP4 enriched in these pathways in low expressed cohort (Fig. 5B,E). We speculated that the reason for this result might be due to too few samples in the training set or similar genes functions.
Drug analysis
We generated drug-gene interaction networks using the DGIdb database. The analysis revealed predictions for a total of 5 small molecule drugs (Table 1). There were 3 small molecule drugs predicted for NCOR2, namely benzbromarone, 9,10-phenanthrenequinone, and chembl312032 (Fig. 6). For SOCS1, 2 small molecule drugs were predicted, namely insulin and aldesleukin (Fig. 6). However, no small molecule drugs that were predicted interacted with DAXX, EFNB1, and RBBP4.
Regulatory network and subcellular localization of AGAP2-AS1
The subcellular localization analysis indicated that AGAP2-AS1 predominantly localized within exosomes and the cytoplasm (Fig. 7A). The AGAP2-AS1 expression between PE and control cohorts was significantly different, and markedly lower in the PE cohort (Fig. 7B). Subsequently, we found the constructed ceRNA network containing AGAP2-AS1, and the regulatory network of AGAP2-AS1 was extracted (Fig. 7C). The regulatory network included 41 mRNAs, 2 miRNAs, and 1 lncRNA, which contained the regulated relationship pairs AGAP2-AS1-hsa-miR-497-5p-SRPRB, and AGAP2-AS1-hsa-miR-195-5p-RPL36 (Fig. 7C). Significant differences were observed in the expression levels of both miRNAs and mRNAs between the PE and control cohorts (Fig. 7D-E). The 2 miRNAs were both up-regulated in PE cohorts, and most mRNAs were also significantly up-regulated in PE cohort (Fig. 7D-E). Correlation analysis showed that AGAP2-AS1 had a significant strong or extremely strong correlation with 27 mRNAs, such as PAFAH1B1, SKI, PNPLA6, etc. (Fig. 7F).
Construction of AGAP2-AS1 regulatory subnetwork. (A) Subcellular localization of AGAP2-AS1. (B) The expression of AGAP2-AS1 between PE and control cohorts. (C) Construction of AGAP2-AS1 regulatory network. Blue circles indicate lncRNA, green circles indicate miRNA, red circles indicate mRNA. (D) The expression of hsa-miR-195-5p and hsa-miR-497-5p between PE and control cohorts. (E) The expression of mRNA between PE and control cohorts. (F) The heatmap of correlation between AGAP2-AS1 and mRNA. (G) GO enrichment analysis of mRNAs. (H) KEGG enrichment analysis of mRNAs. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The results of enrichment analysis of the mRNAs demonstrated that there were 415 GO BP, 23 GO CC, 28 GO MF, and 5 KEGG pathways enriched (Supplementary Tables 3–4), including cerebral cortex radial glia-guided migration, platelet alpha granule lumen, sulfur compound binding, VEGF signaling pathway, etc. (Fig. 7G-H).
Validation of the expression of biomarkers in clinical samples
The expression of biomarkers was further assessed via real time quantitative-Polymerase Chain Reaction (RT-qPCR). DAXX, EFNB1, NCOR2, and SOCS1 were highly expressed and RBBP4 was lowly expressed in the PE cohort (Fig. 8A-E). Nevertheless, the expression levels of DAXX, SOCS1, and RBBP4 did not exhibit substantial variations between the PE and control cohorts (Fig. 8A,D,E). Overall, the expression trends were in line with findings from public database.
RT-qPCR analysis of expression levels of five hub genes in serum and placenta samples between PE and control cohorts. DAXX (A), EFNB1 (B), NCOR2 (C), SOCS1 (D) and RBBP4 (E). The error bar indicates standard error (SE). ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Discussion
PE is a significant pregnancy complication and has emerged as a primary cause of maternal and neonatal deaths worldwide due to its unknown mechanism and poor drug treatment, exploring the potential biomarkers related to its pathogenesis is of important for the prevention and diagnosis of PE29. PE is also associated with maternal and neonatal health problems in pregnant women, such as maternal-associated chronic hypertension, myocardial ischemia, end-stage renal disease, placental abruption, intrauterine fetal growth restriction, neonatal-associated bronchopulmonary dysplasia, and cognitive impairments30. Moreover, PE is associated with maternal and neonatal health problems in pregnant women, such as maternal-associated chronic hypertension, myocardial ischemia, end-stage renal disease, intrauterine fetal growth restriction, neonatal-associated bronchopulmonary dysplasia and cognitive impairment31,32.
In this study, we downloaded PE GeneChip datasets GSE96983, GSE96984 and GSE24129 from the GEO database, and screened 251 DEGs by bioinformatics analysis and validation of the RFalgorithm, followed by downscaling of the differential genes to obtain 17 candidate hub genes, including Ras-related C3 botulinum toxin substrate 1 (RAC1), protein kinase C epsilon (PRKCE), T-lymphoma invasion and metastasis 1 (TIAM1), Janus kinase 1 (JAK1), ephrin B1 (EFNB1), Death domain-associated protein (DAXX), protein tyrosine phosphatase non-receptor type 13 (PTPN13), mitogen-activated protein kinase kinase kinase 5 (MAP3K5), nuclear receptor corepressor 2 (NCOR2), RB binding protein 4 (RBBP4), myosin heavy chain 14 (MYH14), adenylate cyclase 1 (ADCY1), Suppressor of cytokine signaling (SOCS1), Forkhead box O1 (FOXO1), Wnt family member 5 A (WNT5A), Nuclearrespiratoty factor 2 (NF2), and vascular endothelial growth factor A (VEGFA). The 17 candidate keys were analyzed in other datasets GSE96984 and GSE24129 for the expression of candidate hub genes, resulting in 5 hub genes with significant differences and consistent expression pattern, namely DAXX, EFNB1, NCOR2, RBBP4, and SOCS1.
