Abstract
Coat colour formation in mammals is influenced by melanogenesis and pigmentation processes regulated by miRNAs, including miRNA-200a. Although miRNA-200a is differentially expressed in the skin of cashmere goats with varying coat colours, its regulatory mechanism remains unclear. In this study, miRNA-200a target genes were predicted using miRBase and TargetScan, identifying GNAI1 and PLCB4 as the target genes through GO and KEGG analyses. Dual-luciferase assays using wild-type and mutant plasmids confirmed a direct interaction between miRNA-200a and the 3’UTR regions of these genes. RT-qPCR and Western blot analyses demonstrated that the expression levels of miRNA-200a and its target genes differed significantly between black and white goat skin. In HaCaT cells, transfection with miRNA-200a mimics or inhibitors altered GNAI1 and PLCB4 expression at both mRNA and protein levels. To validate these findings in vivo, subcutaneous injection of antagomiR-200a into BALB/c mice significantly reduced melanin content (P < 0.01) and increased the expression of GNAI1 and PLCB4. These results indicate that miRNA-200a modulates skin pigmentation by suppressing GNAI1 and PLCB4, thereby influencing coat colour in cashmere goats. This study provides a foundational understanding for leveraging genetic regulation to enhance coat colour diversity and develop naturally pigmented breeds.
Similar content being viewed by others
Introduction
The Liaoning cashmere goat, a breed in China, is renowned for its exceptional cashmere quality, which surpasses that of high-altitude wild goats due to both artificial and natural selection1. Cashmere, a rare and luxurious animal fibre derived from cashmere goats, is among the most prized animal fibres worldwide. Renowned for its exceptional softness, rarity, and high production costs, cashmere is often referred to as “soft gold”2. Annual global cashmere production is estimated at approximately 4,500 to 5,000 tonnes, with China leading the market, contributing around 60% of total output3,4. However, China’s domestic cashmere production has been declining annually since 2014, resulting in a supply deficit that cannot meet the growing demands of local and international markets. The global cashmere goat population is predominantly concentrated in Asia, with China and Mongolia being the key producers. China hosts an estimated 40 million cashmere-producing goats, whilst Mongolia accounts for approximately 27.6 million goats3. In 2023, the global cashmere clothing market was valued at USD 2.80 billion and is anticipated to expand at a compound annual growth rate of 6.2% between 2024 and 2030, reaching USD 4.24 billion by the end of the forecast period5. The cashmere goat industry plays a vital role in animal husbandry across producing regions, serving as a cornerstone for local economic development4.
Despite its economic and cultural importance, the cashmere industry faces significant challenges. The dominance of white cashmere limits consumer demand for a broader range of colours6. Additionally, the textile dyeing process, including the dyeing of cashmere, is a major contributor to environmental pollution. Synthetic dyes and toxic chemicals used in dyeing often contain hazardous substances such as heavy metals, formaldehyde, and chlorine-based compounds, which contaminate water resources and damage ecosystems7. In response to rising environmental concerns, consumers are increasingly drawn to sustainable and eco-friendly natural fibres like cashmere. Natural-coloured cashmere, which eliminates the need for dyeing, is particularly favoured for its minimal environmental impact and perceived purity8. Developing high-quality cashmere breeds with diverse coat colours has thus become a critical research priority to meet consumer preferences and support sustainable industry practices.
Coat colour diversity is determined by the quantity, ratio, and distribution of melanin in hair follicles. Melanogenesis in hair follicles involves melanocyte activity, the transfer of melanin granules to cortical and medullary keratinocytes via cytoskeletal proteins and exosomal mechanisms9, and the formation of pigmented hair shafts10. Melanocyte activity is regulated by a complex interplay of genetic factors, signalling pathways, e.g., GPCRs like MC1R11, secreted factors, and oxidative stress12,13. Following melanin synthesis, the pigment is transferred to adjacent keratinocytes via cytoskeletal proteins, a critical process for completing pigmentation14,15. This melanin transfer from melanocytes to keratinocytes is a key determinant of coat colour in animals16. Recent research by Bento-Lopes, et al.17 delineated the roles of cytoskeletal proteins and intercellular signalling in melanin granule transfer from melanocytes to keratinocytes. However, the precise mechanisms underlying intercellular melanin transfer remain poorly understood, reflecting the complexity of these cellular processes. Studies such as those by Aplin, et al.18 have begun to illuminate the intricate cellular interactions involved in intercellular communication, which may yield insights into melanin transfer mechanisms in melanocytes and keratinocytes.
