Abstract
Colorectal cancer (CRC) lymph node metastasis (LNM) is a crucial factor affecting the prognosis and treatment outcomes of CRC patients. It has been confirmed that altered glycosylation is a key event during CRC lymphatic metastases. Sialylation is one of the most significant glycosylation alterations in tumors. However, the predictive role of sialylation and sialylated protein in CRC remains elusive, especially in CRC-LNM. In this study, we explored and identified 1102 sialylated glycoproteins in CRC-LNM using metabolic labeling strategy and proteomics analysis. Combined with comprehensive analysis with bioinformatics and machine learning algorithms, we screened 25 prognostic sialylation-related genes (SRGs) to construct a new molecular phenotype (LRSRGs-Phenotype) and a prognostic SRG signature (LRSRGs-related Gene Signature) in CRC. Then, we further confirmed that patients in different phenotypes had different prognosis, molecular biological characteristics, immune cell infiltration and could be closely linked to three previously reported immune phenotypes: immune-excluded (Phenotype A), immune-desert (Phenotype B), and immune-inflamed (Phenotype C). Besides, we evaluated and validated the LRSRGs-related gene (ACADM, EHD4, FLOT1, GPC1, GSR, LRRC8A, NGFR, SDHB, and SEC61G) signature and found the risk score was an independent risk factor for CRC prognosis. CRC patients in different risk groups had different somatic mutation, tumor microenvironment and immunotherapy response. Finally, we also identified the potential therapeutic agents for CRC patients in different risk groups. In conclusion, we explored the key sialylated glycoproteins, which may play a key role in tumor LNM and clinical outcomes. And constructed the LRSRGs-phenotype and signature with prognostic and therapeutic predictive value in CRC, hoping to provide reliable scientific basis for future treatments in CRC patients.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the second most commonly diagnosed cancer (more than 1,900,000 cases) and the third most lethal (more than 930,000 cases) worldwide in 2020, and these numbers still appear an increasing trend annually1. Many CRC patients are diagnosed with advanced stages, such as lymph node metastasis (LNM) and distant metastasis2. It is well documented that CRC patients with LNM typically represent poor prognosis with a higher possibility of metastasis and recurrence3. Despite the advances in comprehensive treatments, including surgery, chemoradiotherapy, targeted therapy and immunotherapy, recurrence or metastasis remains challenging to manage, leading to worse prognosis4. Since lymphatic metastasis is considered an intermediate event between primary lesions and distant metastases, it is crucial to elaborate on the underlying metastasis mechanisms and identify applicable biomarkers or proteins for metastasis, especially for lymph node metastasis.
With the rapid development of scientific techniques and instruments, more and more researchers have been focusing on elucidating the relationship between post-translational modification and diseases, such as glycosylation. Glycosylation has been found to be involved in many indispensable physiological processes, including protein folding, degradation, secretion, intracellular signaling, cell adhesion, and motility5,6,7. Moreover, it has been shown to play a pivotal role during multiple tumorigenesis and progression7, such as brain cancer8, prostate cancer9, gastric cancer10, pancreatic cancer11, hepatocellular cancer12and colorectal cancer13,14. Notably, altered glycosylation is also considered a key events during lymphatic metastases15.
Among different forms of glycosylation, the most important alteration in tumor is sialylation, which is the process of transferring sialic acid (SA) from GMP-SA to the glycoprotein (membrane and secretory proteins) by sialyltransferases, consisting of a family of 20 different enzymes13. The dysregulation of sialylation is considered a hallmark of cancer and plays a pivotal role in tumorigenesis, development, and metastasis. Therefore, it is closely related to the prognosis of cancer patients16,17. However, what key sialylated glycoproteins are involved in and how sialylation regulates the process of LNM in CRC are still obscure.
In this study, we chose two CRC cell lines with different metastatic abilities derived from the same patients: SW480 from primary colon adenocarcinoma, and SW620 from lymph node metastasis18,19. Using metabolic labeling strategy and proteomics analysis combined with comprehensive analysis with bioinformatics, we explored and identified the key sialylated glycoproteins, which may play a key role in tumor LNM and clinical outcomes. Furthermore, we conducted convincing validation in various online datasets, CRC cell lines and tumor specimens, hoping to provide a reliable scientific basis for future treatments in CRC patients.
Materials and methods
Cell culture
The human SW480 and SW620 cell lines were purchased from National Collection of Authenticated Cell Cultures (NCACC, Shanghai, China), and cultured and maintained in Leibovitz’s L15 Medium (HyClone, Logan, Utah, USA) supplemented with 10% fetal bovine serum (FBS), under a humidified atmosphere at 37℃ without CO2 addition.
Transwell assay
Cell migratory and invasive ability was examined with Transwell (BD BioCoatTM control inserts, 8.0-mm inserts; BD Biosciences, Bedford, Massachusetts, USA). For migration, 1 × 105 cells in serum free medium were cultured in the upper chamber while medium containing 20% FBS was added to the lower chamber. After incubated 60 h, cells under the surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Then, the cells that crossed the membrane were photographed and numbered under optical microscope. For invasion, the bottom of the chamber was coated with Matrigel diluted at 1:5 with serum free medium.
Wound healing assay
Cells were seed in 6-well plates and an identical wound was made across the center after cells at 90–100% confluence, which then were cultured without fetal bovine serum. The pictures were captured at initial time and 24 h.
Metabolic labeling
As we described previously, SW480 or SW620, about 4 million cells were cultured and labeled with the Ac4ManNAz (ManNAz, 100 mM) or ManNAc (100 mM) for 60 h. To perform click chemistry, the cells were then incubated with Alkyne 555 and catalyst (CuSO4, NaVc, TBTA). Confocal microscope (Nikon, Tokyo, Japan) and flow cytometry (BD Biosciences, San Jose, California, USA) were used to confirm the labeling efficiency.
Flow cytometry analysis
As previously described20, cells were detached by trypsin containing 1 mM EDTA, and labeled with 1 µg/ml Alkyne 555 (Invitrogen, Eugene, Oregon, USA) for 30 min at RT temperature. Then, the cells were analyzed by flow cytometry (BD Biosciences, San Jose, California, USA).
Protein enrichment and mass spectrometry analysis
As we previously presented13, 1 × 107 metabolic labeled cells were used for the click reaction with biotinylated alkyne and glycoprotein enrichment. The biotin-labeled cells were disrupted by lysis buffer and then incubated with streptavidin-agarose resin for 12 h at RT temperature. Finally, the beads were pelleted by centrifugation and washed, reduced and alkylated followed by on bead trypsin digestion. The released peptide was conducted on a Q Exactive Plus (QE+) Orbitrap mass spectrometer with an Easy-nLC 1000. The human proteins database was referred for further analysis (Uniprot Complete Proteome – taxonomy Homo sapiens, release 2016.08) using MaxQuant software v. 1.5.5.1). The details were presented in supplementary materials.
Acquisition of data
The RNA sequencing (RNA-seq) data and relevant clinical information of CRC samples in the training cohort (TCGA-COAD and TCGA-READ) were obtained from the Cancer Genome Atlas (TCGA) database by the “TCGAbiolinks” R package21. Meanwhile, the RNA-seq data and corresponding clinical characteristics of CRC samples in the external validation cohort (GSE39582) were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/)22. Furthermore, the somatic mutation data of CRC samples in the training cohort was obtained from the website (https://portal.gdc.cancer.gov/). The sialylation-related genes (SRGs) were obtained from our results of mass spectrometry analysis.
Screening of LNM-related SRGs
The RNA-seq data in TCGA datasets was normalized into the transcripts per million (TPM) data and the log conversion was further performed. Then the RNA-seq data of SRGs was screened to perform subsequent analyses. The “limma” R package was applied to perform the differentially expressed gene (DEG) analysis between the LNM and no-LNM samples, screening out the LNM-related SRGs (LRSRGs)23. The “ggplot2” R package was used to visualize these results. Univariate Cox regression analysis was utilized to screen out the prognostic LRSRGs, which acted as the candidate genes for subsequent establishment of sialylation-related molecular subtype and prognostic gene signature.
Construction of a LRSRGs molecular phenotype
Based on the expression levels of the prognostic LRSRGs, the unsupervised clustering analysis was applied to construct the LNM- and sialylation-related molecular phenotypes in CRC. The ‘ConsensusClusterPlus’ R package was used to determine the optimal LNM- and Sialylation-related clustering numbers and phenotypes of CRC samples in the training cohort by the consensus clustering algorithm24. The ‘Rtsne’ and ‘ggplot2’ R packages were utilized to perform the Principal Component Analysis (PCA) analysis to show the LRSRGs expression difference among different molecular phenotypes.