Previous research reveals that the DAXX gene mRNA is a newly identified histone chaperone playing a role in the regulation of chromatin architecture and that DAXX is associated with trophoblast differentiation during placental development, with significantly elevated expression in patients with PE33,34. Sabri N et al. demonstrated that tumor suppressor Speckle-type POZ protein regulates tumor progression by modulating the polyubiquitination activity of DAXX35. Additionally, treatment with the HPV16 E7 protein can increase the expression of DAXX protein. Conversely, disrupting DAXX expression and using agonists for JNK can diminish the inhibitory effects of HPV16 E7 on TNF-α-induced apoptosis. This indicates that the DAXX/JNK pathway might be implicated in the anti-apoptotic function of HPV16 E7 36. Currently, no literature has been found linking NCOR2 with preeclampsia. We are the first to identify NCOR2 as a biomarker associated with PE, thereby expanding research in this field and providing new therapeutic targets. In our study, the similarity scores of DAXX and NCOR2 genes were greater than 0.6, indicating that these two genes are strongly similar and both are upregulated in PE.
EFNB1 (Ephrin-B1) is expressed in chorionic capillary endothelial cells during early and mid-gestation. Inhibition of EFNB1 suppresses HTR8/SVneo cell invasiveness through downregulation of MMP2 and MMP9; and impairs spiral artery remodeling by decreasing placental growth factor expression, leading to PE pathogenesis37,38. Previous studies have indicated that EFBN1 plays a vital role in promoting autophagy in colonic epithelial cells, thereby contributing to the maintenance of intestinal homeostasis and the regulation of blood pressure in humans39,40.
There are no reports have detected the association of RBBP4 with the pathogenesis of PE. However, it has been surfaced that RBBP4 is involved in the overlapping functions of the regulation of cell proliferation, apoptosis, and the deposition of histone H3.3 during the preimplantation development of mouse embryos, which is thought to be a crucial component in the pathogenesis of PE41,42,43. Liu H et al. showed that SOCS1 negatively regulates the JAK/STAT1 signaling pathway and affects placental trophoblast invasiveness, and the assessment of clinical specimens revealed elevated levels of SOCS1 and IFN-γ expression in patients diagnosed with PE compared to those in the healthy control cohort44. Mayor-Lynn K et al. showed that SOCS1 can alter normal placental function by regulating cellular activity, leading to the development and progression of PE45. Our study analysis demonstrated a significant upregulation of EFNB1 and SOCS1, as well as a notable downregulation of RBBP4, in PE, corroborating the aforementioned observations.
The DGIdb database predicts the interaction of drugs with the hub gene and finds 5 potential small molecule drugs for the treatment of eclampsia, which provides new ideas for the treatment of eclampsia. We identified a total of five target drugs 9,10-PHENANTHRENEQUINONE, HEMBL312032, BENZBROMARONE, INSULIN, ALDESLEUKIN, which could be influential in the future management of individuals suffering from PE. INSULIN has been reported to be involved in the pathogenesis of PE, and its distinct process might be linked to the crucial function of insulin signaling in the differentiation, longevity, and effector actions of immune cells, primarily through the standard activation of the PI3K/Akt/mTOR pathway46,47. Hyperinsulinemia, characterized by insulin resistance or aggressive insulin treatment, might directly contribute to immune cell impairment, which is a key factor in the onset of PE48,49.
We extracted the AGAP2-AS1-related regulatory sub-network from ceRNA network and performed Gene Set Enrichment Analysis (GSEA) analysis. AGAP2-AS1, a 1567 nucleotide lncRNA situated on chromosome 12q14.1, has been identified as being unusually expressed in multiple diseases, such as cancer50,51. Decreased expression of AGAP2-AS1 in PE has been previously reported, whereas overexpression of AGAP2-AS1 in cells enhances the growth, invasion, and movement of trophoblast cells and hinders the programmed cell death of these cells.AGAP2-AS1 stabilizes trophoblast cells and reduces apoptosis by sponging the miR-574-5p Promoter-Depressor Protein Jun Dimerization Protein 2 (JDP2), and our findings are consistent with previous reports28. In brain tumor research, AGAP2-AS1 is involved in the regulation of the Wnt signaling pathway through its interaction with miR-15a/b-5p, thereby influencing the pathogenesis, diagnosis, prognosis, and treatment of brain tumors52. Another study on renal cell carcinoma (RCC) revealed a different pathway, where IGF2BP3 stabilizes AGAP2-AS1, allowing it to competitively bind with miR-9-5p, consequently upregulating the expression of Thrombospondin-2 (THBS2) and activating the PI3K/AKT signaling pathway. This process induces M2 polarization of macrophages and promotes the progression of RCC53. Additionally, in studies on glioblastoma (GBM), AGAP2-AS1 in exosomes (Exo-AGAP2-AS1) was found to inhibit the function of miR-486-3p through a sponging mechanism, thereby regulating the expression of Transforming Growth Factor β1 (TGF-β1) in myeloid-derived suppressor cells (MDSCs). This regulatory mechanism promotes the development of GBM cells and provides new potential targets for GBM treatment54. We followed up with GSEA analysis, and we found that AGAP2-AS1 was mainly enriched in the VEGF pathway as well as the EGFR pathway and MAPK pathway. Previously, it has been reported in the literature that VEGF is involved in the formation of gestational diabetes and peeclampsia55,56. MAPK has been used as an important pathway for stabilizing trophoblast cells and an important mechanism in the pathogenesis of PE57. Our study is consistent with previous reports. By correlation analysis, we found that AGAP2-AS1 may affect the pathogenesis of PE by regulating genes such as TRIM35 to influence the expression of DAXX, EFNB1, NCOR2, RBBP4 and SOCS1.
