Abstract
Allergic asthma in children is typically associated with house dust mites (HDM) as the key allergen. Nevertheless, the diagnostic rate remains below 60% due to the absence of specific symptoms and diagnostic markers, which hinders the implementation of targeted personalized therapies. This study investigates immunological features of asthma with house dust mite (HDM) sensitisation in children, aiming to uncover diagnostic markers at single-cell resolution. The cohort comprised 8 children with physician-diagnosed asthma (age range: 4–11 years), stratified into groups based on HDM sensitization status. Single-cell RNA sequencing of peripheral blood mononuclear cells (PBMCs) was conducted, employing Seurat for cell identification and differential gene expression analysis. Enrichment analyses and LASSO regression identified signature genes related to cellular origin, with protein–protein interaction networks elucidating cellular communication differences between groups. A total of 11 distinct cell types were identified, with classical monocytes and monocytes being the predominant cell types that differentiated the two groups. Among these, 12 genes were up-regulated, and 40 down-regulated, mainly involving MHC-II complex and antigen presentation pathways, as validated by Gene Ontology and Gene Set Enrichment Analysis. The machine learning model accurately predicted cellular groupings, evidenced by an area under the curve of 0.83. Enhanced communication signals in HDM allergy cases involved monocytes, contrasting with reduced interactions in naive CD8 + cells. HLA-DR and HLA-DP were identified as the primary hallmark receptors, and the innate immunity differences with non-dust mite allergic asthma were characterized by 18 genes including top candidates MT-ND4 and RPS3A. Individuals with HDM-sensitized asthma exhibited altered expression of MHC-II complex genes in their PBMCs and distinct gene expression patterns in antigen-presenting cells, highlighting the critical role of HLA-DR and HLA-DP in the HDM allergen presentation.
Similar content being viewed by others
Introduction
Asthma effects approximately 334 million individuals globally, with 43% of adults and 14% of children afflicted, and its prevalence is increasing worldwide1,2,3,4. While genetic variations contribute to heterogeneous responses to allergens and environmental triggers5,6,7, diagnostic disparities persist, particularly in developed countries where advanced clinical workflows (e.g., symptom evaluation, pulmonary function tests, and bronchodilator response assessments) enhance detection rates2,4,8. Allergic asthma is frequently linked to type 2 inflammation and is regulated by type 2 helper T lymphocytes (Th2) and respiratory epithelium. A fundamental process involves epithelial-derived cytokines and ILC2s initiating T2 asthma by stimulating dendritic cell (DC) activation and airway phenotypic alterations, which then lead to their migration to secondary lymphoid tissues (SLT) for allergen presentation to naive T cells and the induction of a T2 profile. Myeloid DCs (mDCs), plasmacytoid DCs (pDCs), and monocyte-derived DCs are central to this process9.
Childhood asthma, distinct from adult-onset disease, often manifests before age three, with dynamic phenotypes and stronger atopic predisposition10,11,12. The diagnosis of asthma in children is further complicated by their limited ability to articulate symptoms with clarity. Over 80% of pediatric cases are allergic8, frequently linked to house dust mite (HDM) sensitization13. HDM allergens, including Dermatophagoides pteronyssinus (DP) and Dermatophagoides farinae (DF), release proteases such as Der p1 and Der p2, which disrupt airway epithelial integrity, amplify Th2 responses, and trigger IgE-mediated inflammation14,15.
HDM-induced immune pathways and exacerbation triggers are critical to understanding asthma pathogenesis16,17. HDM allergens activate dendritic cells (DCs) and monocytes, leading to impaired MHC-II antigen presentation. This dysregulation contributes to chronic airway inflammation and reduced immune tolerances16,17. Additionally, HDM proteases disrupt epithelial barriers, allowing allergens to penetrate and activate innate immune sensors, such as formyl peptide receptor 2 (FPR2), which drives the release of Th2-promoting cytokines like IL-33 s17. These mechanisms amplify Th2 responses, exacerbating asthma symptoms.
Despite progress in identifying genetic risk variants on allergic asthma2,18,19, mechanisms underlying allergen-specific sensitization, particularly HDM-driven immune dysregulation, remain poorly characterized. Single-cell RNA sequencing (scRNA-seq) enables high-resolution profiling of cellular heterogeneity20,21,22. This enhanced resolution provides researchers with a more detailed and nuanced understanding of cell types, subtypes, and states.
Using the 10X Genomics platform, we compared peripheral blood mononuclear cells (PBMCs) from HDM-sensitized and non-sensitized children with asthma, identifying immune cell signatures and molecular markers unique to HDM-associated disease. To our knowledge, this is the first study to apply scRNA-seq for elucidating immune pathways in childhood asthma with HDM sensitization.
Methods
PBMC collection
Participants were recruited from the paediatric outpatient clinic of Shenzhen Bao’an Women’s and Children’s Hospital. Inclusion criteria comprised: (1) asthma diagnosis confirmed by the Guidelines for the Diagnosis and Optimal Management of Asthma in Children (2016)10, including recurrent respiratory symptoms (e.g., wheezing, dyspnea) and positive bronchial provocation test results; (2) age 4–11 years; (3) asthma medications (e.g., inhaled corticosteroids, bronchodilators) use ≤ 6 months and discontinuation ≥ 4 weeks prior to enrollment; and (4) stable, well-controlled asthma with documented absence of exacerbations and no guideline-indicated medications during the preceding 4 weeks. For children aged < 5 years, diagnosis relied on ≥ 3 annual episodes of respiratory symptoms with documented improvement post-bronchodilator therapy. Exclusion criteria included comorbidities (e.g., atopic dermatitis, food allergies), parasitic infections (excluded via stool microscopy and serum parasitic antigen testing), and acute infections within the preceding 4 weeks. Allergic status was confirmed by measuring total IgE and specific IgE to Dermatophagoides pteronyssinus (DP), Dermatophagoides farinae (DF), and Blomia tropicalis (BT)23,24. PBMCs were isolated from the whole blood of eight patients using EDTA-vacutainers (BD) and cryopreserved in RPMI1640 with 40% FCS and 10% DMSO. Following thawing and a one-hour incubation, the cells were washed twice in a medium supplemented with 0.04% BSA and subsequently processed for scRNA-seq (Fig. S1).