Molecular and clinical significance of the LRSRGs molecular phenotypes
The ‘survival’ and ‘survminer’ R packages were used to calculate and visualize the prognostic difference among different molecular phenotypes. The ‘ComplexHeatmap’ R package was applied to show the variations in the clinical characteristics and the expression levels of 25 prognostic LRSRGs among different molecular phenotypes25. Using the single-sample gene set enrichment analysis (ssGSEA) algorithm in the ‘GSVA’ R package, the molecular significance of different molecular phenotypes was explored26. In addition, the ssGSEA method was also applied to estimate the immune cell infiltration of CRC individuals among different molecular phenotypes.
Establishment and validation of a prognostic LNM- and sialylation-related gene signature
Consistent with the previous methods27,28, the least absolute shrinkage and selection operator (LASSO) regression was applied to further screen hub prognostic LRSRGs by the ‘glmnet’ R package. The ‘igraph’ and ‘reshape2’ R packages were used to establish correlation network of these hub genes. The protein-protein interaction (PPI) network and Cytoscape software were applied to display the interaction of proteins among the proteins coding of the hub genes based on the STRING database (http://www.string-db.org/). Then, the multivariate Cox regression analysis based on these hub genes was applied to establish the prognostic LRSRGs signature. The regression coefficients of these genes and their corresponding mRNA expression levels were used to calculate the risk scores of each sample. The specific formula of risk score was as follows: risk score = ∑(mRNA expression levels × corresponding regression coefficients). The cut-off value of risk score (median value) was utilized to divide CRC individuals into high- and low-risk subgroups. Then, the PCA analysis and survival curves were also performed by the same methods as previously introduced27.
Analysis of independent prognostic factors and construction of nomogram prediction model
In order to assess the predictive value of the prognostic LRSRGs signature, the ‘timeROC’ R package was applied to perform the time-dependent Receiver Operating Characteristic (ROC) curves. In addition, the univariate and multivariate Cox regression analyses were also applied to evaluate the independent prognostic value of the signature. Furthermore, based on the ‘rms’ R package, the nomograms and their corresponding nomogram calibration curves of CRC individuals in the training and validation cohorts were established to predict the survival probability of CRC individuals in 1, 3, and 5 years.
Functional enrichment analysis
Functional enrichment analysis was performed as previous methods following previously established methods28. The ‘clusterProfiler’ R package and the OmicShare tools (https://www.omicshare.com/tools) was applied to perform the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses based on the DEGs between the high- and low-risk groups29. In addition, to further explore the significantly enriched pathways of these DEGs, Gene Set Enrichment Analysis (GSEA) was also conducted using the GSEA software (https://www.gsea-msigdb.org/gsea/login.jsp/). Furthermore, the ‘maftools’ R package was applied to analyze the somatic mutation data and calculate the tumor mutation burden (TMB) values of CRC individuals. The Cancer Therapeutics Response Portal (CTRP) and the Pharmaceutical Regulatory Information System (PRISM) derived drug response data were applied to identify the potential therapeutic agents for CRC individuals in high-risk group.
Tumor Microenvironment Analysis
Consistent with the previous methods27, Tumor Immune Estimation Resource (TIMER), ssGSEA and CIBERSORT methods were applied to explore the potential relationship between the risk score and the tumor microenvironment (TME). Furthermore, the expression levels of the genes reported as immune checkpoints were compared between different risk groups.
RNA isolation and real-time PCR
As previously described20. Total RNA was extracted from cultured cells using RNA isolater (Vazyme, China). Subsequently, 1 µg of total RNA underwent reverse transcription to generate complementary DNA (cDNA) using HiScript III RT SuperMix for qPCR with gDNA wiper (Vazyme, China). Then, Real-Time PCR was conducted with ChamQTM Universal SYBR qPCR Master Mix (Vazyme, China). The cycling protocol involved an initial denaturation step at 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 60 s, and a final extension step at 72 °C for 5 min. All primers were synthesized by Tsingke (Beijing, China) and are listed in Table S1. The mRNA expression levels of the genes were determined using the 2 − ΔΔCt method and normalized to GAPDH.
Western blot
As previously described20. Cells were lysed with RIPA. After centrifugation and extraction, equal amounts of protein was separated in SDS-PAGE, transferred to PVDF, probed with corresponding antibody and finally visualized by ECL kit.
Immunohistochemistry (IHC)
A cancer tissue array comprising 43 paired normal and tumor CRC samples was obtained from Ruijin Hospital (Shanghai, China). IHC was conducted following standard protocols, with optimal sections of tissue specimens selected and deparaffinized. IHC was performed as the following antibodies: GSR (18257-1-AP, Proteintech), ACADM (A4567, ABclonal), and GPC1 (16700-1-AP, Proteintech). Each tissue-array was scored by three independent pathologists using a semi-quantitative method based on the German semi-quantitative scoring system30.
Statistical analysis
All the statistical analyses were conducted by the R software (version: 3.6.0) in this study. All P values of statistical data were based on two-sided statistical tests. P < 0.05 was considered to be statistically significant.
Results
Labeling and enrichment of sialylated membrane proteins in SW480 and SW620 cells
The flow chart of this study was illustrated in Figure S1. SW480 cells isolated from primary tumor and SW620 cells isolated from lymph-node metastasis of the same colon cancer patient have been a useful patient-matched model for studying colorectal cancer treatment in vitro. We first checked and confirmed their migration and invasion ability by transwell assay. As shown in Fig. 1A, the invasive ability of SW620 cells was significantly higher than that of SW480 cells. Notably, due to the critical role of sialylation in tumor progression, which is mainly driven by 20 sialyltransferases (STs), we checked the mRNA expression levels of these 20 STs in both SW480 and SW620 cells. And we found that ST6NAC2, ST6NAC3, ST6NAC5, ST6NAC6, ST8SIA2, ST8SIA6, ST3GAL1, ST3GAL4, ST3GAL5, and ST3GAL6 were highly expressed in SW480 cell. In contrast, ST8SIA1, ST8SIA3, ST8SIA4, ST8SIA5, ST6GAL1, and ST3GAL3 were highly expressed in SW620 cell. Overall, the results demonstrated that 80% (16 out of 20) of the STs were differentially expressed (Fig. 1B). Subsequently, we aimed to identify the targeted sialylated proteins in both cell lines. Figure 1C demonstrated a schematic representation of our MS strategy. Ac4ManNAz was added to the culture medium and used to incorporate azidos to tag the sialic acid-linked glycans and the sialylated proteins while ManNAc was used as a control. Biotin-alkyne could specifically recognize and react with azido-tagged proteins on the cell surface, which were then enriched with streptavidin beads (Fig. 1D). IF and FACS were used to confirm the labeling efficiency, showing that compared with the ManNAc group, labeling signals were obviously stronger in cells incubated with Ac4ManNAz in both SW480 and SW620 cells (Fig. 1E). In addition, we checked the enrichment and selectivity efficiency using Western blot and Silver staining. As shown in Fig. 1F-G, the Ac4ManNAz group showed a higher signal of streptavidin 680 and more proteins than the ManNAc group.
Labeling and enrichment of sialylated membrane proteins in SW480 and SW620 cells. (A) Compare the differences in invasion and migration of SW480 and SW620 cell lines. (B) mRNA Relative expression levels of 20 sialyltransferases (STs) in SW480 and SW620 cell lines. (C) Schematic flowchart of this part study design to identify sialylated membrane proteins in SW480 and SW620 cells. (D) Metabolic labeling with Ac4ManNAz and ManNAc (negative control) was performed on SW480 and SW620 cells, followed by confocal microscopy to observe fluorescence-labeled sialylated proteins. AF555 (red) and DAPI (blue) were used to visualize polysaccharides containing azide groups and cell nuclei respectively. (E) Flow cytometric analysis of SW480 and SW620 cells following treatment with Ac4ManNAz or ManNAc. Cells were labeled using fluorescent probes and analyzed using PE-A signal. Histograms depict three cell populations: control cells (gray), cells treated with ManNAc (black), and cells treated with Ac4ManNAz (red). (F) Detection of sialylated glycoproteins in lysates of SW480 and SW620 cells. (G) Silver staining was performed to detect sialylated glycoproteins in lysates of SW480 and SW620 cells. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
MS analysis and identification
Following MS analysis of the elution from cells incubated with Ac4ManNAz, we identified 1102 proteins by querying the UniProt Database, including 712 proteins from SW480 cells and 995 proteins from SW620 cells (Table S2). GO analysis (Cellular Component) of all the proteins revealed that the majority of them were associated with extracellular exosome, membrane, focal adhesion, and cell-cell adherens junctions, indicating effective enrichment of membrane proteins.
