Abstract
The aetiology of Alzheimer’s disease (AD) and Parkinson’s disease (PD) are unknown and tend to manifest at a late stage in life; even though these neurodegenerative diseases are caused by different affected proteins, they are both characterized by neuroinflammation. Links between bacterial and viral infection and AD/PD has been suggested in several studies, however, few have attempted to establish a link between fungal infection and AD/PD. In this study we adopted a nanopore-based sequencing approach to characterise the presence or absence of fungal genera in both human brain tissue and cerebrospinal fluid (CSF). We observed the presence of small fungal burden DNA in two AD brains and a control case (extensive amyloid angiopathy). This approach would be well-placed to investigate potential links between microbial infection and neurodegenerative disease.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) and Parkinson’s Disease (PD) are two of the most common neurodegenerative diseases in the world. Patients with AD or PD display a loss of neurons in the central nervous system (CNS), however, the mechanism by which this occurs has yet to be fully understood1. AD is the leading cause of dementia, characterised by progressive cognitive impairment, memory deficits, and loss of independence2, with over 520,000 people in the UK being affected by AD-caused dementia3. Parkinsonism is a clinical syndrome characterized by slowness of movement, rigidity, tremor, and postural instability4. Causes of Parkinsonism may include idiopathic PD and Lewy body dementia (LBD), with PD being the most common form of Parkinsonism4 and the second most common type of degenerative dementia in patients aged over 65 years5. Currently, identified risk factors include age, diabetes, hypertension, head injury, chronic kidney disease, depression, chronic obstructive pulmonary disease, stroke, genetic susceptibility, and environmental factors. With prevalence of AD, PD, and dementia steadily increasing, investigating further risks underpinning the aetiology of these diseases is crucial as they remain largely unknown2,6,7,8.
Recently, the potential role of microbial infection as an AD/PD risk factor or mediator of progression has gained attention due to the recognition of inflammation as a prominent feature of AD/PD. Additionally, evidence for elevated levels of proinflammatory cytokines in AD and PD patients and increased AD or PD risk conferred by variants in numerous immune-related genes has been shown9,10,11. It has also been suggested that Herpes virus type 1 (HSV1) and Chlamydia pneumoniae, and various types of spirochaete are associated with AD12. Moreover, a recent study highlighted the association between hospital related infections, such as urinary tract and respiratory infections, with dementia in the elderly population13. Mone et al.14 have compared bacterial communities in AD and non-AD brains and found differences in bacterial microbiomes, in which the brain microbiome starts off as a healthy microbiome and over time the microbiomes community changes until it becomes associated with AD.
However, in contrast to viruses and bacteria, the role of fungi in AD/PD and dementia has received little attention. The potential involvement of fungi should not be overlooked, as many species are capable of infecting the CNS and neuropathology may be mediated by subsequent inflammation15. More direct evidence supporting the potential role of fungal infection in AD/PD originates from a series of studies identifying fungal proteins and DNA in AD/PD patients’ brains and CSF16,17,18,19,20,21,22. In addition, a study has shown that certain fungal species such as Saccharomyces cerevisiae, Malassezia restricta and Candida albicans, could be associated with certain brain tumors23.
Although these findings appear promising, the presence of fungal proteins and DNA in AD/PD brains and CSF is insufficient for establishing the role of infection in the cause or progression of AD/PD as it is unknown whether the fungi were viable and actively contributing to disease pathology24. Furthermore, fungal material was also detected in some control individuals19,20,21, which may be expected as increased blood-brain barrier (BBB) permeability is also associated with normal healthy ageing25 and there is the possibility of brain microbiomes being present24. For these reasons, it is necessary to develop methods to further establish the potential involvement of fungi in the CNS of AD/PD patients and to establish whether the fungi are potentially pathogenic, as well as contributing to neurodegeneration via inflammatory pathways. Our findings suggest that there is a low, but detectable, fungal burden in some human brain samples, regardless of disease status.
Methods
Mice tissue
Murine brains were obtained from the Armstrong-James laboratory (Department of Infectious Diseases, Imperial College London). All mouse experiments were approved by the United Kingdom Home Office and the Imperial College London Animal Ethics Committee and performed in accordance with the project license PPL PF9534064. Heterozygote Cftrtm1Unc-Tg(FABPCFTR)1Jaw/J mice (originally obtained from Jackson laboratories) were bred in house. All mice were housed in individually vented cages with free access to autoclaved food and water. All methods are reported in accordance with ARRIVE guidelines.
Ex-vivo brain tissue from naive mice was used to optimize the DNA/RNA extraction method, quantitative polymerase chain reaction (qPCR), polymerase chain reaction (PCR) and nanopore sequencing. Briefly, to harvest brain tissue mice were euthanized by cervical dislocation, confirmed through severing the femoral artery. Heads were removed and the scalp was resected. A sagittal incision was carried out from the base of the skull to the tip of the snout. The cranium was then resected, and brains were removed en bloc and cryopreserved at -80 °C until required.