The diagnosis was made by an experienced deputy chief physician of pediatric based on clinical history, physical examination, and guideline criteria10.This included evaluating recurrent respiratory symptoms (e.g., wheezing, dyspnea) and verifying improvement post-bronchodilator therapy. Two trained nurses performed blood collection, spirometry, and bronchial provocation tests (BPT). One nurse performed the procedures, while the other nurse independently recorded the results to ensure the accuracy and consistency of data collection. The methods were in accordance with the ethical standards of the Declaration of Helsinki and its subsequent amendments. Written informed consent was obtained from all participants by their parents/legal guardians. This study was reviewed and approved by the Medical Ethics Committee of Shenzhen Baoan Women’s and Children’s Hospital, Jinan University (approval number: LLSCHY–2021–01–03).
scRNA-seq, QC, and data preprocessing
scRNA-seq was conducted using the 10 × Chromium controller (10X Genomics). The processed PBMC samples were loaded into discrete channels of a 10 × chip. The 10 × Genomics system, in conjunction with v2 or v3 reagents, enabled the capture of individual cells and the generation of sequencing libraries. Subsequent sequencing was conducted using a 150 bp paired-end kit on the Illumina NovaSeq 6000 platform at oeBiotech (Shanghai) Co.
CellRanger v5.0.0 software was utilized with the default settings to demultiplex data, produce FASTQ reads, align reads to the hg19 reference genome, and compute unique molecular identifier (UMI) barcodes, gene expression levels, and mitochondrial UMI percentages for each cell. Following the removal of doublets using DoubletFinder, only high-quality cells were retained for subsequent analysis by using the Seurat R package (V4.4.0)25. Cells were subjected to a rigorous quality control procedure and screened according to the upper and lower quartiles of the distribution of RNA features, and cells with more than 10% of mitochondrial genes were systematically excluded (Fig. S2a). The top 2,000 highly variable features were selected for linear dimensionality reduction via principal component analysis (PCA), with a focus on the top 20 principal components (Fig. S2b,c). A t-distributed stochastic neighbour embedding (t-SNE) dimensionality reduction was employed to achieve a clustering visualisation.
The non-HDM patients and HDM patients were assigned to groups C and T, respectively. Patient T05, a 5-year-old girl with a prolonged asthma exacerbation accompanied by high fever, exhibited the highest serum IgE levels, along with polysensitization to additional allergens (e.g., birch pollen) and CT-confirmed non-infectious status. The clustering results of T05 showed significant heterogeneity compared to the T group (Fig. S2d) and were therefore separately classified into the O group.
Transcriptome dimensionality reduction and cell clustering
The cluster markers were determined by utilizing FindAllMarkers within the Seurat package, with a criterion of positive expression threshold of 0.585 (|log(foldchange)|). The ten most highly expressed genes for each cluster were selected based on fold-change and the maximum difference in cell ratio between clusters. The cell types of the clusters were identified using the cell annotation tool CellMarker2.0, based on the respective cluster markers26,27.
Differential expression: mapping, enrichment, and interaction
The differential expression genes (DEGs) among various cell types within the T-C, O-T, and O-C control pairs were identified utilizing FindAllMarkers. The identified DEGs were filtered based on a threshold of 0.25 and an adjusted p-value of 0.05 and were subsequently visualised using the scRNAtoolVis package28. Gene Ontology (GO) term analysis and KEGG Gene Set Enrichment Analysis (GSEA) for DEGs were conducted using the Clusterprofiler and fgsea R packages29,30,31,32. The protein–protein interaction network (PPI) of DEGs was reconstructed using the String database (version 12.0)33,34,35,36, and further refined using the Cytoscape software (version 3.10)37.
Cell–cell communication inferring
Cell–cell communication was assessed using the CellChat software (version 1.1.0)38, which requires calculation of relative expression levels of a set of universally encoded genes in all transcriptomes relative to the total number of mapped reads. Expression levels were then averaged within each sample group to provide a comprehensive examination of the dynamics of cell-to-cell interactions. This strategy facilitates the identification of key signalling molecules and provides a comprehensive insight into their expression profiles across different cell subsets.
Classification model for identifying asthma patients with HDM sensitisation
A predictive model was developed to distinguish cells originating from HDM or non-HDM samples using least absolute shrinkage and selection operator (LASSO) Cox regression implemented in the glmnet R package39,40,41. The DEGs between groups T and C were used for model training, while all the cells were randomly divided into a training set (70%) and a test set (30%). The model was optimised by selecting variables and determining the optimal lambda through a binomial error distribution and a tenfold cross-validation test, with the aim of minimising classification error. Genetic traits under the optimal lambda were then examined the potential for improving prediction accuracy.
Statistics
All statistical analyses were implemented based on the R (v4.2.0). By default, p \(<\) 0.05 was deemed to be statistically significant.