MicroRNAs (miRNAs) are emerging as key regulatory factors in coat colour determination. These molecules bind to the 3’UTR regions of mRNAs to suppress transcription or inhibit protein translation, thereby influencing pigmentation processes19,20. Previous research has identified differential expression of miRNA-200a in the skin of cashmere goats with varying coat colours21, although its specific role in pigmentation remains unclear. We hypothesise that miRNA-200a regulates coat colour diversity in cashmere goats by modulating melanin synthesis and transfer through its target genes, thereby influencing pigmentation. This study investigates the regulatory role of miRNA-200a in pigmentation by predicting its target genes using bioinformatics, validating these relationships both in vitro and in vivo, and analysing its effects on melanin production. The findings aim to provide theoretical support for employing genetic regulation to enhance coat colour diversity in animals.
Results
Expression of miRNA-200a, GNAI1, and PLCB4 in vivo
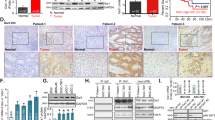
KEGG analysis revealed the involvement of GNAI1 and PLCB4 in melanogenesis (hsa04916) (Fig. 1A). The in vivo expression of miRNA-200a was significantly higher in the black skin group compared to the white skin group, with levels increased by 60.4% (P < 0.01; Fig. 1B; Tables S1, S2). The mRNA expression levels of the target genes GNAI1 and PLCB4 were significantly lower in the black skin group than in the white skin group, by 82.3% and 58.8%, respectively (P < 0.01). This finding was further validated at the protein level (Fig. 1C; Fig. S1), where the protein contents of GNAI1 and PLCB4 were both lowered in black skin compared to white skin. Together, these results suggest a regulatory role of miRNA-200a in pigmentation through its impact on the expression of GNAI1 and PLCB4.
In vivo expression analysis of miRNA-200a and its target genes GNAI1 and PLCB4 in cashmere goat skin pigmentation. (A) KEGG pathway analysis demonstrating involvement of GNAI1 and PLCB4 (highlighted in red) in melanogenesis (pathway ID: hsa04916). Pathway map adapted from KEGG (Kanehisa et al., 2023); (B) mRNA expression levels of target genes GNAI1 and PLCB4 in black and white skin of cashmere goats; (C) Protein expression of target genes GNAI1 and PLCB4 in black and white skin of cashmere goats. M: marker; 1: black (black group); 2: white (white group). **, P < 0.01; ****, P < 0.0001.
Target site prediction and Dual-Luciferase validation
The interactions between miRNA-200a and its target genes, GNAI1 and PLCB4, were predicted and experimentally validated. Binding sites for miRNA-200a on the 3’UTR regions of GNAI1 and PLCB4 were predicted using TargetScan (Fig. 2A). These interactions were further validated through dual-luciferase reporter assays, where significant reductions in luciferase activity were observed in wild-type (WT) constructs of GNAI1 and PLCB4 when miRNA-200a mimics were used (P < 0.01). No significant reduction in luciferase activity was detected in the mutant-type (MT) constructs or empty vector controls (Fig. 2B). These results confirm that GNAI1 and PLCB4 are directly regulated by miRNA-200a through binding to their 3’UTR regions.
miRNA-200a regulation of GNAI1 and PLCB4 in HaCaT cells
The effects of miRNA-200a on the expression of GNAI1 and PLCB4 were examined in HaCaT cells. Significant changes in the mRNA and protein levels of these target genes were observed following the overexpression or inhibition of miRNA-200a. RT-qPCR analysis revealed that overexpression of miRNA-200a decreased the mRNA levels of GNAI1 and PLCB4 by 60.5%, whilst inhibition of miRNA-200a increased their expression by 84.8% (Fig. 3A and B). Similarly, western blot analysis showed that the protein levels of GNAI1 and PLCB4 were reduced by miRNA-200a overexpression and elevated by its inhibition (Fig. 3C; Fig. S2). These results demonstrate that the expression of GNAI1 and PLCB4 is regulated by miRNA-200a at both the transcriptional and protein levels.