Human tissue
This project has been approved by Imperial College Healthcare Tissue Bank (ICHTB). Human tissue and CSF were obtained from the ICHTB, the Multiple Sclerosis and Parkinson’s Tissue Bank (PB), and the London Neurodegenerative Diseases Brain Bank (LNDBB) (Table 1). REC3 Wales has confirmed the approval of the ethics for the research project. The data obtained in this work was analysed anonymously. ICHTB is supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London. ICHTB is approved by Wales REC3 to release human material for research (22/WA/0214). PB is approved by Wales REC3 to release human material for research (18/WA/0238). The project is being undertaken solely within the UK and ethical approval for the project is provided under the London Neurodegenerative Diseases Brain Bank Research Tissue Bank ethics approval 18/WA/0206 from Research Ethics Committee for Wales. All research was performed in accordance with the relevant guidelines/regulations and informed consent was obtained from all participants and/or their legal guardians.
DNA extraction
Five different DNA extraction methods were tested in parallel (Supplementary File S1). Amongst these methods, the DNeasy PowerSoil Pro Kit was selected as it provided better performance (Supplementary File S1). DNA was extracted from 25 mg brain samples using the DNeasy PowerSoil Pro Kits (Qiagen, Germany, 47014) following the manufacturer’s protocol with the following modifications: a program with six repeats of bead beating at 4.5 m/s was performed for 45 s using the FastPrep-24™ 5G (Thermo Fisher). Samples were incubated for two minutes on ice after the third repeat. The solution C6 was replaced with elution buffer AE (19077) and the elution volume was decreased to 36 µL with an incubation step of 10 min. Nuclease-free water and PBS buffer were extracted with the samples to control for potential reagent contamination.
Handling brain and CSF samples in the laboratory
To avoid sample contamination, the handling of human brain and CSF samples, as well as DNA/RNA extractions, were performed in a safety cabinet (Class 2) equipped with a HEPA filter. The safety cabinet, pipettes, tube racks, and other lab consumables were subjected to a treatment of UV light for 30 min before the handling of the samples. In addition, all pipettes and working surfaces were decontaminated with DNA AWAY decontaminant and 10% ChemGene. Disposable sterile blades were used to perform the slicing of the human brains and sterile filtered tips were used to handle the CSF samples. All Eppendorf, PCR tubes, and beads (for bead beating) used in this study were DNA/RNA free to avoid the introduction of contaminants. All DNA and RNA extractions had negative controls (nuclease free-water and PBS) and kit controls.
Cerebrospinal fluid (CSF)
CSF was extracted by using the DNeasy Blood and Tissue Kit (Qiagen, 69504) with the following modifications: 500 µL of CSF was centrifuged for 10 min at 13,000 rpm and the supernatant (300 µL) was discarded. CSF was then resuspended with remaining fluid (200 µL). The volume of buffer AE was reduced to 36 µL.
RNA extraction and reverse transcription qPCR (rt-qPCR)
RNA was extracted from 25 mg brain samples using the ZymoBIOMICS™ RNA Miniprep Kit (Zymo Research R2001) following the manufacturer’s protocol with the following modifications: three repeats of bead beating at 4.5 m/s was performed for 45 s using the FastPrep-24™ 5G (Thermo Fisher) and the final elution volume was decreased to 40 µL. Extracted RNA were reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, 4368814) followed by qPCR (FungiQuant Table 2).
Fungal mock community and spiking mice brains
Aspergillus fumigatus, Candida auris and Cryptococcus neoformans were cultured in Sabouraud Dextrose Agar (SDA) containing chloramphenicol at concentration of 100 mg/L. A. fumigatus was incubated for three days at 45 °C, and C. auris and C. neoformans were incubated for five days at 37 °C. After the incubation period, A. fumigatus, C. auris, and C. neoformans were sub-cultured into T25 Nunc EasYFlasks containing SDA. The harvesting of fungal spores for all species was performed by adding 10 ml of sterile 0.1% Tween into the T25 flasks, followed by vigorous agitation and finally filtration through a 10 ml sterile syringe filled with glass wool. Spore counting was performed using a Hemocytometer Counting Chamber (Neubauer-improved bright line Marienfeld, Germany). The spore suspensions of each species were diluted to 2 million spores/ml. A combined 1 million spore (absolute number) mock community containing equal proportions of A. fumigatus, C. auris and C. neoformans was made. To mimic the presence of fungi in infected brains, 25 mg mice brain samples were spiked with 1 ml of 10-fold serial dilutions of the mock community from 10,000 spores to 1 spore (absolute number). Spiked brains were vortexed for 15 s and centrifuged at 13,000 s rpm for 5 min. Supernatant was removed and mice brains were subjected to DNA purification procedures using the DNeasy PowerSoil Pro Kits (Qiagen, Germany, 47014). DNA purification was followed by qPCR and nanopore sequencing.