Research has found that lncRNAs in biological fluids, as promising biomarkers, have shown significant emerging relevance in the risk stratification of pregnancy-related complications (PRC), including preeclampsia (PE). Studies on the expression patterns of lncRNAs and their potential clinical applications hold diagnostic, prognostic, and therapeutic value, paving the way for innovative approaches to improve prenatal care and the prognosis of pregnant women and fetuses58. Additionally, Xiao-Hong Wei and others have discovered that the long non-coding RNA (lncRNA) DUXAP8 is upregulated in the placental tissues of patients with preeclampsia and is significantly correlated with multiple clinical indices. This finding reveals the crucial role of DUXAP8 in regulating the biological behavior of trophoblasts through the FAM134B-mediated endoplasmic reticulum (ER) phagocytosis process, thus providing new theoretical support and perspectives for exploring the pathogenesis of preeclampsia (PE)59.However, further studies are needed to determine how LNC RNA affects the development of PE. Currently, although there is no direct evidence to suggest an interaction between lncRNA AGAP2-AS1 and the five core genes, existing research has revealed that the downregulation of lncRNA AGAP2-AS1 could act as a key inhibitory factor in preeclampsia (PE) by competitively inhibiting JDP2 at the post-transcriptional level through miR-57428. Given the direct or indirect association between these five core genes and preeclampsia, it is reasonable to speculate that there might be some as yet undefined but significant connections between AGAP2-AS1 and these core genes. This potential mechanism warrants further investigation, and our current findings lay a theoretical foundation and direct the path for future scientific exploration.
Conclusion
In this study, we established a ceRNA regulatory network and identified hub genes of PE, namely DAXX, EFNB1, NCOR2, RBBP4 and SOCS1, which provided new ideas for the study of the pathogenesis of PE. However, this study is limited by its small sample size. This may weaken the statistical power and introduce sample bias, thereby introducing uncertainties or errors in the research results, which reduces the credibility of the conclusions. To overcome this limitation, we plan to enhance the statistical power in future research by increasing the sample size. This will help to reduce potential biases and errors, ensuring the scientific integrity and rigor of the experimental design, thereby improving the reliability and scientific validity of the research findings.We will follow up with an in-depth study on how LncRNA regulates the relationship between AGAP2-AS1 and DAXX, EFNB1, NCOR2, RBBP4 and SOCS1.
Materials and methods
Data sources
The datasets related to PE (GSE96983, GSE96984, and GSE24129) were retrieved from the GEO repository, accessible through the https://www.ncbi.nlm.nih.gov/. The samples of all three data sets are placental samples. The dataset GSE96983 consists of three samples from individuals with PE and four samples from healthy controls. The dataset GSE96984 comprises three samples from PE subjects and four samples from control subjects. The GSE96983 and GSE96984 datasets were from the same cohort of patients, with patients aged 26, 28, 29, 30, 31, 32, and 33, respectively. The mRNA sequencing data (mRNA-seq) and lncRNA sequencing data (lncRNA-seq) were downloaded from the GSE96984. The miRNA sequencing data (miRNA-seq) was downloaded from the GSE96983. The mRNA-seq was downloaded from the GSE24129 to perform validation, and the GSE24129 included 8 PE samples and 8 control samples60. In the GSE24129 dataset, all placenta biopsy samples originated from collections following caesarean section procedures. To ensure that the delivery process did not potentially affect the gene expression profiles of the tissue samples, placentas from women who had never experienced a delivery were purposely selected as the source of the samples.