Identification of in vitro expression of miRNA-200a and its target genes GNAI1 and PLCB4. (A) mRNA expression of GNAI1 in HaCaT cells transfected with miRNA-200a mimics/inhibitors/NC; (B) mRNA expression of PLCB4 in HaCaT cells post-transfection; (C) Protein expression levels of GNAI1 and PLCB4 in HaCaT cells post-transfection. 1: inhibitor group; 2: NC group; 3: mimics group. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Effects of AntagomiR-200a on melanin content in BALB/c mice
The effects of AntagomiR-200a on melanin content and the expression of GNAI1 and PLCB4 in BALB/c mice were evaluated. A significant reduction in melanin content was observed in the skin of mice injected with AntagomiR-200a compared to the control group (P < 0.01; Fig. 4A). Immunohistochemical analysis indicated that the proteins GNAI1 and PLCB4 were primarily localised in the sebaceous glands (Fig. 4B). RT-qPCR analysis revealed that miRNA-200a expression levels were significantly 75% lower in the AntagomiR-200a group than in the control group (P < 0.001; Fig. 5A; Table S3). Conversely, the mRNA levels of GNAI1 and PLCB4 were significantly increased following AntagomiR-200a injection by 44.6% and 50.8%, respectively (P < 0.01; Fig. 5B; Table S3). Western blot analysis showed that the protein levels of GNAI1 and PLCB4 were also significantly elevated in the AntagomiR-200a group (Fig. 5C; Fig. S3). These results suggest that inhibition of miRNA-200a leads to a decrease in melanin content and an upregulation of GNAI1 and PLCB4.
Expression of miRNA-200a and its target genes in the skin of BALB/c mice injected with antagomiR-200a. (A) mRNA expression levels of miRNA-200a after antagomiR-200a injection; (B) mRNA expression levels of target genes GNAI1 and PLCB4 after antagomiR-200a injection; (C) Protein expression levels of GNAI1 and PLCB4 in mouse skin. 1: AntagomiRNA-200a group; 2: DEPC group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Materials and methods
Experimental animals
Ten 2-year-old adult male cashmere goats (five white and five black) were purchased from the Bairin Cashmere Goat Breeding Centre in Jilin Province, China. Skin tissues (1 cm2) were collected from the scapular region and immediately preserved in liquid nitrogen. Twenty 7–8-week-old male BALB/c mice (weighing 20–22 g) were obtained from the Experimental Animal Centre of Jilin University in Jilin Province, China. All animal experiments were performed in accordance with the relevant guidelines and regulations. This study was conducted in accordance with the ARRIVE guidelines (https://arriveguidelines.org). The experimental protocols were approved by the Animal Welfare and Research Ethics Committee of Jilin University (Approval No. KT202402385).
Cell culture
The human embryonic kidney cell line HEK-293 T obtained from Wuhan Sunncell Biotech Co., Ltd. (https://www.sunncell.com.cn; Wuhan, China) was employed in this study. These cells, originally derived from primary embryonic kidney tissue transformed with adenovirus 5 (Ad5) DNA fragments, exhibit stable immortalisation through expression of the SV40 large T antigen. Owing to their high transfection efficiency and minimal endogenous interference, HEK-293 T cells represent an optimal system for dual-luciferase reporter assays.
For cell culture, cryopreserved HEK-293 T cells were thawed in a 37 °C water bath for 90 s, centrifuged at 140 × g for 5 min (LD5-2B, Beijing Rebel Medical Equipment Co., Ltd., China), and resuspended in 1 mL DMEM (Procell, China). The cells were then transferred to culture flasks and maintained in DMEM supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (P/S; HyClone, China) at 37 °C under 5% CO₂ until reaching 70–80% confluence.
Human immortalised keratinocyte cells (HaCaT) were obtained from Bluefcell (Shanghai) Biotechnology Development Co., Ltd. (http://www.bluefcell.com; Shanghai, China) and cultured in high-glucose DMEM (supplemented with 10% FBS and 1% P/S). Subsequent procedures were performed the same as those for HEK-293 T cells, except trypsin digestion was extended to 10 min.