Identification of the LNM-related SRGs (LRSRGs)
530 differentially sialylated membrane (secretory) proteins and corresponding encoding genes (SRGs) were identified using the criteria of SW480-specific protein expression, SW620-specific protein expression, or LOG Value Ratio > 1 (Table S3). Subsequently, 470 overlapping SRGs were identified between the 19,712 genes in the TCGA-COREAD transcriptome dataset and the 530 differentially sialylated SRGs, and these SRGs were selected for further analysis. 68 LRSRGs were identified using the DEG analysis (false discovery rate < 0.05) between primary non-lymph node metastatic CRC tissues (N0) and primary lymph node metastatic CRC tissues (N1-2) in the TCGA-COREAD datasets (Fig. 2A). According to the enrichment analysis, these LRSRGs were mainly enriched in the metabolism- and tumorigenesis-related pathways, such as Carbon metabolism, TCA Cycle, Ferroptosis, and Focal adhesion (Fig. 2B). 25 LRSRGs were correlated with overall survival (OS) using the univariate Cox analysis (P < 0.05) in TCGA-COREAD datasets (Fig. 2C). Based on the hazard ratio (HR) values, the following genes were identified as protective factors: ACADM, AGMAT, CCT2, EHD4, EIF4A3, FH, GSR, LRRC59, PPA2, RAN, SAMM50, SDHB, SFXN1, TM7SF3, and VDAC3. Conversely, the following genes were identified as risk factors: ESYT2, FLOT1, GPC1, LRRC8A, NGFR, NRP2, PRNP, RPL30, SEC61G, and VAT1.
Identification of the LNM-related SRGs (LRSRGs) and LRSRGs Molecular Phenotypes in CRC. (A) Volcano map of the LNM-related SRGs between primary non-lymph node metastatic CRC tissues (N0) and primary lymph node metastatic CRC tissues (N1-2) in the TCGA-COREAD datasets. (B) KEGG analysis of the LRSRGs. (C) Forest plot to show the results of the univariate cox regression analysis between LRSRGs expression levels and prognosis. (D) The consensus matrix thermogram defines three molecular phenotypes (k = 3) and shows their correlation area by unsupervised learning. (E) The principal component analysis (PCA) of LRSRGs shows a remarkable difference in transcriptomes among the three subtypes. (F) The Kaplan-Meier curves for OS of LRSRGs molecular phenotypes in TCGA. (G) The heatmap shows the differences in clinical features and mRNA expression levels of LRSRGs among the three phenotypes.
Construction and evaluation of the LRSRGs molecular phenotypes in CRC
We identified three novel molecular phenotypes by unsupervised learning analysis based on 25 LRSRGs associated with OS (Fig. 2D). The PCA analysis showed that the three phenotypes could be distinguished by the expression levels of these LRSRGs (Fig. 2E). Patients in cluster C had a better prognosis than those in cluster A and cluster B (Fig. 2F). Significant differences in the expression levels of LRSRGs and the clinical characteristics of CRC patients among different phenotypes were observed using the heatmap (Fig. 2G). The clinical and GSVA analyses were performed to further explore the clinical significance and biological discrepancy among the three phenotypes. The results showed that cluster A and cluster B had worse TNM stages and could be related to MSI status and CMS stage (Fig. 3A). The results of GSVA analysis demonstrated that the biological discrepancy mainly involved the metabolism-, immune- and tumorigenesis-related pathways, such as Carbon metabolism, TGF-beta signaling pathway, Focal adhesion, and MAPK signaling pathway (Fig. 3B). In addition, we calculated the degree of immune cell infiltration in each CRC patient by ssGSEA algorithm to further observe the variations in immune cell infiltration among the LRSRGs molecular phenotypes. Patients in cluster A and cluster C had higher infiltration degrees of immune cells than those in cluster B, which indicated that the three phenotypes could be closely linked to three previously reported immune phenotypes: immune-excluded (cluster A), immune-desert (cluster B), and immune-inflamed (cluster C) (Fig. 3C).
Clinical and biological significance of LRSRGs molecular phenotypes in CRC. (A) Stacked plots to display clinicopathological characteristics among different phenotypes. (B) The heatmaps show the results of GSVA analysis of the differentially expressed genes among the three phenotypes. (C) The boxplot shows the differences in ssGSEA scores of immune cell infiltration among the three phenotypes. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Establishment of a prognostic LRSRGs-related gene signature
25 OS-associated LRSRGs were screened to establish a prognostic signature using the LASSO Cox regression analysis. A 9-gene signature (ACADM, EHD4, FLOT1, GPC1, GSR, LRRC8A, NGFR, SDHB, and SEC61G) was filtered out by the minimum value of lambda (λ) (Fig. 4A-B). The coefficients and the differentially expressed levels of these genes were shown in the bar plots (Fig. 4C-D). The correlation of mRNA expression levels among these genes was shown by the circle plot (Fig. 4E). The protein-protein interaction (PPI) networks indicated that GSR, ACADM, and SDHB were hub genes (Fig. 4F-G). The somatic mutation profiles of these 9 genes were shown in Fig. 4H-I. In line with the formula mentioned above in the “methods” section, the risk score of each patient was calculated as follows: (−0.0230) × expression of ACADM + (−0.1209) × expression of EHD4 + 0.1494 × expression of FLOT1 + 0.0654 × expression of GPC1 + (−0.1057) × expression of GSR + 0.1880 × expression of LRRC8A + 0.2076 × expression of NGFR + (−0.0762) × expression of SDHB + 0.2699 × expression of SEC61G. Then, the relationship between risk score and clinical characteristics of CRC patients was explored. The results demonstrated that risk score was positively correlated with T stage, N stage, M stage, TNM stage, and CMS stage. Patients with poor prognosis and venous invasion had higher risk scores. Consistent with the above results, patients in cluster C had the lowest risk scores (Figure S2).
Establishment of a prognostic LRSRGs-related gene signature. (A-B) Charts to show the Log Lambda value and the 9 prognostic LRSRGs-related genes with non-zero coefficient. (C) Bar plot to show the coefficient of each gene. (D) Boxplot to show the differences in the expression levels of each gene between normal and tumor tissues in CRC. (E) The network plot to show the correlation among these 9 genes. (F-G) PPI network constructed of by STRING to indicate the interactions among these 9 genes. (H) The doughnut chart shows the locations of CNV alterations in these 9 genes on 23 chromosomes. (I) Waterfall chart to show the mutation status of each genes in CRC. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Evaluation and validation of LRSRGs-related gene signature
Based on the median value of risk scores in the TCGA training group, patients in both training and testing datasets were classified into low-risk and high-risk groups. To evaluate the value of LRSRGs-related gene signature in CRC patients, the Kaplan-Meier survival curves showed that patients with high risk scores could have a worse prognosis than patients with low risk scores in both training and testing groups (Fig. 5A, C). Then, the time-dependent ROC curves were performed to evaluate the sensitivity and specificity of the risk scores in predicting prognostic features. According to the areas under the curve (AUC), the risk scores showed the potential ability to predict the OS in TCGA training (1-year AUC = 0.730, 3-year AUC = 0.714, 5-year AUC = 0.730) and GSE39582 testing (1-year AUC = 0.628, 3-year AUC = 0.621, 5-year AUC = 0.599) groups (Fig. 5B, D).
Evaluation of LRSRGs-related gene signature and construction of the nomogram prediction model. (A-D) Kaplan-Meier survival curve for the OS between the high- and low-risk group in the TCGA training dataset (A) and GSE39582 testing dataset (C). AUC of time-dependent ROC curve for the risk score in the TCGA (B) training dataset and GSE39582 (D) testing dataset. (E-H) The forest plots to show the results of the univariate and multivariate Cox regression analysis regarding OS in the TCGA (E-F) training and the GSE39582 (G-H) testing datasets. (I-L) Nomograms for predicting 6-year survival in the TCGA (I) training dataset and GSE39582 (K) testing datasets. Calibration curves for the nomogram predicting 2–6 years survival in the TCGA (J) training dataset and GSE39582 (L) testing datasets.