Creation of the competing endogenous RNA (ceRNA) network
Firstly, the gene expression matrix of three PE samples and four control samples in the GSE96984 dataset was analyzed by the voom method in the “limma” R package (version 3.48.3) (|log2FC| > 0.5 and P < 0.05)61, and DE-mRNAs and DE-lncRNAs between the PE groups and the control groups were identified. Subsequently, DE-miRNAs between PE and control were analyzed in the GSE96983 dataset (|log2FC| > 0.5 and P < 0.05). Finally, volcano plots were drawn using the “ggplot2” R package (version 3.3.5)62 to show DE-lncRNAs, DE-miRNAs and DE-mRNAs, and heatmaps of differential expression were drawn using the “pheatmap” R package (version 1.0.12)63. The target miRNAs of DE-lncRNAs were predicted by StarBase (http://starbase.sysu.edu.cn) and miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw), respectively, and the screened miRNAs were taken to intersect to obtain candidate miRNAs, and its intersection with DE-miRNAs was taken to obtain overlapping miRNAs. Similarly, the target mRNAs of DEmiRNAs were predicted by StarBase (http://starbase.sysu.edu.cn) and miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw), respectively, and the screened mRNAs were intersected to obtain candidate mRNAs, whose intersection with DE-mRNAs was taken to obtain overlapping mRNAs. Subsequently, the Cytoscape software (version 3.9.1) was used to integrate the interactions of DElncRNAs and overlapping miRNAs and overlapping mRNAs, to construct the lncRNAs-miRNAs-mRNAs regulatory network with overlapping miRNAs regulating both DElncRNAs and overlapping mRNAs as a complete ceRNA. Finally, the overlapping mRNAs were enriched (P < 0.05 and counts ≥ 1) by the “ClusterProfiler” R package (version 4.0.2)64 to look for common functions and related pathways. The top 10 GO-enriched functions and KEGG-enriched pathways were plotted using the “ggplot2” R package (version 3.3.5)65,66,67.
Screening of biomarkers
The STRING database was utilized to construct the PPInetwork of the identified mRNAs, considering only interactions with a confidence score higher than 0.9. Then, the BottleNeck, EcCentricity, Closeness, Betweenness, and Stress in Cytoscape were utilized to identify hub genes. The hub genes were identified by intersecting the top 30 genes obtained from five different algorithms. The hub genes underwent additional screening based on Receiver Operating Characteristic (ROC) curves. Candidate genes were identified among the hub genes based on an area under the curve (AUC) value exceeding 0.7. Subsequently, the expression levels of candidate genes in both PE and control groups were examined using datasets GSE96984 and GSE24129. The candidate genes that were significantly different between PE and control cohorts and had the same expressive trend in both datasets were used for subsequent analysis and noted as biomarkers. The functional similarity among the biomarkers was assessed via “GOSemSim” R package (version 2.18.1)68,69. Lastly, the correlation between the identified biomarkers was determined using the Spearman correlation coefficient.
Functional analysis
To investigate the enrichment pathways associated with the biomarkers, we conducted GSEA using the “clusterProfiler” R package (version 4.0.2) along with org.Hs.eg.db (version 3.13.0)64. The reference gene sets used for analysis were obtained by downloading the “C2.cp.kegg.v7.5.1.symbols.gmt” and “C2.go.v7.5.entrez.gmt” datasets. Based on the median expression value of the identified biomarkers, the samples from GSE96984 dataset were categorized into cohorts with high and low expression levels, and differential analysis was carried out. The all genes were ranked via log fold change (logFC). The threshold value were |normalized enrichment score (NES)| > 1, NOM P < 0.05, and q < 0.25.
Prediction of small molecule drugs
Each biomarker was utilized as a keyword to predict potential small molecule drugs that interact with the biomarkers in the Drug-Gene Interaction Database (DGIdb, available at https://dgidb.org). Subsequently, the biomarker-drug network was visualized using Cytoscape software (version 3.7.2)70.
Creation of regulatory network and subcellular localization of AGAP2-AS1
Firstly, the subcellular localization of AGAP2-AS1 was performed through lncLocator Database (www.csbio.sjtu.edu.cn/bioinf/lncLocator). Then, the AGAP2-AS1 expression between PE and control cohorts was compared via Wilcox test. The regulatory network of AGAP2-AS1 was extracted from the constructed ceRNA network. The expression of AGAP2-AS1-regulated miRNAs and mRNAs between PE and control were analyzed. Additionally, the correlation of AGAP2-AS1 with AGAP2-AS1-regulated mRNAs was calculated by Pearson. The 0.0 < |r| < 0.2 was extremely weak or no correlation, 0.2 ≤ |r| <0.4 was weak correlation, 0.4 ≤ |r| < 0.6 was moderate correlation, 0.6 ≤ |r| < 0.8 was strong correlation, and 0.8 ≤ |r| < 1.0 was extremely strong correlation. Afterwards, the enrichment analysis were carried out for mRNAs via “ClusterProfiler” R package (version 4.0.2) (P < 0.05 and count ≥ 1)64.
Differential gene correlation analysis with LncRNA regulated genes
In order to delve deeper into the association between lncRNA regulated genes and DE-mRNA in PE, we performed a correlation analysis.Genes with significant positive correlation between LncRNA and DE-mRNA were selected for correlation analysis. Spearman’s correlation analysis was employed to assess the correlation between the expression of DE-mRNA and the data obtained from GEO datasets on the expression of regulatory lncRNAs.