Target gene prediction and selection
Bioinformatic prediction of miRNA-200a target genes was performed using TargetScan (https://www.targetscan.org/vert_80)22, miRDB (https://mirdb.org)23, and miRbase (https://www.mirbase.org)24. Functional enrichment analysis through DAVID 6.7 (https://david.ncifcrf.gov/home.jsp)25 identified GNAI1 and PLCB4 as pigmentation-associated genes26, with significant enrichment in relevant Gene Ontology (GO) terms and KEGG pathways (hsa04916, melanogenesis)27,28,29. Further validation using UniProt (https://www.uniprot.org)30 and KEGG (https://www.genome.jp/kegg)27,28,29 databases confirmed their roles in pigmentation pathways. This integrative approach, combining transcriptomic and proteomic analyses, aligns with established methodologies for elucidating gene regulatory networks, as demonstrated by Bo, et al.31.
RT-qPCR analysis
Total RNA was extracted from cashmere goat skin tissues using the TRIzol method32. RNA concentrations and optical density (OD) values were measured using a nucleic acid analyser. cDNA was synthesised from RNA using an InnovaGene Biotech kit (AR121-Mix). RT-qPCR was performed for miRNA-200a and its target genes according to the fluorescence quantification kit protocol (SQ121-01). Three parallel experiments were conducted using the SYBR Green method. The RT-qPCR programme consisted of an initial denaturation step at 94 °C for 2 min, followed by 40 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 40 s, and extension at 72 °C for 25 s. The reaction was terminated under the system’s built-in dissolution conditions. Internal controls were U6 for miRNA-200a and GAPDH for GNAI1 and PLCB4. The designed primers are shown in Table 1 and were synthesised by Kumei Biotech Co., Ltd. (Changchun, China). Reference sequences for primer design were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov), and the RT-primer for miRNA-200a was designed using miRNA Design software v1.01 (Vazyme Biotech Co., Ltd; https://bio.vazyme.com/), following the manufacturer’s recommended parameters.
Western blot analysis
Proteins were extracted from cashmere goat skin tissues and HaCaT cells, and protein concentrations were determined using a BCA Protein Assay Kit (Solarbio, Beijing, China). Equal amounts of protein were separated via SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked at 37 °C for 90 min with a blocking solution and incubated overnight at 4 °C with primary antibodies against GNAI1 and PLCB4 (Santa Cruz Biotechnology, USA) at a 1:1,000 dilution. After washing, membranes were incubated with Goat anti-Rabbit IgG secondary antibodies (Santa Cruz Biotechnology, Beijing, China) at 37 °C for 2 h. ECL reagent was used for visualisation.
Dual-Luciferase reporter assay
The psi-CHECK2 plasmid was used to construct miRNA-200a target gene dual-luciferase expression vectors. Wild-type and MT primers were designed with XhoI and NotI restriction sites flanking the binding site sequences of miRNA-200a and its target genes (Table 2). Synthesised target sequences were ligated into the psi-CHECK2 vector to create psi-CHECK2-WT and psi-CHECK2-MT constructs. HEK-293 T cells were seeded in 96-well plates at a density of 1.4 × 105 cells/well and co-transfected with 0.2 µg psi-CHECK2-WT or psi-CHECK2-MT and 0.5 µg miRNA-200a mimics or negative control (NC) using Lipofectamine 2000 (Thermo, Beijing, China). Luciferase activity was measured 24 h post-transfection using the Dual-Luciferase Reporter Assay system (Promega, WI, USA).
Validation of miRNA-200a’s role in HaCaT cells
HaCaT cells were transfected with miRNA-200a mimics, inhibitors, and NC33. RNA and proteins were extracted to measure GNAI1 and PLCB4 expression levels via RT-qPCR and western blot (WB). Results confirmed miRNA-200a’s ability to regulate its target genes at both transcriptional and translational levels.
Animal experiment
BALB/c mice were randomly assigned to experimental and control groups, with 10 mice per group. The experimental group received a subcutaneous injection of 0.2 OD antagomiR-200a (Shanghai, China), whilst the control group was injected with ultrapure water treated with diethylpyrocarbonate (DEPC) and sterilised under high temperature and pressure. Injections were administered once daily for seven consecutive days.
For euthanasia, the mice were placed in a transparent, airtight chamber (glass on all sides) connected to a carbon dioxide gas cylinder to induce asphyxiation. Following euthanasia, a 2 cm² skin sample was collected from the scapular region of each mouse for melanin content determination, immunohistochemical analysis, RT-qPCR, and WB analysis.