Results
scRNA-seq identified discrete cell subpopulations and individuals responsive to HDM allergy
A total of 8 children (5 HDM-sensitized, 3 non-HDM-sensitized) aged 4–11 years (mean = 5.75) were included in this study. Table 1 summarizes the clinical characteristics of the cohort, including age, total IgE (tIgE), HDM-specific IgE (sIgE), and treatment status. The HDM-sensitized cohort (n = 5) exhibited elevated total IgE (tIgE) levels (mean tIgE = 1181 IU/ml; > 1000 IU/ml in four cases), consistent with allergic sensitization, alongside HDM-specific IgE (sIgE) levels > 40 U/ml to DP and DF, confirming an HDM-driven allergic asthma phenotype. In contrast, the control group (n = 3) demonstrated significantly lower tIgE levels (mean = 168.4 IU/ml; < 300 IU/ml in all cases) and undetectable DP/DF-specific IgE. PBMCs were isolated and subjected to 10X Genomics sequencing. After quality control, 5202 to 7881 cells were detected in each sample, with an average UMIs per cell ranging from 3618 to 5093, an average number of genes per cell ranging from 1385 to 1753, and an average percentage of mitochondrial UMIs per cell ranging from 0.0560 to 0.0890. A total of 36,601 cells and 32,254 genes were retained for subsequent analysis. The top 20 principal components were selected for dimensionality reduction based on the quality report (Fig. S2).
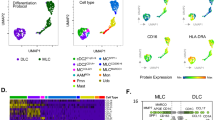
Following PCA reduction, tSNE grouped the cells into 25 clusters (Fig. 1a). The samples from the disparate groups exhibited a high degree of alignment across all clusters (Fig. 1b), while clusters 12, 14, and 16 were exclusive to patient T05 (Fig. 1c, left). Utilising the top 10 markers from each cluster, 11 cell types were annotated using the Cellmarker2.0 database (Fig. 1c, right). NK cells, naive CD8 + T cells, gamma-delta T cells, and naive CD4 + T cells were predominant in both control and test groups, except in patient T05. These cell types constituted approximately 80% of the total cell count in each patient. The representation of B cells, monocytes, classical monocytes, megakaryocytes, and dendritic cells was reduced in the HDM group. The percentages of classical monocytes, monocytes, and megakaryocytes were found to be statistically significantly different between the two groups (Fig. 1d). The cellular composition of T05 was distinct and characterised by a dominance of B cells and effector CD8 + memory T cells (Tem cells), with Tem cells being almost unique to this patient. The proportion of T lymphocytes in T05 was significantly lower at 23% compared to the other patients, with rates ranging from 76 to 80%.
The scRNA-seq profile of PBMCs from eight patients with allergic asthma (a) Clustering PBMCs using t-SNE dimensionality reduction. Twenty-five distinct clusters were discovered, each with a unique color. (b) Distribution of cells by sample group. Red: HDM-sensitized (group T); Blue: non-HDM-sensitized (group C); Green: patient T05 (group O) (c) Left: Cell clusters unique to group O, highlighted in green. Right: The percentage of each sample’s 11 primary cell types. Eleven major cell types comprise the 25 cell clusters: Naive CD8 + T cell, Naïve CD4 + T cell, Natural Killer cell, Gamma-delta T cell, B cell, Classical monocyte, Effector CD8 + memory T cell (Tem CD8 +), Mucosal-Associated Invariant T cell (MAIT cell), Monocyte, Megakaryocyte, and Dendritic. (d) The proportions of 11 different cell types in the groups that are HDM-allergic (T) and non-HDM-allergic (C), respectively. Wilcoxon’s test was employed to ascertain if the difference was significant. "*" indicates p < 0.05, and “ns” indicates not significant.
Differential expression analysis reveals signature genes responsive to HDM antigens
Altered expression levels of 52 genes were found between the HDM-sensitized and non-HDM-sensitized groups, with 12 genes upregulated and 40 downregulated (Fig. 2a, Table S1). GO term analysis indicated that the DEGs in the HDM-sensitized group are primarily associated with cell surface receptor-related processes (Fig. 2b), with notable involvement of HLA-DRA, CD74, and HLA-DPB1 in the GO term network. The GSEA based on the KEGG pathway revealed a significant enrichment of DEGs in the antigen processing and presentation pathway, with a particular emphasis on the role of MHC-II in the ASTHMA pathway (NES: 1.76, p < 0.05) (Fig. 2c). The PPI network demonstrated interactions between the DEGs (Fig. 2d), with cytohubba analysis identifying hub genes such as GZMH, GZMA, GZMB, GNLY, NKG7, CCL5, PRF1, KLRD1, KLRK1, and CD247, which were predominantly downregulated in the HDM-sensitized group and expressed mainly in atypical T cell types (NK, gamadelta T, and Tem) (Fig. 2e). Furthermore, several of these hub genes, including NKG7, CCL5, GZMA, PRF1, and CD247, were highly expressed in another atypical cell type, MAIT cells.
Analysis of differential expression gene (DEGs) in asthma patients with HDM sensitized (T) and non-HDM sensitized (C). (a) Volcano plot of 52 differential expression genes, with 40 down-regulated and 12 up-regulated in group T. (b) Network of Gene Ontology (GO) term by enrichment analysis of DEGs. The outer circular arrangement of nodes represents GO terms, the inner matrix arrangement of nodes represents DEGs, and the edges denote the genes participating in that particular pathway. Green nodes represent pathways, red nodes represent up-regulated genes, and blue nodes represent down-regulated genes. Pink edges represent biological processes (BP), blue edges represent molecular functions (MF), and orange edges represent cellular components (CC). Node sizes represent the enrichment score. (c) GSEA analysis of KEGG pathway. NES stands for Normalized Enrichment Score. (d) Protein–protein interaction (PPI) network of DEGs. Blue nodes: down-regulated genes; Pink nodes: up-regulated genes. Yellow rings: hub genes identified by cytohubba. Node sizes represent the link degree. (e) Expression of hub genes in 11 cell types. Colors indicate the average expression and node sizes represent the percentage of cells expressing that gene.