Sample collection and RT-qPCR
A total of 8 PE patients and 8 controls at Affiliated Hospital of Guizhou Medical University from January 2023 to June 2023 were enrolled in this study.All PE patients were diagnosed according to the 2020 guidelines for hypertensive disorders of pregnancy issued by the American College of Obstetricians and Gynecologists (ACOG)2. This study did not involve human in vivo experiments or human transplantation-related studies and has been conducted in accordance with the Declaration of Helsinki, which has been obtained from all participants and informed consent.The exclusion criteria were as follows: pregnant women under the age of 18, smoking, alcohol addiction, history of diabetes, anemia, dyslipidemia, autoimmune diseases, malignancy, or gastrointestinal surgery. After the termination of pregnancy, fetal side placental tissues were immediately collected under sterile conditions, approximately 10*10 mm in size, washed with saline, and rapidly frozen at -80 °C. And the Serum and placenta samples were collected to perform RT-qPCR. This study was approved by Ethics Committee of Affiliated Hospital of Guizhou Medical University, Approval Number 2022(410). All participants provided their informed consent by signing a consent form prior to their involvement in the study. The RNA was extracted by TRIzol (Ambion, Austin, USA) in accordance with the instructions provided by the manufacturer. SureScript-First-strand-cDNA-synthesis-kit (Servicebio, Wuhan, China) was utilized to perform reverse transcription analysis. The RT-qPCR analysis was performed using the 2xUniversal Blue SYBR Green qPCR Master Mix (Servicebio, Wuhan, China). The primer sequences used for the analysis can be found in Table 2. The expression levels of biomarkers were quantified using the 2−ΔΔCt method, with normalization to the mRNA levels of GAPDH71.
Data statistics
Statistical analysis was conducted using R software (version 4.2.0) (https://www.r-project.org) and GraphPad Prism v9.0. Differences between two cohorts were analyzed using ANOVA and Wilcoxon’s test for multiple cohorts. The difference in OS between cohorts was estimated using a log-rank test and Kaplan-Meier (K-M) analysis. Pearson’s correlation test was utilized to determine the associations among subtypes, clinicopathological features, risk scores, immune checkpoint expression, methyltransferases, and levels of immune infiltration. The obtained results were deemed statistically significant at a significance level of P < 0.05.
Data availability
The datasets used and analyzed in the current study are available from the GEO database (https://www.ncbi.nlm.nih.gov/gds) [GSE96983, GSE96984 and GSE24129], StarBase (Http://starbase.sysu.edu.cn), miRTarBase Database (Http://mirtarbase.mbc.nctu.edu.tw), DGIdb database (https://dgidb.org), lncLocator Database (www.csbio.sjtu.edu.cn/bioinf/lncLocator).
Abbreviations
- PE:
-
Preeclampsia
- ceRNA:
-
Competing endogenous RNA
- GEO:
-
Gene Expression Omnibus
- DE-mRNAs:
-
Differentially expressed message RNAs
- DE-lncRNAs:
-
Differentially expressed long non-coding RNAs
- DE-miRNAs:
-
Differentially expressed microRNAs
- PPI:
-
Protein-protein intersection
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- lncRNAs:
-
Non-coding RNAs
- mRNA-seq:
-
mRNA sequencing
- lncRNA-seq:
-
lncRNA sequencing
- miRNA-seq:
-
miRNA sequencing
- GSEA:
-
Gene Set Enrichment Analysis
- NES:
-
Normalized enrichment score
- GO:
-
Gene Ontology
- CC:
-
Cellular component
- BP:
-
Biological process
- MF:
-
Molecular function
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- RT-qPCR:
-
Real time quantitative-polymerase chain reaction
References
Magee, L. A. et al. Guideline 426: Hypertensive disorders of pregnancy: Diagnosis, prediction, prevention, and management. J. Obstet. Gynaecol. Can. 44, 547–71e1. https://doi.org/10.1016/j.jogc.2022.03.002 (2022).
Gestational hypertension and preeclampsia: ACOG Practice Bulletin Summary, Number 222. Obstet. Gynecol. 135, 1492–1495. (2020). https://doi.org/10.1097/aog.0000000000003892
Ives, C. W., Sinkey, R., Rajapreyar, I., Tita, A. T. N. & Oparil, S. Preeclampsia-pathophysiology and clinical presentations: JACC state-of-the-art review. J. Am. Coll. Cardiol. 76, 1690–1702. https://doi.org/10.1016/j.jacc.2020.08.014 (2020).
Armaly, Z., Jadaon, J. E., Jabbour, A. & Abassi, Z. A. Preeclampsia: Novel mechanisms and potential therapeutic approaches. Front. Physiol. 9, 973. https://doi.org/10.3389/fphys.2018.00973 (2018).
Hao, S. et al. Changes in pregnancy-related serum biomarkers early in gestation are associated with later development of preeclampsia. PLoS One. 15, e0230000. https://doi.org/10.1371/journal.pone.0230000 (2020).
Kristensen, J. H., Basit, S., Wohlfahrt, J., Damholt, M. B. & Boyd, H. A. Pre-eclampsia and risk of later kidney disease: Nationwide cohort study. Bmj. 365, l1516. https://doi.org/10.1136/bmj.l1516 (2019).
Rana, S., Lemoine, E., Granger, J. P. & Karumanchi, S. A. Preeclampsia: Pathophysiology, challenges, and perspectives. Circ. Res. 124, 1094–1112. https://doi.org/10.1161/circresaha.118.313276 (2019).
Klinge, C. M. Non-coding RNAs: Long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr. Relat. Cancer. 25, R259–r82. https://doi.org/10.1530/erc-17-0548 (2018).