Analysis of independent prognostic factors and construction of the nomogram prediction model
To explore whether the risk score was an independent prognostic factor of CRC patients, univariate and multivariate Cox regression analyses were performed. In the training group, the results of the univariate Cox regression analysis showed that the risk score (P < 0.001, HR = 3.820, 95%CI = 2.778 − 5.253) and other clinical parameters, including T stage (P = 0.005, HR = 3.266, 95%CI = 1.421 − 7.508), N stage (P < 0.001, HR = 2.925, 95%CI = 1.890 − 4.528), M stage (P < 0.001, HR = 4.225, 95%CI = 2.696 − 6.623) and TNM stage (P < 0.001, HR = 3.421, 95%CI = 2.178 − 5.372), were significantly associated with OS (Fig. 5E). In addition, the multivariate Cox regression analysis demonstrated that the risk score (P < 0.001, HR = 3.139, 95%CI = 2.176 − 4.530) and M stage (P = 0.003, HR = 2.268, 95%CI = 1.329 − 3.872) were the independent prognostic factors of OS (Fig. 5F). Consistent with the above results, the risk score was also an independent prognostic factor of OS in the testing group (Fig. 5G-H). To enhance the prediction of survival probability for CRC patients at 2, 4, and 6 years, we incorporated various clinicopathological characteristics such as T Stage, N Stage, M Stage, TNM Stage, and risk group to construct a nomogram prediction model in both the training and testing groups (Fig. 5I, K). Calibration curves were subsequently generated to demonstrate the accuracy of our predictions for 2, 4, and 6 years (Fig. 5J, L), illustrating a robust predictive performance.
Functional enrichment analysis of LRSRGs-related gene signature
To further explore the downstream biological functions and pathways correlated with the novel signature, GO and KEGG enrichment pathway analyses were performed using the DEGs between the low- and high-risk groups in the training group. Biological Process (BP), Cell Component (CC), and Molecular Function (MF) were all covered in the GO enrichment analysis. The results showed that metabolic process, immune system process, and cell proliferation were enriched for BP; organelle, membrane, and protein-containing complex for CC; binding, catalytic activity, and transcription regulator activity for MF (Figure S3A). Furthermore, the results of KEGG pathway analysis indicated enrichment in pathways such as ECM receptor interaction, focal adhesion, cell cycle, metabolic pathways, etc. (Figure S3B-C). Meanwhile, ECM receptor interaction, focal adhesion, intestinal immune network, and fatty acid metabolism were enriched by the GSEA (Figure S3D).
Analysis of somatic mutation
The tumorigenesis of CRC was closely related to somatic mutations in patients. To unveil the relationship between the risk score and somatic mutations, simple nucleotide variation data of the low- and high-risk groups in the TCGA training dataset was analyzed. APC (77%), TP53 (69%), TTN (53%), KRAS (41%), and SYNE1 (26%) were the top five genes with the highest mutation frequencies in the high-risk group whereas APC (76%), TTN (53%), TP53 (51%), KRAS (42%), and PIK3CA (34%) exhibited highest mutation frequencies in the low-risk group (Fig. 6A-B). TP53 showed a relatively higher mutation frequency in the high-risk group, and PIK3CA showed a relatively higher mutation frequency in the low-risk group (Fig. 6C-D). The differences in gene mutation distributions between the high- and low-risk groups were shown in the forest plot (Fig. 6E). Furthermore, TMB was calculated and analyzed in each patient and the results indicated that patients in the low-risk group had significantly higher TMB levels (Fig. 6F).
Analysis of Somatic Mutation differences in LRSRGs-related Gene Signature. (A-B) Onco-plots of the somatic mutation between the high-risk (A) and low-risk (B) groups in the TCGA dataset. (C-D) Lollipop charts of the mutated TP53 and PIK3CA gene in the high-risk (C) and low-risk (D) groups. (E) Forest plot of the differentially somatic mutation. (F) Boxplot for the TMB scores between the high-risk and low-risk groups. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Analysis of tumor microenvironment and immunotherapy response
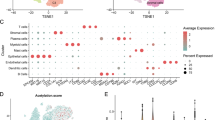
Based on the above functional enrichment analysis results, the immune process and pathway were significantly different between the high- and low-risk groups (Figure S3). To better assess the relationship between risk score and tumor microenvironment, TIMER, ssGSEA, and CIBERSORT analyses of each patient were performed. The results of TIMER analysis indicated that the 9 LRSRGs were closely related to the infiltration status of immune cells, including B Cell, CD4+ T Cell, CD8+ T Cell, Macrophage, Neutrophil, and Dendritic Cell, especially for ACADM, GSR, GPC1 and NGFR genes (Figure S4). In addition, the results of ssGSEA analysis demonstrated that patients in the low-risk group had a higher infiltration rate of most immune cells with dominant anti-tumor effects, such as B cell, CD4+ T cell, CD8+ T cell, Dendritic cell, Neutrophil, and Type 17 T helper cell (Fig. 7A). The immune cell infiltration degree of B cell, CD4 + T cell, and CD8 + T cell was confirmed to be negatively correlated with risk scores (Fig. 7B). Consistent with the aforementioned findings, the results of CIBERSORT analysis also revealed up-regulation of T cell CD4 naive, T cell CD8, T cell follicular helper, and Dendritic cell in the low-risk group, whereas macrophages M0 were significantly down-regulated (Fig. 7C). The correlation between the infiltration abundance of 22 immune cells and the expression levels of 9 LRSRGs was shown in the heatmap (Fig. 7D). Furthermore, to estimate the predictive ability of the 9-gene signature for immunotherapy response, the correlation between risk score and common immune checkpoints (ICPs) was explored, and the gene expression levels of most ICPs were significantly up-regulated in the low-risk group (Fig. 7E).
Analysis of Tumor Microenvironment and Immunotherapy Response. (A) Boxplot for the distribution of 28 tumor-infiltrating immune cells between the high-risk and low-risk groups using ssGSEA method. (B) Lollipop chart for the correlation between risk scores and the infiltration degree of immune cells. (C) The boxplots for the comparison of the 22 immune cells between the high-risk and low-risk groups by CIBERSORT analysis. (D) Heatmap to show the correlation between risk scores and the infiltration degree of immune cells by CIBERSORT analysis. (E) The boxplot to show the results of the different expression levels of ICPs between the high- and low-risk groups. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Identification of potential therapeutic agents for CRC patients with high risk scores
In order to identify the potential therapeutic agents for CRC patients with high risk scores, two different datasets, CTRP and PRISM, were applied. The drug response analysis between high risk score patients (top 10%) and low risk score patients (bottom 10%) was performed with lower AUC values. The correlation analysis between AUC value and risk scores was performed to screen key compounds based on the CTRP (Figure S5A) and PRISM datasets (Figure S5B). The results demonstrated that three CTRP-derived compounds (including ML210, ML162, and 1S,3R−RSL−3) and three PRISM-derived compounds (including dasatinib, romidepsinand YM−155) could be the potential therapeutic agents.
Validation of the expression levels of the 9 LRSRGs
The overreliance on bioinformatics methodologies may result in erroneous experimental outcomes with false positive implications. Thus, we further verify the mRNA expression levels of the 9 LRSRGs in Fig. 8A, demonstrating that most of the 9 LRSRGs were differentially expressed in SW480 and SW620 cells except FLOT1 and LRRC8A. Based on the results of Western blot analysis in Fig. 8B, the relative protein expression levels of GSR, ACADM, and GPC1 in SW480 and SW620 cells were also analyzed and found to be consistent with our conclusions above. Furthermore, the protein expression levels and clinical significance of hub LRSRGs, including GSR, ACADM, and GPC1 were further verified in the tissue-arrays of our center by IHC (Fig. 8C). The results showed that the expression of GSR and ACADM all increased in tumor tissues, while GPC1 expression was decreased. Moreover, the expression levels of these three proteins varied between primary CRC tissues without lymph node metastasis and those with lymph node metastasis.