Melanin samples were prepared by enzymatically hydrolysing the skin tissue with papain, and melanin content was measured at a wavelength of 490 nm34. Immunohistochemical analysis was conducted to determine the localisation of the target genes GNAI1 and PLCB4 in skin tissue, using an antibody dilution ratio of 1:200. Additionally, total RNA and protein were extracted from the skin samples of both groups for RT-qPCR and WB analyses.
Statistical analysis
Statistical analysis was performed using the software SPSS 19.0. Comparisons between the experimental group and the control group were conducted using a t-test. GraphPad Prism software was used for visualising and evaluating results. Statistical significance is represented by symbols: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Discussion
This study highlights the pivotal role of miRNA-200a in regulating skin pigmentation in cashmere goats by targeting and downregulating the expression of GNAI1 and PLCB4, two genes critical for melanin synthesis and transfer. These findings provide novel insights into the molecular mechanisms underlying coat colour variation and position miRNA-200a as a promising genetic target for enhancing coat colour diversity.
MicroRNAs (miRNAs) are well-established regulators of gene expression, with significant roles in melanogenesis. For instance, Sun, et al.35 demonstrated that miRNAs, such as miR-130b, play critical roles in modulating cellular processes through targeted gene regulation. Similarly, miRNAs like miRNA-27 and miRNA-193b have been shown to influence coat colour by modulating components of the WNT signalling pathway36,37. As a member of the miRNA-200 family, miRNA-200a participates in various biological processes, including tumour progression, infection-related pathways, and cellular regulation, such as cell adhesion and meiotic processes38. Studies by Chang, et al.39 have further identified exosomal miRNAs, including miRNA-200a, as biomarkers for disease states, underscoring their multifunctionality. For instance, Du, et al.40 showed that environmental factors, such as PM2.5 exposure, activate miRNA-200a-3p, leading to endoplasmic reticulum stress and impaired hepatic glucose metabolism—effects mitigated by melatonin through miRNA-200a-3p suppression. In the disciplines of transcriptomics and melanin molecular study, it has been demonstrated that miRNA-200a is linked to the development of animal hair colour. For instance, miRNA-200a has been demonstrated to regulate WNT5 A and FZD4, thereby contributing to pigmentation21. Studies using RNA sequencing technologies to examine gene expression variations across different geographical areas and phenotypic species41,42,43 have found that miRNA-200a is linked to the development of alpaca hair colour. Furthermore, its differential expression in brown and white alpaca skin has been verified by qPCR tests44. Based on the above theory, this study provides theoretical evidence for artificial coat colour modulation by further investigating the mechanism of miRNA-200a and target genes, as well as their influence on the synthesis of melanin in cashmere goat skin.
In vivo expression analysis revealed significantly higher levels of miRNA-200a in black cashmere goat skin compared to white skin, coupled with reduced GNAI1 and PLCB4 expression at both mRNA and protein levels. This inverse relationship was confirmed via dual-luciferase reporter assays, which demonstrated direct binding of miRNA-200a to the 3′UTR regions of these genes. Together, these findings indicate that miRNA-200a regulates pigmentation by suppressing its target genes.
GNAI1, a G protein-coupled receptor inhibitor, has not been widely studied in pigmentation. However, our findings suggest it may influence melanocyte activity and melanin transfer by modulating the follicular microenvironment. Whilst previous research associated GNAI1 primarily with diseases like colitis-associated cancer and uveal melanoma45,46, our study highlights its potential role in melanogenesis. For example, Muir, et al.47 identified GNAI1 variants as key regulators in developmental processes, implying potential involvement in melanocyte activity and melanin synthesis. Overexpression of GNAI1 following AntagomiR-200a treatment reduced melanin content in BALB/c mice, confirming a regulatory relationship between miRNA-200a and GNAI1 expression. Interestingly, differential expression of GNAI1 and miRNA-200a in goat skin with varying coat colours supports their functional interplay, with miRNA-200a suppressing GNAI1 expression to influence pigmentation.