López-Camarillo, C. et al. Deciphering the long non-coding RNAs and MicroRNAs coregulation networks in ovarian cancer development: An overview. Cells. 10 https://doi.org/10.3390/cells10061407 (2021).
Huang, Z., Zhou, J. K., Peng, Y., He, W. & Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer. 19, 77. https://doi.org/10.1186/s12943-020-01188-4 (2020).
Greco, S., Gaetano, C. & Martelli, F. Long noncoding competing endogenous RNA networks in age-associated cardiovascular diseases. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20123079 (2019).
Li, P., Duan, S. & Fu, A. Long noncoding RNA NEAT1 correlates with higher disease risk, worse disease condition, decreased miR-124 and miR-125a and predicts poor recurrence-free survival of acute ischemic stroke. J. Clin. Lab. Anal. 34, e23056. https://doi.org/10.1002/jcla.23056 (2020).
Fok, E. T., Davignon, L., Fanucchi, S. & Mhlanga, M. M. The lncRNA connection between cellular metabolism and epigenetics in trained immunity. Front. Immunol. 9, 3184. https://doi.org/10.3389/fimmu.2018.03184 (2018).
Zhang, Q., Wang, Z., Cheng, X. & Wu, H. lncRNA DANCR promotes the migration an invasion and of trophoblast cells through microRNA-214-5p in preeclampsia. Bioengineered. 12, 9424–9434. https://doi.org/10.1080/21655979.2021.1988373 (2021).
Xu, Y. et al. A novel regulatory mechanism network mediated by lncRNA TUG1 that induces the impairment of spiral artery remodeling in preeclampsia. Mol. Ther. 30, 1692–1705. https://doi.org/10.1016/j.ymthe.2022.01.043 (2022).
Matsubara, K., Matsubara, Y., Uchikura, Y. & Sugiyama, T. Pathophysiology of Preeclampsia: The role of exosomes. Int. J. Mol. Sci. 22 https://doi.org/10.3390/ijms22052572 (2021).
Zhang, H. et al. Mir-30-5p-mediated ferroptosis of trophoblasts is implicated in the pathogenesis of preeclampsia. Redox Biol. 29, 101402. https://doi.org/10.1016/j.redox.2019.101402 (2020).
Wu, D. et al. Long noncoding RNA 00473 is involved in preeclampsia by LSD1 binding-regulated TFPI2 transcription in trophoblast cells. Mol. Ther. Nucleic Acids. 12, 381–392. https://doi.org/10.1016/j.omtn.2018.05.020 (2018).
Penkala, I. et al. lncRHOXF1, a long noncoding RNA from the X chromosome that suppresses viral response genes during development of the early human placenta. Mol. Cell. Biol. 36, 1764–1775. https://doi.org/10.1128/mcb.01098-15 (2016).
Mattick, J. S. et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell. Biol. 24, 430–447. https://doi.org/10.1038/s41580-022-00566-8 (2023).
Nemeth, K., Bayraktar, R., Ferracin, M. & Calin, G. A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. https://doi.org/10.1038/s41576-023-00662-1 (2023).
Shang, R., Lee, S., Senavirathne, G. & Lai, E. C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 24, 816–833. https://doi.org/10.1038/s41576-023-00611-y (2023).
Mas, A. M. & Huarte, M. Long noncoding RNA signatures as cancer biomarkers. J. Clin. Oncol. 41, 3059–3062. https://doi.org/10.1200/jco.23.00381 (2023).
Kimura, M., Kothari, S., Gohir, W., Camargo, J. F. & Husain, S. MicroRNAs in infectious diseases: Potential diagnostic biomarkers and therapeutic targets. Clin. Microbiol. Rev. e0001523. https://doi.org/10.1128/cmr.00015-23 (2023).
Mora-Palazuelos, C. et al. The role of ncRNAs in the immune dysregulation of preeclampsia. Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms242015215 (2023).
Lv, Y. et al. Roles of microRNAs in preeclampsia. J. Cell. Physiol. 234, 1052–1061. https://doi.org/10.1002/jcp.27291 (2019).
Zhang, H. W. & Shen, L. K. LncRNA AGAP2-AS1 in cancer: An insufficiently explored and controversial research area. Dig. Liver Dis. https://doi.org/10.1016/j.dld.2023.11.008 (2023).
Xu, Y. et al. Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. J. Cell. Mol. Med. 24, 4557–4568. https://doi.org/10.1111/jcmm.15113 (2020).
Chappell, L. C., Cluver, C. A., Kingdom, J. & Tong, S. Pre-eclampsia. Lancet. 398, 341–354. https://doi.org/10.1016/s0140-6736(20)32335-7 (2021).
Gupta, S., Petras, L., Tufail, M. U., Rodriguez Salazar, J. D. & Jim, B. Hypertension in pregnancy: What we now know. Curr. Opin. Nephrol. Hypertens. 32, 153–164. https://doi.org/10.1097/mnh.0000000000000857 (2023).
Turbeville, H. R. & Sasser, J. M. Preeclampsia beyond pregnancy: Long-term consequences for mother and child. Am. J. Physiol. Ren. Physiol. 318, F1315–f26. https://doi.org/10.1152/ajprenal.00071.2020 (2020).