Among the 12 PBMC cell types, DEGs were not identified in 5 of them, mainly naïve CD8 + T cells, megakaryocytes, dendritic cells, and Tem CD8 + cells. The number of DEGs obtained ranged from 10 (classical monocyte) to 107 (gamadelta T cell) (Fig. 3a). In particular, SHNG5 and RPS3A, which are associated with ribosomal and mitochondrial RNA, were identified in all cell types. In addition, 14 other genes were found to function predominantly in ribosomes and mitochondria in multiple cell types (Fig. 3b)42. Enrichment associated with protein-binding components of the GO term, particularly the structural components of ribosomes, has been observed in several cell types except B-cells. Moreover, GO terms associated with MHC proteins were predominantly enriched in gamadelta T cells, B cells, and classical monocytes, suggesting involvement in immune receptor binding and activation pathways (Fig. 3c). HLA-DR, HLA-DQ, and HLA-DP are the primary antigen-presenting complexes within the MHC-II system. The expression levels of the genes associated with HLA-DR and HLA-DP were higher than those associated with HLA-DQ, with HLA-DR showing particularly strong expression (Fig. 3d). Specially, significant elevation of gene expression levels associated with HLA-DQ and HLA-DP was observed in the O group compared to the T group.
Differential expression analysis of different cell types in the HDM-sensitized group (T) and the non-HDM-sensitized allergic group (C). (a) DEG distribution in different cell types. The X-axis represents the 7 cell types in which DEGs were found, and the Y-axis represents fold changes in DEGs, with red dots and blue dots representing genes that are up-regulated and down-regulated in group T, respectively. (b) DEGs intersecting between cell types. Only DEGs that appear in more than four cell types are shown. The blue dots represent down-regulated genes, whereas the red upper triangles represent up-regulated genes. (c) DEG-enriched GO word intersection. The Y-axis displays the GO terms, while the X-axis displays the various cell types. The generation is shown by the size of the dots, while the color denotes the corrected p-value. (d) The expression of MHC-II-related genes is associated with HLA-DR, HLA-DQ and HLA-DP.
Regression model accurately distinguishes between cells of different patient origins
To identify potential biomarkers, DEGs between groups T and C were filtered using a LASSO regression model. Of the initial 52 DEGs, 37 were selected for model development (Fig. S3). Of these genes, 18 exhibited regression coefficients exceeding zero, with MT-ND4 and RPS3A displaying the highest coefficients (≥ 0.6, Fig. 4a). The area under the curve (AUC) of the model to discriminate between T and C groups of cells was 0.83 (Fig. 4b). Moreover, the accuracy, recall, and F1 score metrics all exceeded 0.75 in both the training and test datasets (Fig. 4c).
Performance of LASSO regression model for discriminating cells between the HDM-sensitized group (T) and the non-HDM-sensitized group (C). (a) The coefficients of thirty-seven signature genes filtered by LASSO regression. (b) ROC curves for classification by the LASSO model on the training and test datasets. (c) Performance evaluation of the LASSO model on the test and training datasets.
Inter-group differences in PBMC intercellular communications
CellChat was employed to compare cellular communication patterns between groups T and C to determine changes in PBMC cell–cell interactions due to mite allergies. The analysis showed no significant overall changes in the total number of interactions or the strength of interactions (Fig. 5a, Tem CD8 + cells excluded). However, at the single-cell level, a notable increase in HDM-related intercellular communications was observed in monocytes, while a decrease was noted in naive CD8 + T cells when compared to non-HDM patients (Fig. 5b, left panel). Regarding interaction strength, interactions involving gamma-delta T cells were weakened, whereas those involving B cells, classical monocytes, monocytes, and dendritic cells were strengthened (Fig. 5b, right panel). Given the importance of MHC-II in previous findings, we conducted further analysis to examine alterations in the levels of MHC-II in both afferent and efferent directions within the two cell groups. Our results indicated that there was an increase in efferent signal intensity solely in classical monocytes and B cells. Conversely, only the intensity of the afferent signal exhibited an increase in naive CD4 + T cells. It is noteworthy that both efferent and afferent signal intensities were elevated in dendritic cells and monocytes (Fig. 5c). Consequently, the MHC-II signal flow within these cells was visualised (Fig. 5c, bottom right).
Comparison of cell–cell communication between different cell types in the HDM-sensitized group (T) and the non-HDM-sensitized group (C). (a) Comparison of interaction number and strength between T and C groups. (b) Differences in the number and intensity of interactions between different cell types in groups T and C. The red and blue lines represent upward and downward changes in the T group, respectively. The arrow’s direction indicates where the interaction begins and ends. (c) Changes in signaling pathways in several cell types in group T versus group C. Cell types in the blue box (classical monocytes and B cells) only increase the intensity of efferent interactions in the MHC-II pathway. Cell types in the orange box (dendritic cells and monocytes) increase the intensity of both efferent and afferent interactions in the MHC-II pathway. Cell types in the green box (naïve CD4 + T cells) only increase the intensity of afferent interactions in the MHC-II pathway. The circle plot in the lower right corner depicts the direction of MHC-II characteristic pathway transmission among the five cell types. The colors represent distinct cell types, while the arrows indicate the direction in which the signal is transmitted.
Distinctive gene expression of PBMC in the severe asthmatic patient
The patient T05 exhibited distinct characteristics compared to other HDM patients in the TSNE dimensionality reduction analysis (Fig. 1). To investigate this variation and comprehend the molecular diversity, DEG analysis was performed on groups O and T. The distribution of DEGs was visualised in a volcano plot, which revealed 476 downregulated and 534 upregulated genes in the PBMCs of T05 (Fig. 6a). The downregulated genes were primarily associated with transcription-translation processes, while the upregulated genes were linked to electron chain transfer processes (Fig. 6b). The "antigen processing and presentation" pathway emerged prominently in the KEGG GSEA results with the highest normalized enrichment score. The “ASTHMA” pathway also appears in the enrichment results, featuring genes such as HLA-DRB1, HLA-DRB5, HLA-DPA1, HLA-DQB1, HLA-DPB1, and HLA-DRA (Fig. 6c). A subset of 35 genes was identified as common between the DEG sets of groups T and C, and groups O and T. The majority of these genes exhibited the lowest or highest expression levels in the T or O group, as opposed to the C group (Fig. 6d). Moreover, 29 out of the 35 shared DEGs were found to overlap with the 37 features selected in the LASSO model (Fig. 6e, Table S2).