Quantitative polymerase chain reaction (qPCR) assays (FungiQuant)
To detect fungal DNA and cDNA in extracted mice and human brain and CSF samples, qPCR was performed using probes and primers (Table 2) targeting a highly conserved region of the fungal 18S ribosomal DNA (rDNA). Sequences were obtained from Liu et al.26, and FungiQuant qPCR settings are shown in Table 2.
A standard curve was generated for each 96-well plate using 10- fold serial dilutions of Mycobiome Genomic DNA Mix (ATCC, USA, MSA-1010), containing equal proportions of A. fumigatus, C. neoformans, Trichophyton interdigitale, Penicillium chrysogenum, Fusarium keratoplasticum, Candida albicans, Candida glabrata, Malassezia globosa, Saccharomyces cerevisiae, and Cutaneotrichosporon dermatis, ranging from one million to one genome copy equivalence. qPCR on each sample was conducted in triplicate with additional controls with no template (all reaction components excluding DNA) and positive controls (mice brain spiked with A. fumigatus, C. auris and C. neoformans). Data was analysed using the Design and Analysis Software v2.6.0 (Applied Biosystems). In the data analysis, default thresholds were used and the average quantification cycle (Cq) of nuclease-free water, PBS, or the no template control, whichever amplifies earlier, was used as a cut-off to reduce false positive results generated by reagent contamination. Samples were defined as positive for fungi when ≥ 2/3 triplicates demonstrate amplification26,27 with Cq values lower than the average Cq of nuclease-free water, PBS, or the no template control.
PCR barcoding for MinION nanopore sequencing
Prior to sequencing, initial PCR was performed on DNA extracted human brain and CSF to amplify fungal targets and attach sequencing barcodes. Primers targeting the fungal ribosomal operon from the V3 region of 18S rDNA to D3 region of 28S rDNA (Fig. 1; Table 2) were used. Sequences were obtained from D’Andreano and Francino28 and PCR settings are shown in Table 2. Negative control templates (nuclease free water) were included in all PCR runs.
Schematic representation of the fungal ribosomal operon region to be sequenced. Forward (blue arrow) and reverse (orange arrow) primers targeting the fungal ribosomal operon and their sequences are shown. Oxford Nanopore universal tag sequences preceding the 18S and 28S rDNA-specific primer sequences are underlined in blue. The resulting ~ 3.5 kb amplicon covers hypervariable internal transcribed spacer regions 1 and 2 (ITS1 and ITS2), as well as variable domains V3–V9 of the 18S rDNA gene and D1–D3 of the 28S rDNA gene, which can refine taxonomic assignment. Figure modified from D’Andreano and Francino28. Presence of PCR products was assessed using 0.5% agarose (Sigma-Aldrich, USA, A9539) gel electrophoresis with SYBR Safe DNA Gel Stain (Invitrogen, USA, S33102), and 1 kb DNA ladders (New England Biolabs, N3232S). Amplicons were subsequently purified using the Genomic DNA Clean and Concentrator®-10 (D4011). Amplicon quantification and size assessment were performed using TapeStation 2200 (Agilent Technologies, G2965A) and the D5000 ScreenTape System (Agilent Technologies, 5067-5588, 5067-5589).
Barcoding PCR was performed using the PCR Barcoding Expansion 1–12 kit (Oxford Nanopore Technologies, EXP-PBC001). Products from the initial PCR were adjusted to a final volume of 48 µL using nuclease-free water. Only products with sufficiently high concentrations were adjusted to a final concentration of 4.17 nM. Each 100 µL reaction contained 48 µL of PCR product, 2 µL of 10 µM PCR barcode, and 50 µL of PCR mix consisting of 20 µL of 5× Phusion HF Buffer, 2 µL of 10 mM dNTP mix, 1 µL of Phusion Hot Start II High-Fidelity DNA Polymerase, and 27 µL of nuclease-free water. PCR conditions were initial denaturation at 98 °C for 30 s, followed by 15 cycles of 98 °C for 10 s, 62 °C for 15 s, 72 °C for 2 min, and final extension at 72 °C for 10 min. Amplicons were purified using the Genomic DNA Clean & Concentrator®-10 (D4011) and quantified using the broad-range double-stranded DNA assay kit (Invitrogen, USA, Q32853) on the Qubit 2.0 Fluorometer (Invitrogen, USA, Q32866) and TapeStation 2200.