PLCB4 is a well-characterised mediator of intracellular calcium signalling, essential for melanogenesis. Elevated PLCB4 expression in white cashmere goat skin suggests that it acts as a negative regulator of melanin synthesis, contributing to the lighter coat colour. This aligns with findings from Liu48, who identified PLCB4 as a key gene influencing seasonal coat colour changes in Arctic foxes. In this study, RNA-seq analysis revealed 1,177 differentially expressed genes, including PLCB4, enriched in the melanin synthesis pathway. Li49 collected skin tissues and hair samples from Dazu black goats, Inner Mongolia cashmere goats, and their first-generation hybrids (F1) to measure melanin content and analyse the expression patterns of coat colour-related miRNAs and their target genes in the skin of different goat breeds. The study found that melanin content varied among the skin and hair of different breeds, with PLCB4 expression significantly higher in the skin of Dazu black goats compared to F1 hybrid goats. Furthermore, PLCB4 expression in F1 hybrid goats showed an age-dependent trend, increasing initially and then declining, indicating that PLCB4 plays a role in regulating coat colour formation in goats.
In line with these observations, our study shows that downregulation of miRNA-200a increases PLCB4 expression, reducing melanin content and resulting in lighter pigmentation. Subcutaneous injection of AntagomiR-200a in mice elevated PLCB4 expression and decreased skin melanin levels, confirming the involvement of PLCB4 in the melanin synthesis pathway. This is supported by prior research linking PLCB4 to diseases such as blue naevi, blue malignant melanoma50, and primary leptomeningeal melanocytoma51. Furthermore, PLCB4 mutations have been implicated in uveal melanoma46, although they are absent in Chinese uveal melanoma cases52. Interestingly, Itoh, et al.53 found that overexpression of miRNA-200a and miRNA-141-3p in B16-4 A5 cells inhibited melanin synthesis and reduced tyrosinase activity. Our results expand on this by demonstrating that miRNA-200a overexpression in HaCaT cells suppresses PLCB4 expression, increasing melanin content and darkening pigmentation. Together, these findings highlight the regulatory role of miRNA-200a and PLCB4 in pigmentation and coat colour variation.
HaCaT cells are spontaneously immortalised human epidermal keratinocytes that retain their capacity for stratified differentiation, forming three-dimensional structures resembling native epidermis. These cells play a functional role in modulating tyrosinase activity and melanosomal transport in co-cultured melanocytes. In this study, we established a miRNA-200a overexpression model in HaCaT cells and quantitatively assessed its suppressive effects on GNAI1 and PLCB4 expression using qPCR and WB analysis. This experimental system effectively recapitulates the physiological mechanism through which miRNA-200a regulates cutaneous pigmentation via target gene modulation within the epidermal microenvironment.
Our in vivo experiments using AntagomiR-200a confirmed miRNA-200a’s regulatory role in pigmentation. The observed reduction in melanin content and increased expression of GNAI1 and PLCB4 in treated mice highlights miRNA-200a’s functional significance in controlling coat colour. These findings establish a foundation for exploring genetic mechanisms underlying pigmentation and open new avenues for developing strategies to enhance coat colour diversity in cashmere goats and other animals. For instance, Cai, et al.54 demonstrated the potential of bio-responsive miRNA delivery systems for targeted therapies, offering exciting prospects for leveraging miRNA-200a to modulate pigmentation.
However, several questions remain unanswered. The precise molecular pathways through which GNAI1 and PLCB4 affect melanogenesis require further elucidation. Additionally, the interaction between genetic regulators like miRNA-200a and environmental factors, including UV exposure and diet, warrants investigation. Future studies should employ high-throughput sequencing and CRISPR-Cas9 gene editing to map the broader gene network regulated by miRNA-200a. Chen, et al.55 demonstrated the utility of transcriptomic mapping in uncovering gene regulatory networks, offering a framework for advancing our understanding of miRNA-200a’s role in pigmentation.
The applicability of these findings to other animal species also warrants investigation. Comparative studies across breeds and species could reveal conserved and divergent roles of miRNA-200a in pigmentation, providing insights into its evolutionary significance. These efforts could ultimately contribute to sustainable breeding practices and enhance coat colour diversity in cashmere goats and other species.
Conclusions
This study demonstrates that miRNA-200a regulates skin pigmentation in cashmere goats by targeting GNAI1 and PLCB4. By modulating the expression of these genes, miRNA-200a influences melanin synthesis and transfer, resulting in coat colour variations. These findings enhance our understanding of the molecular mechanisms underlying coat colour diversity and open avenues for genetic improvement in livestock and other animals.