Phipps, E. A., Thadhani, R., Benzing, T. & Karumanchi, S. A. Pre-eclampsia: Pathogenesis, novel diagnostics and therapies. Nat. Rev. Nephrol. 15, 275–289. https://doi.org/10.1038/s41581-019-0119-6 (2019).
Novakovic, B., Evain-Brion, D., Murthi, P., Fournier, T. & Saffery, R. Variable DAXX gene methylation is a common feature of placental trophoblast differentiation, preeclampsia, and response to hypoxia. Faseb j. 31, 2380–2392. https://doi.org/10.1096/fj.201601189RR (2017).
Drané, P., Ouararhni, K., Depaux, A., Shuaib, M. & Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265. https://doi.org/10.1101/gad.566910 (2010).
Sabri, N. et al. Reduction of oligomer size modulates the competition between cluster formation and phase separation of the tumor suppressor SPOP. J. Biol. Chem. 299, 105427. https://doi.org/10.1016/j.jbc.2023.105427 (2023).
Ding, S. et al. HPV16 E7 protein antagonizes TNF-α-induced apoptosis of cervical cancer cells via Daxx/JNK pathway. Microb. Pathog. 185, 106423. https://doi.org/10.1016/j.micpath.2023.106423 (2023).
Adu-Gyamfi, E. A. et al. Ephrin and eph receptor signaling in female reproductive physiology and pathology†. Biol. Reprod. 104, 71–82. https://doi.org/10.1093/biolre/ioaa171 (2021).
Luo, Q. et al. Ephrin-B2 mediates trophoblast-dependent maternal spiral artery remodeling in first trimester. Placenta. 36, 567–574. https://doi.org/10.1016/j.placenta.2015.02.009 (2015).
Zhang, H. et al. RNF186 regulates EFNB1 (ephrin B1)-EPHB2-induced autophagy in the colonic epithelial cells for the maintenance of intestinal homeostasis. Autophagy. 17, 3030–3047. https://doi.org/10.1080/15548627.2020.1851496 (2021).
Wu, Z. et al. Possible role of Efnb1 protein, a ligand of eph receptor tyrosine kinases, in modulating blood pressure. J. Biol. Chem. 287, 15557–15569. https://doi.org/10.1074/jbc.M112.340869 (2012).
Nunode, M. et al. Mir-515-5p suppresses trophoblast cell invasion and proliferation through XIAP regulation in preeclampsia. Mol. Cell. Endocrinol. 559, 111779. https://doi.org/10.1016/j.mce.2022.111779 (2023).
Xiao, L. et al. Overlapping functions of RBBP4 and RBBP7 in regulating cell proliferation and histone H3.3 deposition during mouse preimplantation development. Epigenetics. 17, 1205–1218. https://doi.org/10.1080/15592294.2021.1999006 (2022).
Tang, R. et al. The gut microbiota dysbiosis in preeclampsia contributed to trophoblast cell proliferation, invasion, and migration via lncRNA BC030099/NF-κB pathway. Mediators Inflamm. 2022, 6367264. (2022). https://doi.org/10.1155/2022/6367264
Liu, H., Wang, W. & Liu, C. Increased expression of IFN-γ in preeclampsia impairs human trophoblast invasion via a SOCS1/JAK/STAT1 feedback loop. Exp. Ther. Med. 21, 112. https://doi.org/10.3892/etm.2020.9544 (2021).
Mayor-Lynn, K., Toloubeydokhti, T., Cruz, A. C. & Chegini, N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod. Sci. 18, 46–56. https://doi.org/10.1177/1933719110374115 (2011).
Ramasubbu, K. & Devi Rajeswari, V. Impairment of insulin signaling pathway PI3K/Akt/mTOR and insulin resistance induced AGEs on diabetes mellitus and neurodegenerative diseases: A perspective review. Mol. Cell. Biochem. 478, 1307–1324. https://doi.org/10.1007/s11010-022-04587-x (2023).
Peterson, C. & Chandler, H. L. Insulin facilitates corneal wound healing in the diabetic environment through the RTK-PI3K/Akt/mTOR axis in vitro. Mol. Cell. Endocrinol. 548, 111611. https://doi.org/10.1016/j.mce.2022.111611 (2022).
van Niekerk, G., Christowitz, C. & Engelbrecht, A. M. Insulin-mediated immune dysfunction in the development of preeclampsia. J. Mol. Med. (Berl). 99, 889–897. https://doi.org/10.1007/s00109-021-02068-0 (2021).
Salsoso, R., Mate, A., Toledo, F., Vázquez, C. M. & Sobrevia, L. Insulin requires A(2B) adenosine receptors to modulate the L-arginine/nitric oxide signalling in the human fetoplacental vascular endothelium from late-onset preeclampsia. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 165993. https://doi.org/10.1016/j.bbadis.2020.165993 (2021).
Chen, J. & Wang, B. AGAP2-AS1: an indispensable lncRNA in tumors. Mini Rev. Med. Chem. 23, 336–342. https://doi.org/10.2174/1389557522666220615154227 (2023).
Bao, Z., Zheng, Q. & Li, L. Oncogenic roles and mechanisms of lncRNA AGAP2-AS1 in human solid tumors. Am. J. Transl Res. 13, 757–769 (2021).