Library preparation and nanopore MinION sequencing
Barcoded PCR products were pooled in 47 µL of nuclease-free water at equal ratios (totalling 1 µg) and library preparation was carried out using the Ligation Sequencing Kit 1D (Oxford Nanopore Technologies, SQK-LSK109) following manufacturer’s protocols. Samples were quantified by Qubit 2.0 and TapeStation 2200. R9.4.1 flow cells (Oxford Nanopore Technologies, FLO-MIN106.001) were primed and quality controlled following manufacturer’s instructions prior to sample loading. Sequencing was performed using MinKNOW v22.03.5 with 36 h runtime. Fast basecalling and barcoding was enabled and reads with Qscore < 8 were filtered out. Reads longer than 500 bp were retained for downstream analysis.
Sequence read processing
The quality of raw sequencing reads was assessed using NanoPlot v1.42.029. Sequencing reads then underwent adapter removal using PoreChop v0.2.430 and subsequent quality filtering using NanoFilt v2.8.031. Sequences were retained if they met the following criteria: minimum quality score = 10, minimum length = 500, maximum length = 4000. Reads were then clustered with 90% identity using VSEARCH v2.27.032 set with the “—cluster-fast” and “—strand both” parameters.
Species assignment
The central sequence was extracted from each cluster using the “split” and “range” functions from SeqKit v2.5.133 to produce representative sequences per cluster. All reads from a cluster were then used to polish the representative sequence to produce a consensus using Medaka v1.11.3 (https://github.com/nanoporetech/medaka). A custom genome database was created by obtaining 9,000 fungal and two human genomes from NCBI34 (date accessed: 23/01/2024), covering 5303 fungal species. These were combined and generated into a searchable database using BLAST35 v2.15.0. The consensus sequence for each cluster was then searched against the custom database using nucleotide BLAST (90% identity, 90% coverage, 1000 maximum target sequences, one maximum high-scoring pairs). A custom Python v3.11.436 script, incorporating the module ‘Pandas’ v1.5.337,38,39, was then generated to assign the appropriate taxon to each cluster and obtain the read counts for each respective cluster. The species with the highest bit-score was compared to other alignments that had a bit-score difference less than 40. Using the species names, it was calculated whether alignments were from a single species, genus or from multiple genera and therefore classified as an “Unknown fungal classification”. The taxonomic assignments were collected and visualised in R v4.0.240 using the ‘ggplot2’ v3.4.441, ‘dplyr’ v1.1.442, ‘readr’ v2.1.543, ‘ggsci’ v3.2.044 and ‘forcats’ v1.0.045.
Results
Detecting fungal DNA in human samples
No fungal DNA was detected upon performing qPCR assays on all human brain samples and CSF samples. Spiked mice brains with the fungal mock community (A. fumigatus, C. auris and C. neoformans) showed amplification ranging from 100 to 1000 spores (spiked into mice brain). It is worth noting that number of spores spiked into the mice brains is not equivalent to the number of genome copies present in Mycobiome Genomic DNA Mix (ATCC, USA, MSA-1010), shown in the Table 3.
When spiking the brain with mock community the Cq Mean value for the mice spiked brain with 100 fungal spores is closer to 1 genome/copy than the 100 genome/copies ATCC DNA standard. The limit of detection (LOD) for genomic DNA standards (MSA-1010) used in this assay is one genome copy.
RNA extracted from human brain samples (Table 1) was transcribed into cDNA and a qPCR assay was performed using the 18S primers mentioned above. No fungal cDNA was detected in any of the brain samples (Table 1).
Species classification
Human brain and CSF samples, mycobiome genomic DNA Mix and “DNA-free” negative controls (BSA and DNA extraction kit control) was sequenced on MinION and analyzed using BLAST to determine which species, if any, are present in AD/PD control brains and CSF. The number of reads for each sample is shown in Table 4.
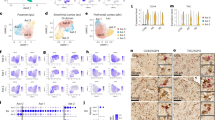
Metabarcoding analysis of samples. (A) Genus-level relative abundance of fungi and human (~ 3.5 kb amplicon region) of sequences taken from patients with/without either Alzheimer’s or Parkinson’s (disease and control), alongside analysis of DNA-free standards (negative) and known mock community standards (positive). X-axis indicates each sample, the first y-axis (left-hand side) indicates relative abundance as a percentage and the second y-axis (right-hand side) indicates total read count. Samples marked with a black triangle are from CSF samples, the remaining are from patient brains. Black circles depicts the number of reads per barcode. (B) Box-plot depicting total read numbers for each condition.
During the analysis of the metabarcoding data it was observed that DNA-free controls had limited read counts (total read counts ranged from 103 to 403, Fig. 2A; Table 4), whereas known mock communities had a higher read count (ranged from 23914 to 56262, Fig. 2A; Table 4). In addition, disease conditioned brains had higher read counts compared to control brains. However, these counts were a fraction of the positive control (Fig. 2B).