Analysis of differentially expressed genes (DEGs) between T05 (O) and patients with HDM sensitisation (T). (a) Volcano plot of DEGs, with 1010 genes significantly altered, 476 genes down-regulated and 534 genes up-regulated in group O. (b) GO term enrichment analysis of DEGs (c) GSEA analysis of KEGG pathways (d) Expression of crossing DEGs in groups T, C, and O. Colors indicate the average expression of each gene in each group, and dot size represents the percentage of expressed cells (e) Intersection of 35 DEGs shared by groups T, C, and O and 37 signature genes chosen by the LASSO model.
Discussion
A more profound comprehension of asthma with HDM sensitisation and its distinctive attributes in comparison to other allergens at the cellular level may assist in the formulation of prevention and management strategies for children. This research explored the distinctions between asthma with HDM sensitisation and asthma without HDM sensitisation in children through scRNA-seq, identifying the immune cell composition and molecular variations between these two groups of patients. Furthermore, the integration of findings on molecular variances and cell interactions enabled the identification of potential signaling pathways and associated receptors that may be pivotal in asthma with HDM sensitisation.
We characterized the immune profile of asthma using scRNA-seq and bioinformatics analysis (Fig. 1a,b). Our results diverge from those reported by Zhong et al.43, who indicated a prevalence of CD4 + T cells over CD8 + T cells in healthy individuals, as we observed a reversal of this trend in individuals with asthma (Fig. 1c). The dysregulation identified in CD4 + and CD8 + T cell populations may play a role in the intricate immunological landscape associated with asthma. Notably, type 2 immunity has been strongly linked to asthma pathogenesis, leading to the differentiation of naive CD4 + T cells into T-helper effector cell subclusters like Th2, Th9, and Th1744,45,46,47. These cells, modulated by interleukin activity, drive the generation of IgE antibodies. Subsequent binding of IgE antibodies to mast cells and basophils triggers the release of allergenic mediators, such as histamine, tryptase, prostaglandins, leukotrienes and cytokines48, induces goblet cell hyperplasia, smooth muscle contraction, and increased vascular permeability, ultimately resulting in uncontrolled immunopathological responses by the respective cell subsets. Given the significant involvement of CD4 + T cells in asthma, we propose that in individuals with asthma, naive CD4 + T cells may have migrated to the airways as T-helper cells, potentially leading to their reduced detection in adequate numbers in PBMCs. Our study definitively confirmed that HDM allergy induces differences in PBMCs concerning monocytes, classical monocytes, and megakaryocytes (Fig. 1d). Monocytes act as precursors to macrophages in specific tissue organs and play a critical role in asthma-related inflammatory and immunological responses49, including antigen presentation. Lander et al. demonstrated that monocytes in the airway mucosa upregulate MHC-II and facilitate the expansion of lung dendritic cells (DCs)50,51. The upregulation of MHC-II and the proliferation of DCs undoubtedly improve the detection and presentation of allergens. The available evidence indicates that different allergens may induce comparable allergic responses. Indeed, HDM sensitisation does not necessarily result in an excessive proliferation and differentiation of immune cells. Rather, alterations in the antigen presentation process appear to be a crucial factor.
Numerous independent studies have consistently verified that T-cell responses to HDM antigens are restricted by HLA-DR52,53,54. The genes that are upregulated in the T-C control are primarily associated with MHC-II functions (Fig. 2a). The MHC-II complex is mainly responsible for presenting extracellular antigens and plays a significant role in allergen-specific IgE responses. In the context of asthma, HLA-II genes are linked to a specific set of MHC II-DR or -DQ alleles55,56, which align with the findings on DEGs. Both GO and KEGG gene set enrichment analyses, in addition to the upregulation of HLA-* genes, emphasize the crucial involvement of MHC-II in individuals with HDM-induced asthma (Fig. 2b,c). A related research study suggested that the restriction of class II HLA may contribute to individual variations in responsiveness to common allergens, with different allergens showing a preference for HLA-DR and HLA-DP genotypes57. We found that genes associated with HLA-DR and HLA-DP were more strongly expressed in patients with HDM sensitisation (Fig. 3d). The alpha and beta chains of HLA-II form heterodimers on antigen-presenting cell surface and facilitate the presentation of antigen peptides for recognition by CD4 T cells. Our findings are in line with this mechanism. No consistent and significant correlation was observed between the HLA-II motif and IgE responses to other allergens such as grass pollen, birch pollen, and cat dander. It indicates a notable association between HDM-specific IgE responses and MHC-II alleles, which may not be the case for immune responses induced by other allergens58. It is reasonable to hypothesize that HLA-DP and HLA-DR are the primary vectors for presenting HDM antigens. In addition, the DEGs-based LASSO model effectively distinguished between cell populations in HDM and non-HDM patients, suggesting that these genes could be representative of the gene expression profile of each patient group. Although the genes significantly contributing to the model may not have a direct relationship with MHC-II, they are associated with asthma, such as GZMB59, which is primarily expressed in cytotoxic T lymphocytes (CTLs), natural killer (NK) cells, and activated CD8 T cells, promoting persistent airway inflammation and remodeling in severe asthma; while the reduction of DUSP2 can reverse neutrophilic airway inflammation and cytokine responses in asthma mouse models60.