Validation of the Expression Levels of the 9 LRSRGs. (A) The boxplot to indicate the different mRNA expression levels of these 9 LRSRGs in SW480 and SW620 CRC cell lines by Real Time PCR. (B) Western blot to validate the different protein expression levels of SW480 and SW620 CRC cell lines. (C) Representative immunohistochemistry images show the different protein expression levels of GSR, ACADM, and GPC1 in different CRC tissues and corresponding normal tissues from the CRC tissue-array of Ruijin hospital. The boxplots display the differences in IHC scores for GSR, ACADM, and GPC1 among different CRC tissues and corresponding normal tissues from the CRC tissue-array. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
According to the current TNM staging system, LNM is the key parameter that separates Stage II and Stage III in CRC patients, which entails different therapeutic and follow-up strategies. The 5-year survival rate decreases from 98.5 to 75.7% and 56.7% for stage I, II, and III CRC patients, respectively31, and one of the most important reasons for this pernicious phenomenon is lymph node involvement. In general, chemotherapy or radiotherapy is necessary for all stage III patients and for high-risk stage II patients who may benefit from this treatment owing to the likelihood of developing recurrence and metastasis32. However, the accurate definition and prediction for CRC patients with LNM is often difficult and challenging. Therefore, effective biomarkers for stratifying CRC patients (low or high risk of LNM) are of great importance to surgery, therapy instruction, and reducing the long-term recurrence rate of CRC patients.
The relationship between sialylation and disease has gained more and more attention in recent decades, especially in cancer33,34. Aberrant sialylation has been reported to be a hallmark of numerous types of cancer and plays a pivotal role in various biological events in cancer cells, such as adhesion, migration, drug resistance, and membrane receptor regulation, which are driven by certain dysregulated enzymes35. It is important to note that substrate proteins are the key effecters for sialylation or other glycosylation. Sialylation of the membrane proteins is driven by ST family consisting of 20 members36. Based on the linkage types of SAs to the terminal glycans, the STs are subdivided into four different subtypes: ST3Gal, ST6Gal, ST6GalNAc, and ST8Sia. However, the role of ST family in tumor remains controversial, either inhibitive or facilitative. Even in the same cancerous disease, CRC, the ST family could demonstrate dynamic changes during CRC progression. Different substrate membrane proteins may be the key factor in this process. Considering that LNM represents a pivotal event in the development and progression of CRC, we examined the association between LNM and substrate membrane proteins, the mRNA expression levels of all 20 sialyltransferases (STs) were initially analyzed in SW480 and SW620 cells to identify potential correlations. Interestingly, 80% (16 out of 20) of the STs exhibited differentially expressed patterns, suggesting a connection between LNM and sialylation proteins. However, we found that 10 STs were highly expressed in SW480, while 6 STs were predominantly expressed in SW620. Similarly, previous studies have shown that STs display differential expression across various cancer types and even among different phenotypes of the same cancer37,38,39. This indicates that while the overall upregulation of cell surface sialylated glycans is a hallmark of tumors and is closely linked to metastasis, the specific STs involved may display distinct expression patterns across various cancers, resulting in different outcomes. Therefore, identifying the specific STs involved in CRC lymph node metastasis and elucidating their roles merits further in-depth investigation in future studies.
Subsequently, we provided and validated core sialylated membrane proteins, which might be crucial during the LNM process of CRC patients. To obtain and identify the sialylated membrane proteins, which are LNM-related, we specifically chose the same patient-derived cell lines, SW480 (from primary tumor) and SW620 (lymph node metastasis). Due to their limited genetic variability and similar background, they are considered a reliable and useful model to investigate genetic and protein changes during CRC progression, metastasis and LNM. By using a metabolic labeling strategy, we successfully labeled sialylated membrane proteins with the Ac4ManNAz (100 mM) in SW480 and SW620 cell lines, which were further enriched for MS and proteomics analysis demonstrating a list of proteins that were differentially sialylated between SW480 and SW620. Innovatively, we integrated the TCGA dataset of lymphatic metastases in CRC and the results of MS. 530 differentially sialylated SRGs were identified, followed by 68 LRSRGs screened out. Subsequently, 25 OS-associated LRSRGs were confirmed, which were utilized to construct the LRSRGs Molecular Phenotypes (Cluster A-C). Our findings revealed that LRSRGs molecular phenotypes were related to clinical characteristics, prognosis, and biological discrepancy of CRC patients. Patients in cluster C had poor prognosis and unfavorable clinical characteristics than those in cluster A or B, which strengthens the clinical value of our newly defined LRSRGs molecular phenotypes. With advancements in algorithms and the development of bioinformatics technologies, an increasing number of tumor prediction models are emerging. In our study, a novel prognostic 9-gene signature, including ACADM, EHD4, FLOT1, GPC1, GSR, LRRC8A, NGFR, SDHB, and SEC61G, was constructed using lasso Cox analysis, which was externally validated in several independent datasets.
To deepen the understanding of the functions of these 9 genes in CRC, their mRNA expression levels were analyzed, and their primary molecular functions were summarized. According to the hazard ratio (HR) values, SEC61G, NGFR, LRRC8A, FLOT1, and GPC1 were categorized as risk-associated genes, whereas ACADM, SDHB, GSR, and EHD4 were deemed protective genes. GSR, namely glutathione reductase, mainly maintains high levels of reduced glutathione in the cytosol by catalyzing the reduction of glutathione disulfide to glutathione40, which is required for the proliferation and progression of several cancers. Yan et al41. showed that GSR could mediate drug resistance by regulating redox homeostasis in glioblastoma cells. Glutathione reductase phosphorylation was necessary for AMPKα1-mediated survival advantage of tumor in colorectal cancer42. Serum level of glutathione reductase was also dysregulated in cancers43. Acyl-CoA dehydrogenase medium-chain (ACADM) is responsible for catalyzing the first step of mitochondrial fatty acid oxidation. Yam et al44. indicated that ACADM acted as a tumor suppressor, downregulation of which increased the aggressiveness of hepatocellular carcinoma. However, the role of ACADM in different cancers is still controversial44,45,46. Succinate dehydrogenase (SDH) is necessary for the oxidation of succinate to fumarate, which also promotes the engagement of electron transportation and is a component of the TCA cycle47. Succinate dehydrogenase-B (SDHB) is the core catalytic subunit of SDH. It has been reported that SDHB gene mutation is correlated to pheochromocytoma and paraganglioma48. Dysregulated SDHB expression has been reported to be involved in different cancers49,50,51,52. Flotillin-1 (FLOT1) is mainly involved in membrane organization, molecular signal transduction and material transport53. Several studies have indicated that FLOT1 is overexpressed in multiple cancers and is closely related to patients’ prognosis54. EHD4 is indicated to regulate membrane reorganization/tubulation and endosomal transport, which acts as an important regulator of EHD1-mediated endosomal recruitment and fission55. However, there is a paucity of studies on the EHD4 in cancers. Deng et al. indicated EHD4 as the mediator in regulating miR-4701-3p and miR-4793-3p on CRC cell apoptosis56. Nerve growth factor receptor (NGFR), also known as tumor necrosis factor receptor superfamily member 16 (TNFRSF16) and CD271, is upregulated in various cancers, such as hypopharyngeal cancer, melanoma and breast cancer, and plays crucial roles in tumor proliferation and metastasis57. Researchers have found the presence of N-/O-glycosylated sites in CD27158. Tamai et al. established a novel anti-glycosylated CD271 monoclonal antibody clone G4B1, which could specifically detect CD271 with sialic acid and O- glycan modifications, demonstrating promising specificity in identifying dysplasia and cancer cells in squamous cell carcinoma59. Glypican-1 (GPC1) is a cell surface proteoglycan that is upregulated in a variety of solid tumors, while lowly expressed in most adult normal tissues60,61, which could be a better target for stroma and cancer cell-targeted therapy62,63. GPC1 has been reported with N-glycosylation at Asn-79 and Asn-116, through which the protein expression and HS substitution are regulated64,65. There are also other N-/O-glycosylation sites predicted by Uniprot. However, little is known about the type of glycosylation or sialylation on the above 7 proteins, and the pathological significance of them is still largely unknow.
Utilizing the 9-gene signature, we observed that CRC patients classified into the high-risk group exhibited markedly reduced overall survival (OS) rates across both the training and subsequent testing cohorts. The multivariate Cox regression analyses further confirmed that the risk score derived from the signature independently predicted OS. Furthermore, we constructed a nomogram that integrates the stage, M stage, and risk scores of the signature, offering a reliable approach for clinicians to predict OS in CRC patients. Recently, several studies have reported that sialylation is related to tumor metastasis, particularly hyposialylation empowers chemo-evasive circulating tumor cell clusters66,67. Additionally, Natoni et al. discovered that sialylation is crucial in controlling lymphocyte proliferation and influencing lymph node metastasis in tumors68. Our study produced similar outcomes in which significantly elevated risk scores were identified in patients with metastasis (M1), lymph node metastasis (N1 and N2), and venous invasion. Furthermore, functional analysis of the differentially expressed genes (DEGs) between high-risk and low-risk patients revealed enrichment in pathways such as focal adhesion, ECM receptor interaction, and fatty acid metabolism, all of which play critical roles in tumor progression.