Our nanopore approach was able to correctly identify all ten genera present in the mycobiome genomic standards (barcode 11; Fig. 2A): Alternaria, Aspergillus, Candida, Cryptococcus, Cutaneotrichosporon, Fusarium, Malassezia, Nakaseomyces (Nakaseomyces glabratus previously known as Candida glabrata), Penicillium, Saccharomyces and Trichophyton (Supplementary Table S1). In addition, we were also able to detect the three genera, Aspergillus, Candida and Cryptococcus, spiked into the mice brain with a relative abundance of 73.3% for Candida and 26.4% for Cryptococcus (barcode 19, Fig. 2A) (Supplementary Table S1). However, the number of reads for Aspergillus were extremely small, with only three reads for barcode 11 (genomic standards) and four reads for barcode 19 (spiked mice brain with fungal mock community). This could possibly be explained by the high variability within the ITS region; despite this variability making the ITS region an excellent fungal DNA barcode region, it can also create shortcomings due to the large sequence divergences. For instance, ~ 42% of full-length ITS sequences in the International Nucleotide Sequence Database Collaboration (INSDC) do not include the full species names and 29% are only annotated as belonging to the fungal Kingdom (or labelled as uncultured fungus)46. In addition, there is a great proportion of fungal sequences in the databases that are incorrectly annotated due to either changes in the taxonomic classification over the years or due to simple human error47.
The only human brain samples that contained a higher number of reads were barcodes 5 (AD brain, 10171 reads), 6 (control brain, 7543 reads) and 8 (AD brain, 12307 reads) (Fig. 2A represented by the black circular dot and Supplementary Table S1). Figure 3 depicts the relative abundances for barcodes 5, 6 and 8 with the exclusion of the human reads, allowing for a closer focus on the fungal burden in those samples. The total number of reads for barcodes 5, 6 and 8 was 10,149, 7543 and 12,307, respectively (Supplementary Table S1); however, when removing human DNA from the analysis we obtained a relative abundance (fungi only) of 57.8%, 6.2%, and 10% for barcodes 5, 6 and 8 respectively. Barcode 5 relative abundance is higher than barcode 6 and 8, as observed in Fig. 3. When looking at barcode 5 it is worth pointing out that 54% (relative abundance) of the reads were classified as “Unclassified Filobasidiaceae” whereas for barcode 8 it is only 8.9% (Supplementary Table S1). A common trend of barcodes 5, 6 and 8 is the presence of members of the genera Cryptococcus, Saccharomyces and Cutaneotrichosporon. In addition, we detected the presence of Candida in barcode 6 and 8 but not in barcode 5.
Discussion
AD and PD are both neurodegenerative diseases associated with the abnormal aggregation of proteins in the CNS such as amyloid-β, tau and α-synuclein48. Despite the fact that both AD/PD have been studied for over a century, the exact causes of these neurodegenerative diseases are still far from being understood49. Several studies have been able to establish a link between bacterial or viral infection and AD/PD50,51, however, few have attempted to establish a connection between fungal infection and AD/PD. Studies conducted by Pisa and Alonso et al.17,18,19,20,21,22 have detected fungal DNA in AD/PD brains belonging to species in the Alternaria, Botrytis, Candida, Cladosporium, Malassezia, Saccharomyces, and Cryptococcus genera, with the latter being the most commonly found genus. Alternaria, Candida, Cladosporium, Malassezia, and Cryptococcus species have been found to be capable of infecting the CNS and are capable of causing fungal menigitis16,42. In fact, there have been reports of cryptococcal meningitis misdiagnosed as AD due to its cause of dementia-like symptoms in patients, who demonstrated loss of memory and independence in performing certain activities as well as slight behavioural changes, with these cognitive deficits reversed following antifungal treatment52,53.
Here, we used standard PCR-based methods alongside nanopore sequencing on the MinION platform to profile the microbial content of healthy and AD/PD brains and CSF. First, we developed and validated a method allowing the detection of fungal DNA in mammalian brain tissue. Primers targeting the fungal ribosomal operon with an amplicon size of 3.5 kb were used, followed by nanopore sequencing, as well as qPCR and RT-qPCR (Fig. 1; Table 2). qPCR and RT-qPCR did not detect fungal DNA in any human brain/CSF samples. The qPCR assay employed in this study allowed the detection low levels of fungal DNA in mice brain (Table 3). However, the number of spores spiked into the mice brains is not proportional to genome copies in the mycobiome standards and there is a ~ 1-fold loss of sensitivity. Possible explanations for this include (i) when counting the number of spores under the microscope there is always some error in accuracy, (ii) some spores may have lost viability and their DNA may have degraded, (iii) the fungal cell wall is resilient to our DNA extraction process and we may have extracted fewer fungal spores than initially counted. Future work would focus on exploring ways to enhance the efficiency of extracting DNA from fungal-infected host tissue.