Cellchat analysis re-emphasized the importance of the MHC-II signaling pathway in HDM-induced asthma development, suggesting that B cells and classical monocytes play a key role in signal output (Fig. 5c), with Naive CD4 + T cells serving as signal recipients, and dendritic cells and monocytes potentially functioning as signal transducers. These cell types, known as antigen-presenting cells (APCs), are able to internalise extracellular debris and present it on their surface for recognition by T cells as potentially harmful non-self antigens61. Consequently, the physical interactions between T cells and antigen-presenting cells are crucial for the initiation of an immune response. Limited research exists on the direct interactions between B cells and monocytes or dendritic cells, warranting further investigation to elucidate their potential direct involvement in asthma.
In the outpatient setting, patients with T05 had more severe symptoms and a stronger IgE response compared to others in the HDM group. The specific IgE assay identified additional allergens, such as BT, indicating a diverse HDM population. Analysis of PBMCs using scRNA-seq revealed a distinct profile for T05, characterized by a predominance of effector memory T (Tem) cells (Fig. 1b). Tem cells, derived from effector T (TE) cells, possess prolonged lifespan, maintain immune memory, and enhance immune function and antigen sensitivity62. TE cells are known to drive type 2 immune responses in allergic asthma, a trait that may also be attributed to Tem cells63. Notably, the genetic disparities between T05 and the HDM cohort surpassed those between HDM and non-HDM patients (Fig. 1), and the expression pattern of DEGs reflected these distinctions (Fig. 6). A progressive relationship was observed between group C, group T, and T05, although the association with asthma severity remains uncertain. The enrichment of shared genes suggested that viral infection may be a contributing factor. Noteworthy findings included the association of Tem cell formation with viral infections and the exacerbation of allergic asthma by viral pathogens64,65,66, implying a history of respiratory viral infections during childhood for patient T05.
Limitations
A limitation of this study is that PBMCs from healthy children were not obtained for comparison in this study due to the limited availability of samples and the associated costs. The inclusion of additional clinical information is hampered by changes in the medical facilities in which the patient is scheduled to visit. Additionally, the conclusions regarding HDM antigen presentation via HLA-DP/DR and dendritic cell processing remain speculative, as they are inferred from transcriptional data rather than direct experimental validation. While consistent with prior studies implicating HLA-II pathways in HDM allergy, functional assays are needed to confirm causality. Although peripheral blood reflects immune status, confounding effects of comorbidities cannot be entirely excluded. Further mechanistic studies is recommended based on our findings.
Conclusions
In summary, this study employed scRNA-seq and bioinformatics to examine the molecular characteristics of children with HDM sensitisation asthma. The analysis demonstrated that the differences observed PBMCs were predominantly evident in antigen-presenting cell. The study results suggest that HDM-associated antigens are recognised and presented by HLA-DR and HLA-DP in the MHC-II signaling pathway of B cells, classical monocytes, and monocytes, and subsequently, these antigens are processed by dendritic cells and monocytes, resulting in the activation of naïve CD4 + T cells and their differentiation into Th2 cells, thereby initiating allergic responses. Moreover, the study identified 37 PBMC signature genes that can accurately differentiate between the two patient groups, thus offering potential diagnostic markers for further research.
Data availability
Data used in this study are freely available upon request to the corresponding author, Dr. Lingyun Zou (lingyun.zou@gmail.com).
Abbreviations
- HDM:
-
House dust mites
- DP:
-
Dermatophagoides pteronyssinus
- DF:
-
Dermatophagoides farinae
- scRNA-seq:
-
Single-cell RNA sequencing
- PBMCs:
-
Peripheral blood mononuclear cells
- tIgE:
-
Total immunoglobulin E
- PCA:
-
Principal component analysis
- t-SNE:
-
T-distributed stochastic neighbour embedding
- DEGs:
-
Differential expression genes
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- PPI:
-
Protein-protein interaction network
- LASSO:
-
Least absolute shrinkage and selection operator
- APCs:
-
Antigen-presenting cells
References
Zar, H. J. & Ferkol, T. W. The global burden of respiratory disease-impact on child health. Pediatr Pulmonol. 49, 430–434. https://doi.org/10.1002/ppul.23030 (2014).
Mims, J. W. Asthma: Definitions and pathophysiology. Int. Forum Allergy Rhinol. https://doi.org/10.1002/alr.21609 (2015).
Vos, T. et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the global burden of disease study 2010. The Lancet. 380, 2163–2196. https://doi.org/10.1016/S0140-6736(12)61729-2 (2012).
To, T. et al. Global asthma prevalence in adults: Findings from the cross-sectional world health survey. BMC Public Health https://doi.org/10.1186/1471-2458-12-204 (2012).
Himes, B. E. et al. Asthma-susceptibility variants identified using probands in case-control and family-based analyses. BMC Med. Genet. https://doi.org/10.1186/1471-2350-11-122 (2010).
Himes, B. E. et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am. J. Human Genet. 84, 581–593. https://doi.org/10.1016/j.ajhg.2009.04.006 (2009).
Dahlin, A. et al. Large-scale, multiethnic genome-wide association study identifies novel loci contributing to asthma susceptibility in adults. J. Allergy Clin. Immunol. 143, 1633–1635. https://doi.org/10.1016/j.jaci.2018.11.037 (2019).
Papi, A., Brightling, C., Pedersen, S. E. & Reddel, H. K. Asthma. The Lancet. 391, 783–800. https://doi.org/10.1016/S0140-6736(17)33311-1 (2018).
Agache, I. et al. Advances and highlights in asthma in 2021. Allergy 76, 3390–3407. https://doi.org/10.1111/all.15054 (2021).
Subspecialty Group of Respiratory Diseases, Society of Pediatrics, Chinese Medical Association, Chinese Journal of Pediatrics Editorial Board. Guideline for the diagnosis and optimal management of asthma in children. Chin. J. Pediatr. 54(3), 167–181 (2016)
Guilbert, T. W., Bacharier, L. B. & Fitzpatrick, A. M. Severe asthma in children. J. Allergy Clin. Immunol Pract. 2, 489–500. https://doi.org/10.1016/j.jaip.2014.06.022 (2014).