Rahmani, F. et al. Interplay between lncRNA/miRNA and Wnt/ss-catenin signaling in brain cancer tumorigenesis. EXCLI J. 22, 1211–1222. https://doi.org/10.17179/excli2023-6490 (2023).
Xu, P. et al. LncRNA AGAP2 antisense RNA 1 stabilized by insulin-like growth factor 2 mRNA binding protein 3 promotes macrophage M2 polarization in clear cell renal cell carcinoma through regulation of the microRNA-9-5p/THBS2/PI3K-Akt pathway. Cancer Cell. Int. 23, 330. https://doi.org/10.1186/s12935-023-03173-5 (2023).
Tian, Y., Gao, X., Yang, X., Chen, S. & Ren, Y. Glioma-derived exosome Lncrna Agap2-As1 promotes glioma proliferation and metastasis by mediating Tgf-beta1 secretion of myeloid-derived suppressor cells. Heliyon. 10, e29949. https://doi.org/10.1016/j.heliyon.2024.e29949 (2024).
Sirico, A. et al. Placental diabesity: placental VEGF and CD31 expression according to pregestational BMI and gestational weight gain in women with gestational diabetes. Arch. Gynecol. Obstet. 307, 1823–1831. https://doi.org/10.1007/s00404-022-06673-3 (2023).
Waller, J. P., Howell, J. A., Peterson, H., George, E. M., Bidwell, G. L. III Elastin-like polypeptide: VEGF-B fusion protein for treatment of preeclampsia. Hypertension. 78, 1888–1901. https://doi.org/10.1161/hypertensionaha.121.17713 (2021).
Jiang, J. & Zhao, Z. M. LncRNA HOXD-AS1 promotes preeclampsia progression via MAPK pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 8561–8568. https://doi.org/10.26355/eurrev_201812_16618 (2018).
Cordier, A. G., Zerbib, E., Favier, A., Dabi, Y. & Darai, E. Value of non-coding RNA expression in biofluids to identify patients at low risk of pathologies associated with pregnancy. Diagnostics (Basel). 14 https://doi.org/10.3390/diagnostics14070729 (2024).
Wei, X. H. et al. Overexpression of long noncoding RNA DUXAP8 inhibits ER-phagy through activating AKT/mTOR signaling and contributes to preeclampsia. Cell. Mol. Life Sci. 81, 336. https://doi.org/10.1007/s00018-024-05385-y (2024).
Nishizawa, H. et al. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod. Biol. Endocrinol. 9, 107. https://doi.org/10.1186/1477-7827-9-107 (2011).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. https://doi.org/10.1093/nar/gkv007 (2015).
Gustavsson, E. K., Zhang, D., Reynolds, R. H., Garcia-Ruiz, S. & Ryten, M. Ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics. 38, 3844–3846. https://doi.org/10.1093/bioinformatics/btac409 (2022).
Wu, C. et al. Analysis of glutathione stransferase mu class 5 gene methylation as a prognostic indicator in low-grade gliomas. Technol. Health Care. https://doi.org/10.3233/THC-231316 (2024).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Camb). 2, 100141. https://doi.org/10.1016/j.xinn.2021.100141 (2021).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–d92. https://doi.org/10.1093/nar/gkac963 (2023).
Yu, G. et al. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 26, 976–978. https://doi.org/10.1093/bioinformatics/btq064 (2010).
Yu, G. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol. Biol. 2117, 207–215. https://doi.org/10.1007/978-1-0716-0301-7_11 (2020).
Otasek, D., Morris, J. H., Bouças, J., Pico, A. R. & Demchak, B. Cytoscape automation: Empowering workflow-based network analysis. Genome Biol. 20, 185. https://doi.org/10.1186/s13059-019-1758-4 (2019).
Raza, W., Guo, J., Qadir, M. I., Bai, B. & Muhammad, S. A. qPCR analysis reveals association of differential expression of SRR, NFKB1, and PDE4B genes with type 2 diabetes mellitus. Front. Endocrinol. (Lausanne). 12, 774696. https://doi.org/10.3389/fendo.2021.774696 (2021).
Funding
Supported by Guizhou Provincial Science and Technology Projects ZK[2024]-227,Science and Technology Fund of Guizhou Provincial Health Commission gzwkj 2023-056,Affiliated Hospital of Guizhou Medical University projects gyfynsfc[2023]-13.
Author information
Authors and Affiliations
Contributions
Fan Lu, Ni Zeng analyzed the data, conducted experiments, recruited patients, collected samples, drafted and revised the manuscript. Xiang Xiao and XingXing Wang collected the samples and analyzed the data. Han Gong analyzed the data. HouKang Lei developed the hypothesis, designed the study, and drafted the manuscript.All the authors have read and agreed to the final version of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by Ethics Committee of Affiliated Hospital of Guizhou Medical University, Approval Number 2022(410). All participants written informed consent by signing a consent form prior to their involvement in the study.
Informed consent
Patient consent has been obtained.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, F., Zeng, N., Xiao, X. et al. Exploring the ceRNA network involving AGAP2-AS1 as a novel biomarker for preeclampsia. Sci Rep 14, 27330 (2024). https://doi.org/10.1038/s41598-024-79224-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-79224-2