Secondly, we used MinION sequencing to detect and quantify fungal species present in brain tissue samples. Our nanopore approach successfully identified all ten genera (barcode 11; Fig. 2A) present in the mycobiome genomic standards: Alternaria, Aspergillus, Candida, Cryptococcus, Cutaneotrichosporon, Fusarium, Malassezia, Nakaseomyces (Nakaseomyces glabratus previously known as Candida glabrata), Penicillium, Saccharomyces and Trichophyton. In addition, our nanopore approach was able to detect the three species (barcode 19) spiked into the mice brain, although for both barcodes (11 and 19) the number of reads for the genus Aspergillus is extremely small. This might be explained due to the large sequence divergences within this region54 and also due to errors in sequences annotations over the years that could lead to false assumptions when it comes to species identification.
Of relevance, our findings suggest the presence of fungal DNA in two AD patients (barcode 5 and 8) and one control brain (barcode 6) with the genera Cryptococcus, Saccharomyces and Cutaneotrichosporon being the 3 most common genera found amongst the three patient samples. This finding is in line with studies conducted by Pisa and Alonso et al.17,18,19,20,21,22. In addition, it is worth noting that barcode 6 was used on a control brain sample from a 90-year-old patient that exhibited an extensive amyloid angiopathy, which is characterized by the accumulation of amyloid β-peptide. The presence of fungal DNA in barcode 6 could be expected as AD disease is associated with the abnormal aggregation of proteins such as amyloid β-peptides. Furthermore, it can be observed that barcodes 5, 6 and 8 have reads classified as members belonging to the family Filobasidiaceae. This family previously contained (before the move to “one fungus, one name”) fungi that are known human pathogens such as Cryptococcus neoformans. C. neoformans is known to be able to capable of crossing the BBB and to cause meningitis55,56. However, we cannot assume that the fungal reads classed as “Unclassified Filobasidiaceae” are Cryptococcus or other species or genera that used to belong that family, since there were changes in taxonomy and there are still possible lapses in the fungal database that were not updated. The lower sensitivity found in our nanopore approach when compared to the qPCR assay (FungiQuant), is further supported by a study where they aimed to detect bacterial biodefense pathogens by using amplicon sequencing. In this study it was found that after 10 min of nanopore sequencing the LODs were at least one to two orders of magnitude lower than qPCR (single plex)57.
In the future to improve sensitivity and confidence of species identification in metabarcoding studies, multiple pipelines and databases may need to be used. In addition, newer machine learning-based classification systems may also be considered to enable the identification of novel species58. Use of fungi-specific primers for PCR also reduces the need for depletion of background or host DNA and can increase detection of species with lower abundances59. This is however accompanied by the drawback of PCR artefacts and biases which affect the estimation of species abundance as the use of universal primers produces different amplification efficiencies in different species59. Varying copy numbers of the ribosomal operon across species may also affect the number of reads obtained from each species and skew abundance estimations59.
In conclusion, we found evidence of the presence of fungal DNA in one healthy human brain (with extensive amyloid angiopathy), as well as in two AD brains by the use of long-read nanopore sequencing. The approach developed during this study to detect low levels of fungal DNA in CNS tissue will be of use for further exploring the microbial aetiology hypothesis for AD and PD.
Data availability
All raw reads have been submitted to the National Center for Biotechnology Information with accession number: PRJNA1048355.Code availabilityAll scripts can be found at https://github.com/harrychown/brains/.
Code availability
All scripts can be found at https://github.com/harrychown/brains/.
References
Ransohoff, R. M. How neuroinflammation contributes to neurodegeneration. Science 353, 777–783 (2016).
Scheltens, P. et al. Alzheimer’s disease. Lancet 397, 1577–1590 (2021).
Alzheimer’s disease | Alzheimer’s Society. https://www.alzheimers.org.uk/about-dementia/types-dementia/alzheimers-disease (2023).
Keener, A. M. & Bordelon, Y. M. Parkinsonism. Semin. Neurol. 36, 330–334 (2016).
Walker, Z., Possin, K. L., Boeve, B. F. & Aarsland, D. Lewy body dementias. Lancet 386, 1683–1697 (2015).
alzheimer_europe_dementia_in_europe_yearbook_2019.pdf. https://www.alzheimer-europe.org/sites/default/files/alzheimer_europe_dementia_in_europe_yearbook_2019.pdf.
Lindestam Arlehamn, C. S., Garretti, F., Sulzer, D. & Sette, A. Roles for the adaptive immune system in Parkinson’s and Alzheimer’s diseases. Curr. Opin. Immunol. 59, 115–120 (2019).