Padem, N. & Saltoun, C. Classification of asthma. Allergy Asthma Proc. 40, 385–388. https://doi.org/10.2500/aap.2019.40.4253 (2019).
Gray-Ffrench, M., Fernandes, R. M., Sinha, I. P. & Abrams, E. M. Allergen management in children with type 2-high asthma. J. Asthma Allergy. 29(15), 381–394. https://doi.org/10.2147/JAA.S276994 (2022).
Singh, M. & Hays, A. Indoor and outdoor allergies. Prim. Care: Clin. Off. Pract. 43, 451–463. https://doi.org/10.1016/j.pop.2016.04.013 (2016).
Gold, D. R. et al. NIAID, NIEHS, NHLBI, and MCAN workshop report: The indoor environment and childhood asthmaimplications for home environmental intervention in asthma prevention and management. J. Allergy Clin. Immunol. 140, 933–949. https://doi.org/10.1016/j.jaci.2017.04.024 (2017).
Wang, Q. et al. Neferine attenuates HDM-induced allergic inflammation by inhibiting the activation of dendritic cell. Inflammation 46(6), 2433–2448. https://doi.org/10.1007/s10753-023-01891-6 (2023).
Smole, U. et al. Serum amyloid A is a soluble pattern recognition receptor that drives type 2 immunity. Nat. Immunol. 21, 756–765. https://doi.org/10.1038/s41590-020-0698-1 (2020).
Seumois, G. et al. Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aba6087 (2020).
Liu, X. et al. Single-cell RNA transcriptomic analysis identifies Creb5 and CD11b-DCs as regulator of asthma exacerbations. Mucosal Immunol. 15, 1363–1374. https://doi.org/10.1038/s41385-022-00556-1 (2022).
Papalexi, E. & Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18, 35–45. https://doi.org/10.1038/nri.2017.76 (2017).
Su, M. et al. Data analysis guidelines for single-cell RNA-seq in biomedical studies and clinical applications. Mil. Med. Res. https://doi.org/10.1186/s40779-022-00434-8 (2022).
Jovic, D. et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin. Transl. Med. https://doi.org/10.1002/ctm2.694 (2022).
Bousquet, J. et al. Allergic rhinitis and its impact on asthma (ARIA) 2008*. Allergy 63, 8–160. https://doi.org/10.1111/j.1398-9995.2007.01620.x (2008).
Huang, H. et al. Evaluation of a fully automated and completely quantitative allergen-specific IgE and total IgE detection assay. Anal. Methods 13, 3940–3946. https://doi.org/10.1039/d1ay01172c (2021).
Hao Y, Hao S, Andersen-Nissen E, III WMM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. (2021) https://doi.org/10.1016/j.cell.2021.04.048
Zhang, X. et al. Cell Marker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728. https://doi.org/10.1093/nar/gky900 (2018).
Hu, C. et al. Cell Marker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51, D870–D876. https://doi.org/10.1093/nar/gkac947 (2022).
Zhang J. scRNAtoolVis: Making ScRNA-seq visualization more easier and better. (2023)
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2, 100141. https://doi.org/10.1016/j.xinn.2021.100141 (2021).
Yu G. Gene ontology semantic similarity analysis using GOSemSim. Springer US. 207–15 (2020). https://doi.org/10.1007/978-1-0716-0301-7_11
Yu, G. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics 34, 3766–3767. https://doi.org/10.1093/bioinformatics/bty410 (2018).
Korotkevich, G., Sukhov, V. & Sergushichev, A. Fast gene set enrichment analysis. 31(4), 608 (2019).
Mering, Cv. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261. https://doi.org/10.1093/nar/gkg034 (2003).
Szklarczyk, D. et al. STRING v11: Proteinprotein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. https://doi.org/10.1093/nar/gky1131 (2018).
Szklarczyk, D. et al. The STRING database in 2021: Customizable proteinprotein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. https://doi.org/10.1093/nar/gkaa1074 (2020).
Szklarczyk, D. et al. The STRING database in 2023: Proteinprotein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. https://doi.org/10.1093/nar/gkac1000 (2022).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Jin, S. et al. Inference and analysis of cell-cell communication using cell chat. Nat. Commun. https://doi.org/10.1038/s41467-021-21246-9 (2021).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Soft. https://doi.org/10.18637/jss.v033.i01 (2010).
Simon, N., Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J. Stat. Softw. https://doi.org/10.18637/jss.v039.i05 (2011).
Tay, J. K., Narasimhan, B. & Hastie, T. Elastic net regularization paths for all generalized linear models. J. Stat. Softw. https://doi.org/10.18637/jss.v106.i01 (2023).
Stelzer, G. et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. https://doi.org/10.1002/cpbi.5 (2016).
Zhong, J. et al. Single-cell RNA sequencing reveals the molecular features of peripheral blood immune cells in children, adults and centenarians. Front Immunol. https://doi.org/10.3389/fimmu.2022.1081889 (2023).
Oliphant, C. J., Barlow, J. L. & McKenzie, A. N. J. Insights into the initiation of type 2 immune responses. Immunology 134, 378–385. https://doi.org/10.1111/j.1365-2567.2011.03499.x (2011).
Wynn, T. A. Type 2 cytokines: Mechanisms and therapeutic strategies. Nat. Rev. Immunol. 15, 271–282. https://doi.org/10.1038/nri3831 (2015).
Shamji, M. H. et al. The role of allergen-specific IgE, IgG and IgA in allergic disease. Allergy 76, 3627–3641. https://doi.org/10.1111/all.14908 (2021).