In addition, we also observed significant differences in immune regulatory processes, such as immune system process enriched for BP, Intestinal immune network enriched for GSEA among different groups (high- or low- risk) or phenotypes (cluster A-C). The ability of immune evasion is an important hallmark of tumors69. It has been known that dysregulated sialylation is involved in immune modulation and tumor immune evasion, and the sialic acid blockade could increase the immune activity towards tumors70. Wang et al. claimed that increased α2,6-sialylation by ST6GAL1 could inhibit T cell proliferation and thus promote the immune escape of hepatocarcinoma cells71. Yu et al. proposed that ST3GAL1-mediated O-linked sialylation of CD55 was a strategy to block CD55-mediated immune evasion72. Our study demonstrated that CRC patients with high-risk scores or in the high-risk group harbored obvious immune-suppressive features as there was a lower infiltration of CD8+ T cells and CD4+ T cells in the high-risk group. TMB refers to the total number of somatic coding mutations. In our research, risk scores demonstrated a positive correlation with TMB, suggesting that patients might benefit from immune checkpoint inhibitor therapies. Moreover, checkpoint inhibitor immunotherapy has been identified as a promising approach to improve patient outcomes. Anti-PD1 immunotherapy is particularly recommended for metastatic CRC patients with MSI-H/dMMR status. Additionally, our study found that the expression levels of checkpoint inhibitors varied between high and low-risk groups. These findings suggest that patients in the low-risk group could be more responsive to immunotherapy.
More significantly, glycosylation has a broad and meaningful clinical application prospect73. In recent years, it has been suggested that the glycosylation status of such traditional biomarkers could provide more useful and valuable messages7,74,75. Studies showed α2–3-sialylated PSA demonstrated excellent performance in discriminating between high-risk prostate cancer and benign hyperplasia patients76,77. Serum sialylated-MUC1 was obviously increased in CRC patients. Besides, decreased serum IgG sialylation levels were observed in CRC patients, which was closely related to poorer prognosis78. These results implicate the potential of targeting sialylation as a promising strategy for treating CRC patients. However, current evidence of therapeutic agents with cancer-specific glycosylation is markedly limited. Thus, we further identified three CTRP-derived compounds (including ML210, ML162, and 1S,3R −RSL−3,) and three PRISM-derived compounds (including dasatinib, romidepsin and YM−155) as potential therapeutic agents for CRC patients with high LRSRGs risk scores, which needs to be further verified.
Our study presents several strengths. First, we identified and validated the relationship between lymph node metastasis (LNM) and sialylated membrane proteins in CRC using both mass spectrometry and molecular biology techniques. Second, we identified 25 prognostic LRSRGs and employed them to define a novel CRC phenotype, termed LRSRGs Molecular Phenotype. This phenotype can be used to distinguish patient prognosis, clinical characteristics, and treatment efficacy through multi-omics approaches. Moreover, we developed a 9-gene signature based on these 25 prognostic LRSRGs and validated its potential clinical value in predicting patient prognosis, molecular biological characteristics, immune cell infiltration, response to immunotherapy, and other potential therapeutic agents. Nonetheless, we recognize that the study has multiple limitations. First, the LRSRGs signature and phenotypes were derived from retrospective data across multiple databases, which requires further prospective studies to substantiate their clinical significance. Second, we only validated the expression of three LRSRGs, and additional work is needed to test the expression levels of the remaining LRSRGs and their correlation with clinical features in CRC patients. Future research should also explore the mechanistic underpinnings of how these LRSRGs influence CRC progression could provide crucial insights into their roles in metastasis. And a comprehensive glycomic and glycoproteomic analysis may be required in the future to establish a robust functional link between the identified sialylated proteins, their biological impact on anti-tumor immunity, the development of an immune-suppressive microenvironment, and their relationship with the expression and activity patterns of the analyzed STs.
Conclusion
In summary, our research identified key sialylated glycoproteins that potentially play a crucial role in tumor LNM and clinical outcomes in CRC. We provided a robust prognostic phenotype, the LRSRGs Molecular Phenotype, and a risk model using a 9-gene signature for CRC patients. Notably, the new phenotype and signature have significant implications for clinical treatments, such as targeted therapy and immunotherapy, offering new insights into the molecular mechanisms and unveiling potential novel therapies for CRC. Targeting sialylation, in particular, appears to be a promising therapeutic approach with substantial potential for developing innovative therapeutic strategies and combination treatments for CRC patients.
Data availability
All the public datasets can be downloaded in the Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/), the University of California Santa Cruz (UCSC, https://xenabrowser.net/datapages/), the Cancer Cell Line Encyclopedia project (CCLE, https://portals.broadinstitute.org/ccle), the Human Protein Atlas (HPA, https://www.proteinatlas.org/) database, and the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database.
Abbreviations
- ACADM:
-
Acyl-CoA Dehydrogenase Medium-Chain
- AUC:
-
Areas Under the Curve
- BP:
-
Biological Process
- CC:
-
Cell Component
- CTRP:
-
Cancer Therapeutics Response Portal
- DEG:
-
Differentially Expressed Gene
- FLOT1:
-
Flotillin-1
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene Ontology
- GPC1:
-
Glypican-1
- GSEA:
-
Gene Set Enrichment Analysis
- HR:
-
Hazard Ratio
- ICPs:
-
Immune Check Points
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LASSO:
-
Least Absolute Shrinkage and Selection Operator
- LNM:
-
Lymph Node Metastasis
- LRSRGs:
-
LNM-Related SRGs
- MF:
-
Molecular Function
- NGFR:
-
Nerve Growth Factor Receptor
- OS:
-
Overall Survival
- PCA:
-
Principal Component Analysis
- PPI:
-
Protein-Protein Interaction
- PRISM:
-
Pharmaceutical Regulatory Information System
- RNA-seq:
-
RNA Sequencing
- ROC:
-
Receiver Operating Characteristic
- SA:
-
Sialic Acid
- SDH:
-
Succinate Dehydrogenase
- SDHB:
-
Succinate Dehydrogenase-B
- SRGs:
-
Sialylation-Related Genes
- STs:
-
Sialyltransferases
- TCGA:
-
The Cancer Genome Atlas
- TIMER:
-
Tumor Immune Estimation Resource
- TMB:
-
Tumor Mutation Burden
- TME:
-
Tumor Microenvironment
- TNFRSF16:
-
Tumor Necrosis Factor Receptor Superfamily Member 16
- TPM:
-
Transcripts Per Million
References
Baidoun, F. et al. Colorectal Cancer epidemiology: recent trends and Impact on outcomes. Curr. Drug Targets. 22, 998–1009. https://doi.org/10.2174/1389450121999201117115717 (2021).
Chen, K., Collins, G., Wang, H. & Toh, J. W. T. Pathological features and prognostication in Colorectal Cancer. Curr. Oncol. 28, 5356–5383. https://doi.org/10.3390/curroncol28060447 (2021).
Benson, A. B. et al. Colon cancer, Version 2.2021, NCCN Clinical Practice guidelines in Oncology. J. Natl. Compr. Canc Netw. 19, 329–359. https://doi.org/10.6004/jnccn.2021.0012 (2021).
Baum, M., Fallowfield, L., Farewell, V., Macbeth, F. & Treasure, T. NICE guidelines: management of colorectal cancer metastases. Br. J. Surg. 107, e357. https://doi.org/10.1002/bjs.11789 (2020).
Fletcher, C. M., Coyne, M. J., Villa, O. F., Chatzidaki-Livanis, M. & Comstock, L. E. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 137, 321–331. https://doi.org/10.1016/j.cell.2009.02.041 (2009).
Magalhães, A., Duarte, H. O. & Reis, C. A. The role of O-glycosylation in human disease. Mol. Aspects Med. 79, 100964. https://doi.org/10.1016/j.mam.2021.100964 (2021).
Pinho, S. S. & Reis, C. A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer. 15, 540–555. https://doi.org/10.1038/nrc3982 (2015).