Tahami Monfared, A. A., Byrnes, M. J., White, L. A. & Zhang, Q. Alzheimer’s disease: Epidemiology and clinical progression. Neurol. Ther. 11, 553–569 (2022).
Long, J. M. & Holtzman, D. M. Alzheimer disease: An update on pathobiology and treatment strategies. Cell 179, 312–339 (2019).
Kurz, C., Walker, L., Rauchmann, B.-S. & Perneczky, R. Dysfunction of the blood-brain barrier in Alzheimer’s disease: Evidence from human studies. Neuropathol. Appl. Neurobiol. 48, e12782 (2022).
Tansey, M. G. et al. Inflammation and immune dysfunction in Parkinson disease. Nat. Rev. Immunol. 22, 657–673 (2022).
Itzhaki, R. F. et al. Microbes and Alzheimer’s disease. J. Alzheimers Dis. 51, 979–984 (2016).
Janbek, J., Frimodt-Møller, N., Laursen, T. M. & Waldemar, G. Dementia identified as a risk factor for infection-related hospital contacts in a national, population-based and longitudinal matched-cohort study. Nat. Aging 1, 226–233 (2021).
Moné, Y. et al. Evidence supportive of a bacterial component in the etiology for Alzheimer’s disease and for a temporal-spatial development of a pathogenic microbiome in the brain. Front. Cell. Infect. Microbiol. 13, 1123228 (2023).
Phuna, Z. X. & Madhavan, P. A closer look at the mycobiome in Alzheimer’s disease: Fungal species, pathogenesis and transmission. Eur. J. Neurosci. 55, 1291–1321 (2022).
Alonso, R., Pisa, D., Rábano, A., Rodal, I. & Carrasco, L. Cerebrospinal fluid from Alzheimer’s disease patients contains fungal proteins and DNA. J. Alzheimers Dis. 47, 873–876 (2015).
Pisa, D., Alonso, R., Juarranz, A., Rábano, A. & Carrasco, L. Direct visualization of fungal infection in brains from patients with Alzheimer’s disease. J. Alzheimers Dis. 43, 613–624 (2015).
Pisa, D., Alonso, R., Rábano, A., Rodal, I. & Carrasco, L. Different brain regions are infected with fungi in Alzheimer’s disease. Sci. Rep. 5, 15015 (2015).
Pisa, D., Alonso, R., Rábano, A., Horst, M. N. & Carrasco, L. Fungal enolase, β-tubulin, and chitin are detected in brain tissue from Alzheimer’s disease patients. Front. Microbiol. 7, 1772 (2016).
Alonso, R., Pisa, D., Aguado, B. & Carrasco, L. Identification of fungal species in brain tissue from Alzheimer’s disease by next-generation sequencing. J. Alzheimers Dis. 58, 55–67 (2017).
Alonso, R., Pisa, D., Fernández-Fernández, A. M. & Carrasco, L. Infection of fungi and bacteria in brain tissue from elderly persons and patients with Alzheimer’s disease. Front. Aging Neurosci. 10, 159 (2018).
Pisa, D., Alonso, R. & Carrasco, L. Parkinson’s disease: A comprehensive analysis of fungi and bacteria in brain tissue. Int. J. Biol. Sci. 16, 1135–1152 (2020).
Narunsky-Haziza, L. et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 185, 3789-3806.e17 (2022).
Link, C. D. Is there a brain microbiome?. Neurosci. Insights 16, 26331055211018708 (2021).
Verheggen, I. C. M. et al. Increase in blood-brain barrier leakage in healthy, older adults. Geroscience 42, 1183–1193 (2020).
Liu, C. M. et al. FungiQuant: A broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 12, 255 (2012).
Charalampous, T. et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 37, 783–792 (2019).
D’Andreano, S., Cuscó, A. & Francino, O. Rapid and real-time identification of fungi up to species level with long amplicon nanopore sequencing from clinical samples. Biol. Methods Protoc. 6, bpaa026 (2021).
De Coster, W. & Rademakers, R. NanoPack2: Population-scale evaluation of long-read sequencing data. Bioinformatics 39, btad311 (2023).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3, e000132 (2017).
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669 (2018).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11, e0163962 (2016).
Sayers, E. W. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–D26 (2022).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinform. 10, 421 (2009).
Van Rossum, G. & Drake, F. L. Python 3 Reference Manual: (Python Documentation Manual Part 2) (CreateSpace Independent Publishing Platform, 2009).
Rossum, G. V. & Drake, F. L. Python 3: Reference Manual (SohoBooks, 2009).
Data Structures for Statistical Computing in Python—SciPy Proceedings. https://proceedings.scipy.org/articles/Majora-92bf1922-00a (2010).