Huber, M. & Lohoff, M. Change of paradigm: CD8+ T cells as important helper for CD4+ T cells during asthma and autoimmune encephalomyelitis. Allergo J Int. 24, 8–15. https://doi.org/10.1007/s40629-015-0038-4 (2015).
Ogulur, I. et al. Type 2 immunity in allergic diseases. Cell. Mol. Immunol. 22(3), 211–242 (2025).
Tomita, K. et al. Identification and characterization of monocyte subpopulations from patients with bronchial asthma. J. Allergy Clin. Immunol. 96, 230–238. https://doi.org/10.1016/s0091-6749(95)70012-9 (1995).
Robays, L. J. et al. Chemokine receptor CCR2 but Not CCR5 or CCR6 mediates the increase in pulmonary dendritic cells during allergic airway inflammation. J. Immunol. 178, 5305–5311. https://doi.org/10.4049/jimmunol.178.8.5305 (2007).
Plantinga, M. et al. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T Helper 2 Cell-mediated immunity to house dust mite allergen. Immunity 38, 322–335. https://doi.org/10.1016/j.immuni.2012.10.016 (2013).
Yssel, H. et al. T cell activation-inducing epitopes of the house dust mite allergen Der p I. Proliferation and lymphokine production patterns by Der p I-specific CD4+ T cell clones. J. Immunol. 148, 738–745. https://doi.org/10.4049/jimmunol.148.3.738 (1992).
van Neerven, R. et al. T cell epitopes of house dust mite major allergen Der p II. J. Immunol. 151, 2326–2335. https://doi.org/10.4049/jimmunol.151.4.2326 (1993).
O’hehir, R. E., Eckels, D. D., Frew, A. J., Kay, A. B. & Lamb, J. R. MHC class II restriction specificity of cloned human T lymphocytes reactive with Dermatophagoides farinae (house dust mite). Immunology 64, 627 (1988).
Li, X. et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J. Allergy Clin. Immunol. 125, 328-335.e11. https://doi.org/10.1016/j.jaci.2009.11.018 (2010).
Kontakioti, E., Domvri, K., Papakosta, D. & Daniilidis, M. HLA and asthma phenotypes/endotypes: A review. Hum Immunol. 75, 930–939. https://doi.org/10.1016/j.humimm.2014.06.022 (2014).
Young, R. P. et al. HLA-DR and HLA-DP genotypes and immunoglobulin E responses to common major allergens. Clin. Experim. Allergy. 24(5), 431–439 (1994).
Stephan, K. et al. Genetic linkage of HLAclass II locus to mite-specific IgE immune responsiveness. Clin. Experim. Allergy. 29, 1049–1054. https://doi.org/10.1046/j.1365-2222.1999.00610.x (1999).
de la Mata, S. H. et al. Cytotoxic CD4+ tissue-resident memory T cells are associated with asthma severity. Med. 4(12), 875–897 (2023).
Wei, C., Wang, Y. & Hu, C. Bioinformatic analysis and experimental validation of the potential gene in the airway inflammation of steroid-resistant asthma. Sci. Rep. https://doi.org/10.1038/s41598-023-35214-4 (2023).
Rawat, K. et al. CCL5-producing migratory dendritic cells guide CCR5+ monocytes into the draining lymph nodes. J. Experim. Med. https://doi.org/10.1084/jem.20222129 (2023).
Schietinger, A. & Greenberg, P. D. Tolerance and exhaustion: Defining mechanisms of T cell dysfunction. Trends Immunol. 35, 51–60. https://doi.org/10.1016/j.it.2013.10.001 (2014).
Miyahara, N. et al. Effector CD8+ T cells mediate inflammation and airway hyper-responsiveness. Nat Med. 10, 865–869. https://doi.org/10.1038/nm1081 (2004).
Jartti, T. & Gern, J. E. Role of viral infections in the development and exacerbation of asthma in children. J. Allergy Clin. Immunol. 140, 895–906. https://doi.org/10.1016/j.jaci.2017.08.003 (2017).
Rudd, P. A. et al. Role of human metapneumovirus and respiratory syncytial virus in asthma exacerbations: Where are we now?. Clin. Sci. 131, 1713–1721. https://doi.org/10.1042/CS20160011 (2017).
Han, J., Takeda, K. & Gelfand, E. W. The role of RSV infection in asthma initiation and progression: Findings in a mouse model. Pulm. Med. 2011, 1–8. https://doi.org/10.1155/2011/748038 (2011).
Acknowledgements
We would like to express our sincerest gratitude to the medical professionals at the paediatric outpatient clinic of Shenzhen Bao’an Women’s and Children’s Hospital for their dedicated involvement in patient enrolment, blood sampling, and clinical supervision.
Funding
This study was supported by the Chongqing medical scientific research project (Joint project of Chongqing Health Commission and Science and Technology Bureau, 2024ZDXM011), the Natural Science Foundation of Chongqing (CSTB2023NSCQ-MSX0237), and the Science and Technology Research Project of Chongqing Municipal Education Commission (KJZD-K202300101).
Author information
Authors and Affiliations
Contributions
LY Zou and XO Hong designed the study, collected all samples and data, and supervised the experiments; LY Zou, B Ye and Kang Chen analyzed the data, performed the visualisation and prepared the manuscript; Kang Chen and XO Hong participated in the statistical analysis. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The present study was approved by Shenzhen Baoan Women’s and Children’s Hospital, Jinan University (approval number: LLSCHY–2021–01–03). The participants’ guardians provided written informed consent in person.
Consent for publication
All authors have reviewed and agreed to the published version of the manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zou, L., Chen, K., Hong, X. et al. Single-cell RNA sequencing reveals immunological link between house dust mite allergy and childhood asthma. Sci Rep 15, 16812 (2025). https://doi.org/10.1038/s41598-025-01538-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01538-6