Veillon, L., Fakih, C., Abou-El-Hassan, H., Kobeissy, F. & Mechref, Y. Glycosylation changes in Brain Cancer. ACS Chem. Neurosci. 9, 51–72. https://doi.org/10.1021/acschemneuro.7b00271 (2018).
Munkley, J., Mills, I. G. & Elliott, D. J. The role of glycans in the development and progression of prostate cancer. Nat. Rev. Urol. 13, 324–333. https://doi.org/10.1038/nrurol.2016.65 (2016).
Wu, J. et al. Tunicamycin specifically aggravates ER stress and overcomes chemoresistance in multidrug-resistant gastric cancer cells by inhibiting N-glycosylation. J. Exp. Clin. Cancer Res. 37, 272. https://doi.org/10.1186/s13046-018-0935-8 (2018).
Lumibao, J. C., Tremblay, J. R., Hsu, J. & Engle, D. D. Altered glycosylation in pancreatic cancer and beyond. J. Exp. Med. 219 https://doi.org/10.1084/jem.20211505 (2022).
Na, K. et al. Potential Regulatory Role of Human-Carboxylesterase-1 glycosylation in Liver Cancer Cell Growth. J. Proteome Res. 19, 4867–4883. https://doi.org/10.1021/acs.jproteome.0c00787 (2020).
Zhang, S. et al. Differential expression of ST6GAL1 in the tumor progression of colorectal cancer. Biochem. Biophys. Res. Commun. 486, 1090–1096. https://doi.org/10.1016/j.bbrc.2017.03.167 (2017).
Fernández-Ponce, C. et al. The role of glycosyltransferases in Colorectal Cancer. Int. J. Mol. Sci. 22 https://doi.org/10.3390/ijms22115822 (2021).
Bindeman, W. E. & Fingleton, B. Glycosylation as a regulator of site-specific metastasis. Cancer Metastasis Rev. 41, 107–129. https://doi.org/10.1007/s10555-021-10015-1 (2022).
Cazet, A. et al. Consequences of the expression of sialylated antigens in breast cancer. Carbohydr. Res. 345, 1377–1383. https://doi.org/10.1016/j.carres.2010.01.024 (2010).
Pearce, O. M. & Läubli, H. Sialic acids in cancer biology and immunity. Glycobiology. 26, 111–128. https://doi.org/10.1093/glycob/cwv097 (2016).
Zhao, L. et al. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut. 59, 1226–1235. https://doi.org/10.1136/gut.2009.202739 (2010).
Leibovitz, A. et al. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 36, 4562–4569 (1976).
Zhang, S. et al. N-myc downstream-regulated gene 1 inhibits the proliferation of colorectal cancer through emulative antagonizing NEDD4-mediated ubiquitylation of p21. J. Exp. Clin. Cancer Res. 38, 490. https://doi.org/10.1186/s13046-019-1476-5 (2019).
Colaprico, A. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71. https://doi.org/10.1093/nar/gkv1507 (2016).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. https://doi.org/10.1093/nar/30.1.207 (2002).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. https://doi.org/10.1093/nar/gkv007 (2015).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 26, 1572–1573. https://doi.org/10.1093/bioinformatics/btq170 (2010).
Gu, Z. & Hübschmann, D. Make interactive Complex heatmaps in R. Bioinformatics. 38, 1460–1462. https://doi.org/10.1093/bioinformatics/btab806 (2022).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 462, 108–112. https://doi.org/10.1038/nature08460 (2009).
Shao, Y. et al. An original ferroptosis-related gene signature effectively predicts the prognosis and clinical status for Colorectal Cancer patients. Front. Oncol. 11, 711776. https://doi.org/10.3389/fonc.2021.711776 (2021).
Shao, Y. et al. Comprehensive Analysis of Ferroptosis-Related Markers for the clinical and biological value in gastric Cancer. Oxid. Med. Cell. Longev. 2021 (7007933). https://doi.org/10.1155/2021/7007933 (2021).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Shao, Y. et al. Impact of cuproptosis-related markers on clinical status, tumor immune microenvironment and immunotherapy in colorectal cancer: a multi-omic analysis. Comput. Struct. Biotechnol. J. 21, 3383–3403. https://doi.org/10.1016/j.csbj.2023.06.011 (2023).
Figueredo, A., Charette, M. L., Maroun, J., Brouwers, M. C. & Zuraw, L. Adjuvant therapy for stage II colon cancer: a systematic review from the Cancer Care Ontario Program in evidence-based care’s gastrointestinal cancer disease site group. J. Clin. Oncol. 22, 3395–3407. https://doi.org/10.1200/jco.2004.03.087 (2004).
Park, J. S. et al. High-risk clinicopathological features and their predictive significance in Korean patients with stage II colon cancer. J. Cancer Res. Clin. Oncol. 142, 2051–2059. https://doi.org/10.1007/s00432-016-2208-2 (2016).
Da Silva, A. et al. N-acetylneuraminate pyruvate lyase controls sialylation of muscle glycoproteins essential for muscle regeneration and function. Sci. Adv. 9, eade6308. https://doi.org/10.1126/sciadv.ade6308 (2023).
Yao, Y. et al. Mucus sialylation determines intestinal host-commensal homeostasis. Cell. 185, 1172–1188e1128. https://doi.org/10.1016/j.cell.2022.02.013 (2022).
Cao, K. et al. Attenuation of Sialylation augments Antitumor Immunity and improves response to Immunotherapy in Ovarian Cancer. Cancer Res. 83, 2171–2186. https://doi.org/10.1158/0008-5472.Can-22-3260 (2023).
Rao, F. V. et al. Structural insight into mammalian sialyltransferases. Nat. Struct. Mol. Biol. 16, 1186–1188. https://doi.org/10.1038/nsmb.1685 (2009).
Wang, P. H. et al. Altered mRNA expression of sialyltransferase in squamous cell carcinomas of the cervix. Gynecol. Oncol. 83, 121–127. https://doi.org/10.1006/gyno.2001.6358 (2001).
Wang, P. H. et al. Altered mRNA expressions of sialyltransferases in ovarian cancers. Gynecol. Oncol. 99, 631–639. https://doi.org/10.1016/j.ygyno.2005.07.016 (2005).
Wu, X. et al. Sialyltransferase ST3GAL1 promotes cell migration, invasion, and TGF-β1-induced EMT and confers paclitaxel resistance in ovarian cancer. Cell. Death Dis. 9, 1102. https://doi.org/10.1038/s41419-018-1101-0 (2018).
Begum, S. S., Das, D., Gour, N. K. & Deka, R. C. Computational modelling of nanotube delivery of anti-cancer drug into glutathione reductase enzyme. Sci. Rep. 11, 4950. https://doi.org/10.1038/s41598-021-84006-1 (2021).
Zhu, Z. et al. Glutathione reductase mediates drug resistance in glioblastoma cells by regulating redox homeostasis. J. Neurochem. 144, 93–104. https://doi.org/10.1111/jnc.14250 (2018).
Wang, Y. N. et al. AMPKα1 confers survival advantage of colorectal cancer cells under metabolic stress by promoting redox balance through the regulation of glutathione reductase phosphorylation. Oncogene. 39, 637–650. https://doi.org/10.1038/s41388-019-1004-2 (2020).
Bilgin, E., Atli, G., Duman, B. B. & Okten, A. I. Evaluation of oxidative stress biomarkers in Brain metastatic and non-metastatic Lung Cancer patients with different cell types. Anticancer Agents Med. Chem. 21, 2032–2040. https://doi.org/10.2174/1871520621666210211163055 (2021).
Ma, A. P. Y. et al. Suppression of ACADM-Mediated fatty acid oxidation promotes Hepatocellular Carcinoma via aberrant CAV1/SREBP1 signaling. Cancer Res. 81, 3679–3692. https://doi.org/10.1158/0008-5472.Can-20-3944 (2021).
Xu, L., Lu, Z., Yu, S., Li, G. & Chen, Y. Quantitative global proteome and phosphorylome analyses reveal potential biomarkers in kidney cancer. Oncol. Rep. 46 https://doi.org/10.3892/or.2021.8188 (2021).
Hsieh, C. H. et al. Quantitative proteomics of Th-MYCN transgenic mice reveals Aurora kinase inhibitor altered metabolic pathways and enhanced ACADM to suppress Neuroblastoma Progression. J. Proteome Res. 18, 3850–3866. https://doi.org/10.1021/acs.jproteome.9b00245 (2019).
Cardaci, S. et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat. Cell. Biol. 17, 1317–1326. https://doi.org/10.1038/ncb3233 (2015).