Team, T. Pandas Development. pandas-dev/pandas: Pandas. Zenodo https://doi.org/10.5281/zenodo.13819579 (2024).
R: The R Project for Statistical Computing. https://www.r-project.org/.
ggplot2: Elegant Graphics for Data Analysis (3e). https://ggplot2-book.org/.
tidyverse/dplyr.tidyverse (2024).
Read Rectangular Text Data. https://readr.tidyverse.org/.
Scientific Journal and Sci-Fi Themed Color Palettes for ggplot2. https://nanx.me/ggsci/.
Tools for Working with Categorical Variables (Factors). https://forcats.tidyverse.org/.
Abarenkov, K. et al. The curse of the uncultured fungus. MycoKeys 86, 177–194 (2022).
Lücking, R. et al. Fungal taxonomy and sequence-based nomenclature. Nat. Microbiol. 6, 540–548 (2021).
Mucke, L. Neuroscience: Alzheimer’s disease. Nature 461, 895–897 (2009).
Zhang, W., Xiao, D., Mao, Q. & Xia, H. Role of neuroinflammation in neurodegeneration development. Signal Transduct. Target Ther. 8, 267 (2023).
Panza, F., Lozupone, M., Solfrizzi, V., Watling, M. & Imbimbo, B. P. Time to test antibacterial therapy in Alzheimer’s disease. Brain 142, 2905–2929 (2019).
Bu, X.-L. et al. A study on the association between infectious burden and Alzheimer’s disease. Eur. J. Neurol. 22, 1519–1525 (2015).
Ala, T. A., Doss, R. C. & Sullivan, C. J. Reversible dementia: A case of cryptococcal meningitis masquerading as Alzheimer’s disease. J. Alzheimers Dis. 6, 503–508 (2004).
Hoffmann, M., Muniz, J., Carroll, E. & De Villasante, J. Cryptococcal meningitis misdiagnosed as Alzheimer’s disease: Complete neurological and cognitive recovery with treatment. J. Alzheimers Dis. 16, 517–520 (2009).
Xu, J. Fungal DNA barcoding. Genome 59, 913–932 (2016).
Kwon-Chung, K. J., Boekhout, T., Wickes, B. L. & Fell, J. W. Systematics of the genus Cryptococcus and its type species C. neoformans. In Cryptococcus: From Human Pathogen to Model Yeast (eds Heitman, J. et al.) 1–15 (Wiley, 2010). https://doi.org/10.1128/9781555816858.ch1.
Kwon-Chung, K. J. et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 4, a019760 (2014).
Player, R. et al. Comparison of the performance of an amplicon sequencing assay based on Oxford Nanopore technology to real-time PCR assays for detecting bacterial biodefense pathogens. BMC Genom. 21, 166 (2020).
Liang, Q., Bible, P. W., Liu, Y., Zou, B. & Wei, L. DeepMicrobes: Taxonomic classification for metagenomics with deep learning. NAR Genom. Bioinform. 2, lqaa009 (2020).
Ciuffreda, L., Rodríguez-Pérez, H. & Flores, C. Nanopore sequencing and its application to the study of microbial communities. Comput. Struct. Biotechnol. J. 19, 1497–1511 (2021).
Acknowledgements
This paper presents independent research funded by a Wellcome Trust Institutional Strategic Support Fund’s Springboard Fellowship Scheme (Imperial College London) awarded to JR (WPIA_PSN105). RL and MCF were supported by a grant from the Wellcome Trust (219551/Z/19/Z) and the MRC (MR/R015600/1). Dr. Tom Williams and Professor Darius Armstrong-James (DIDE, Imperial college) acknowledged for the donation of most of the mice brain samples analysed in this study. In addition, we also would like to acknowledge Dr. Norman van Rhijn and Professor Mike Bromley (Manchester Fungal Infection Group) for the donation of mice brains used during this project. JR has previously received an honoraria from Gilead Sciences. MCF is funded by the CIFAR Fungal Kingdoms program. We wish to thank the Imperial College Healthcare Tissue Bank, the Multiple Sclerosis and Parkinson’s Tissue Bank, and the London Neurodegenerative Diseases Brain Bank for their help and support with this project.
Author information
Authors and Affiliations
Contributions
R.L., M.C.F. and J.R. conceived and designed the study. R.L. and I.U.W. carried out the experimental work. T.J.W. carried out the animal work. R.L., H.C., I.U.W. and J.R. carried out the experimental validation and analysis. R.L., H.C., I.U.W., T.J.W., M.C.F. and J.R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leitao, R., Wan, I.U., Chown, H. et al. Detection of fungal sequences in human brain: rDNA locus amplification and deep sequencing. Sci Rep 14, 31790 (2024). https://doi.org/10.1038/s41598-024-82840-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-82840-7