Data availability
Data are provided within the manuscript or supplementary information files.
References
Tiwari, M., Gujar, G., Shashank, C. G. & Ponsuksili, S. Selection signatures for high altitude adaptation in livestock: A review. Gene 927, 148757. https://doi.org/10.1016/j.gene.2024.148757 (2024).
Zhao, Q. et al. Genomic inbreeding and runs of homozygosity analysis of cashmere goat. Animals 14, 1246. https://doi.org/10.3390/ani14081246 (2024).
Chao, C. Analysis Report on China’s Cashmere Industry, (2024). https://chinecashmere.com/blog/analysis-report-on-chinas-cashmere-industry/
Rong, Y. et al. Genome-wide association study for cashmere traits in inner Mongolia cashmere goat population reveals new candidate genes and haplotypes. BMC Genom. 25, 658. https://doi.org/10.1186/s12864-024-10543-4 (2024).
Mali, S. Cashmere Clothing Market Report 2025 (Global Edition). 250 (Cognitive Market Research, 2024).
Hu, S. et al. KIT is involved in melanocyte proliferation, apoptosis and melanogenesis in the Rex Rabbit. PeerJ e9402, (2020). https://doi.org/10.7717/peerj.9402 (2020).
Khattab, T. A., Abdelrahman, M. S. & Rehan, M. Textile dyeing industry: environmental impacts and remediation. Environ. Sci. Pollut. Res. 27, 3803–3818. https://doi.org/10.1007/s11356-019-07137-z (2020).
Bumin, Z. & Bumin, M. Analysis of consumer preferences in sustainable fashion consumption. J. Innovations Sustain. 8, 01. https://doi.org/10.51599/is.2024.08.03.01 (2024).
Wu, X. & Hammer, J. A. Melanosome transfer: It is best to give and receive. Curr. Opin. Cell Biol. 29, 1–7. https://doi.org/10.1016/j.ceb.2014.02.003 (2014).
Clark, S. A. & Deppmann, C. D. How the stress of fight or flight turns hair white. Nature 577, 623–624. https://doi.org/10.1038/d41586-019-03949-8 (2020).
Hearing, V. J. Determination of melanin synthetic pathways. J. Invest. Dermatology. 131, E8–e11. https://doi.org/10.1038/skinbio.2011.4 (2011).
Balkrishna, A. et al. Melanogrit potentiates melanogenesis by escalating cellular tyrosinase activity and MITF levels via pERK Inhibition. Biosci. Rep. 44 (Bsr20231324). https://doi.org/10.1042/BSR20231324 (2024).
Paus, R., Sevilla, A. & Grichnik, J. M. Human hair graying revisited: principles, misconceptions, and key research frontiers. J. Invest. Dermatology. 144, 474–491. https://doi.org/10.1016/j.jid.2023.09.276 (2024).
Tian, X., Cui, Z., Liu, S., Zhou, J. & Cui, R. Melanosome transport and regulation in development and disease. Pharmacol. Ther. 219, 107707. https://doi.org/10.1016/j.pharmthera.2020.107707 (2021).
Ebanks, J. P., Wickett, R. R. & Boissy, R. E. Mechanisms regulating skin pigmentation: the rise and fall of complexion coloration. Int. J. Mol. Sci. 10, 4066–4087. https://doi.org/10.3390/ijms10094066 (2009).
Tadokoro, R. & Takahashi, Y. Intercellular transfer of organelles during body pigmentation. Curr. Opin. Genet. Dev. 45, 132–138. https://doi.org/10.1016/j.gde.2017.05.001 (2017).
Bento-Lopes, L. et al. Melanin’s journey from melanocytes to keratinocytes: Uncovering the molecular mechanisms of melanin transfer and processing. Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms241411289 (2023).
Aplin, J. D., Myers, J. E., Timms, K. & Westwood, M. Tracking placental development in health and disease. Nat. Reviews Endocrinol. 16, 479–494. https://doi.org/10.1038/s41574-020-0372-6 (2020).
Bartel, D. P. MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233. https://doi.org/10.1016/j.cell.2009.01.002 (2009).
Mazziotta, C. et al. Micrornas modulate signaling pathways in osteogenic differentiation of mesenchymal stem cells. Int. J. Mol. Sci. 22, 1–21. https://doi.org/10.3390/ijms22052362 (2021).