Armstrong, N. et al. SDHB knockout and succinate accumulation are insufficient for tumorigenesis but dual SDHB/NF1 loss yields SDHx-like pheochromocytomas. Cell. Rep. 38, 110453. https://doi.org/10.1016/j.celrep.2022.110453 (2022).
Xiao, Z. et al. SDHB downregulation facilitates the proliferation and invasion of colorectal cancer through AMPK functions excluding those involved in the modulation of aerobic glycolysis. Exp. Ther. Med. 15, 864–872. https://doi.org/10.3892/etm.2017.5482 (2018).
Mason, E. F. et al. Identification of succinate dehydrogenase-deficient bladder paragangliomas. Am. J. Surg. Pathol. 37, 1612–1618. https://doi.org/10.1097/PAS.0b013e318293d83c (2013).
Kuroda, N. et al. Review of succinate dehydrogenase-deficient renal cell carcinoma with focus on clinical and pathobiological aspects. Pol. J. Pathol. 67, 3–7. https://doi.org/10.5114/pjp.2016.59227 (2016).
Saxena, N. et al. SDHB-Deficient cancers: the role of mutations that Impair Iron Sulfur Cluster Delivery. J. Natl. Cancer Inst. 108 https://doi.org/10.1093/jnci/djv287 (2016).
Wei, J., Wang, R., Lu, Y., He, S. & Ding, Y. Flotillin-1 promotes progression and dampens chemosensitivity to cisplatin in gastric cancer via ERK and AKT signaling pathways. Eur. J. Pharmacol. 916, 174631. https://doi.org/10.1016/j.ejphar.2021.174631 (2022).
Deng, Y. et al. Prognostic value of flotillins (flotillin-1 and flotillin-2) in human cancers: a meta-analysis. Clin. Chim. Acta. 481, 90–98. https://doi.org/10.1016/j.cca.2018.02.036 (2018).
Jones, T., Naslavsky, N. & Caplan, S. Eps15 homology domain protein 4 (EHD4) is required for Eps15 homology domain protein 1 (EHD1)-mediated endosomal recruitment and fission. PLoS One. 15, e0239657. https://doi.org/10.1371/journal.pone.0239657 (2020).
Wang, H. et al. SHU00238 promotes Colorectal Cancer Cell apoptosis through mir-4701-3p and miR-4793-3p. Front. Genet. 10, 1320. https://doi.org/10.3389/fgene.2019.01320 (2019).
Morita, S. et al. Humanized anti-CD271 monoclonal antibody exerts an anti-tumor effect by depleting cancer stem cells. Cancer Lett. 461, 144–152. https://doi.org/10.1016/j.canlet.2019.07.011 (2019).
Johnson, D. et al. Expression and structure of the human NGF receptor. Cell. 47, 545–554. https://doi.org/10.1016/0092-8674(86)90619-7 (1986).
Fujii, K. et al. Establishment of a monoclonal antibody against glycosylated CD271 specific for cancer cells in immunohistochemistry. Cancer Sci. 113, 2878–2887. https://doi.org/10.1111/cas.15340 (2022).
Harada, E. et al. Glypican-1 targeted antibody-based therapy induces preclinical antitumor activity against esophageal squamous cell carcinoma. Oncotarget. 8, 24741–24752. https://doi.org/10.18632/oncotarget.15799 (2017).
Russell, P. J. et al. Immunohistochemical characterisation of the monoclonal antibody BLCA-38 for the detection of prostate cancer. Cancer Immunol. Immunother. 53, 995–1004. https://doi.org/10.1007/s00262-004-0527-7 (2004).
Tsujii, S. et al. Glypican-1 is a Novel Target for Stroma and Tumor Cell Dual-Targeting antibody-drug conjugates in Pancreatic Cancer. Mol. Cancer Ther. 20, 2495–2505. https://doi.org/10.1158/1535-7163.Mct-21-0335 (2021).
Lund, M. E., Campbell, D. H. & Walsh, B. J. The role of Glypican-1 in the Tumour Microenvironment. Adv. Exp. Med. Biol. 1245, 163–176. https://doi.org/10.1007/978-3-030-40146-7_8 (2020).
Svensson, G., Hyrenius Wittsten, A., Linse, S. & Mani, K. The structural role of N-linked glycans on human glypican-1. Biochemistry. 50, 9377–9387. https://doi.org/10.1021/bi200218s (2011).
Awad, W. et al. Structural aspects of N-Glycosylations and the C-terminal region in human Glypican-1. J. Biol. Chem. 290, 22991–23008. https://doi.org/10.1074/jbc.M115.660878 (2015).
Rossi, M. et al. PHGDH heterogeneity potentiates cancer cell dissemination and metastasis. Nature. 605, 747–753. https://doi.org/10.1038/s41586-022-04758-2 (2022).
Gvozdenovic, A. & Aceto, N. Metastasis unleashed: Hyposialylation empowers chemo-evasive circulating tumor cell clusters in breast Cancer. Cancer Res. 83, 2811–2812. https://doi.org/10.1158/0008-5472.Can-23-1978 (2023).
Natoni, A. et al. Sialylation regulates migration in chronic lymphocytic leukemia. Haematologica. 108, 1851–1860. https://doi.org/10.3324/haematol.2022.281999 (2023).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell. 144, 646–674. https://doi.org/10.1016/j.cell.2011.02.013 (2011).
Büll, C. et al. Sialic Acid Blockade suppresses Tumor Growth by enhancing T-cell-mediated tumor immunity. Cancer Res. 78, 3574–3588. https://doi.org/10.1158/0008-5472.Can-17-3376 (2018).
Wang, L. et al. α2,6-Sialylation promotes immune escape in hepatocarcinoma cells by regulating T cell functions and CD147/MMP signaling. J. Physiol. Biochem. 75, 199–207. https://doi.org/10.1007/s13105-019-00674-8 (2019).
Lin, W. D. et al. Sialylation of CD55 by ST3GAL1 facilitates Immune Evasion in Cancer. Cancer Immunol. Res. 9, 113–122. https://doi.org/10.1158/2326-6066.Cir-20-0203 (2021).
Leng, W. L., Yao, H., He, J. X. & Liu, X. W. Venturing beyond Donor-Controlled glycosylation: New perspectives toward Anomeric selectivity. Acc. Chem. Res. 51, 628–639. https://doi.org/10.1021/acs.accounts.7b00449 (2018).
Stowell, S. R., Ju, T. & Cummings, R. D. Protein glycosylation in cancer. Annu. Rev. Pathol. 10, 473–510. https://doi.org/10.1146/annurev-pathol-012414-040438 (2015).
Eichler, J. Protein glycosylation. Curr. Biol. 29, R229–r231. https://doi.org/10.1016/j.cub.2019.01.003 (2019).
Mackay, S. et al. Increased α2,3-sialyl N-glycosylated prostate-specific membrane antigen (PSMA) in post-DRE urine is associated with high grade group prostate cancer. Prostate. https://doi.org/10.1002/pros.24724 (2024).
Ferrer-Batallé, M. et al. Comparative study of blood-based biomarkers, α2,3-Sialic acid PSA and PHI, for high-risk prostate Cancer detection. Int. J. Mol. Sci. 18 https://doi.org/10.3390/ijms18040845 (2017).
Zhang, Z., Wuhrer, M. & Holst, S. Serum sialylation changes in cancer. Glycoconj. J. 35, 139–160. https://doi.org/10.1007/s10719-018-9820-0 (2018).
Funding
This work was supported by National Natural Science Foundation of China (82103207 and 82373237) and National Major Science and Technology Infrastructure of Translational Medicine (Shanghai) (TMSK-2021- 503).
Author information
Authors and Affiliations
Contributions
Conceptualization, S.Z. and B.F.; methodology, Y.S. and M.Y.; software, Y.S.; validation, M.Y., L.Z. and L.Z.; formal analysis, X.Y.; investigation, X.Y.; resources, L.Z.; data curation, Y.S. and L.Z.; writing—original draft preparation, Y.S. and M.Y.; writing—review and editing, J.S and B.F.; funding acquisition, S.Z and B.F. All authors have read and agreed to the published version of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shao, Y., Yu, M., Zhang, L. et al. In-depth analysis of lymph node metastasis-related sialylated protein profiling and their clinical and biological significance in colorectal cancer using mass spectrometry and multi-omics technologies. Sci Rep 14, 28535 (2024). https://doi.org/10.1038/s41598-024-79893-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-79893-z