Li, J. et al. MicroRNA-200a regulates skin pigmentation by targeting WNT5A and FZD4 in cashmere goats. Res. Vet. Sci. 147, 68–73. https://doi.org/10.1016/j.rvsc.2022.03.020 (2022).
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4. https://doi.org/10.7554/eLife.05005 (2015).
Chen, Y. & Wang, X. MiRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 48, D127–D131. https://doi.org/10.1093/nar/gkz757 (2020).
Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–d162. https://doi.org/10.1093/nar/gky1141 (2019).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. https://doi.org/10.1038/nprot.2008.211 (2009).
Slominski, A., Tobin, D. J., Shibahara, S. & Wortsman, J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 84, 1155–1228. https://doi.org/10.1152/physrev.00044.2003 (2004).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–d592. https://doi.org/10.1093/nar/gkac963 (2023).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–462. https://doi.org/10.1093/nar/gkv1070 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
UniProt Consortium. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–d531. https://doi.org/10.1093/nar/gkac1052 (2023).
Bo, C. et al. Integrative transcriptomic and proteomic analysis reveals mechanisms of silica-induced pulmonary fibrosis in rats. BMC Pulm. Med. 22, 13. https://doi.org/10.1186/s12890-021-01807-w (2022).
Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. https://doi.org/10.1016/0003-2697(87)90021-2 (1987).
Wu, S. et al. MiR-27a regulates WNT3A and KITLG expression in cashmere goats with different coat colors. Animal Biotechnol. 32, 205–212. https://doi.org/10.1080/10495398.2019.1675683 (2021).
Yudovsky, D. & Pilon, L. Rapid and accurate estimation of blood saturation, melanin content, and epidermis thickness from spectral diffuse reflectance. Appl. Opt. 49, 1707–1719. https://doi.org/10.1364/ao.49.001707 (2010).
Sun, R. et al. Therapeutic targeting miR130b counteracts diffuse large B-cell lymphoma progression via OX40/OX40L-mediated interaction with Th17 cells. Signal. Transduct. Target. Therapy. 7, 80. https://doi.org/10.1038/s41392-022-00895-2 (2022).
Wu, S., Zhang, W., Shen, D., Lu, J. & Zhao, L. PLCB4 upregulation is associated with unfavorable prognosis in pediatric acute myeloid leukemia. Oncol. Lett. 18, 6057–6065. https://doi.org/10.3892/ol.2019.10921 (2019).
Xiang, B. et al. MiR-19 3b regulated the formation of coat colors by targeting WNT10A and GNAI2 in cashmere goats. Animal Biotechnol. 34, 796–804. https://doi.org/10.1080/10495398.2021.1998089 (2023).
Teng, Y. et al. MiRNA-200a/c as potential biomarker in epithelial ovarian cancer (EOC): evidence based on miRNA meta-signature and clinical investigations. Oncotarget 7, 81621–81633. https://doi.org/10.18632/oncotarget.13154 (2016).
Chang, S. N. et al. Association between exosomal miRNAs and coronary artery disease by next-generation sequencing. Cells 11. https://doi.org/10.3390/cells11010098 (2022).
Du, Z. et al. Melatonin alleviates PM(2.5)-induced glucose metabolism disorder and lipidome alteration by regulating Endoplasmic reticulum stress. J. Pineal Res. 73, e12823. https://doi.org/10.1111/jpi.12823 (2022).
Tiwari, M. et al. Omics strategies for unveiling male fertility-related biomarkers in livestock: A review. Gene Rep. 35 https://doi.org/10.1016/j.genrep.2024.101928 (2024).
Tiwari, M. et al. Deciphering genomic basis of unique adaptation of Ladakhi cattle to Trans-Himalayan high-altitude region of Leh-Ladakh in India. Gene 942 https://doi.org/10.1016/j.gene.2025.149251 (2025).
Tiwari, M., Sodhi, M., Sharma, M., Sharma, V. & Mukesh, M. Hypoxia related genes modulate in similar fashion in skin fibroblast cells of Yak (Bos grunniens) adapted to high altitude and native cows (Bos indicus) adapted to tropical climate during hypoxia stress. Int. J. Biometeorol. 68, 1675–1687. https://doi.org/10.1007/s00484-024-02695-5 (2024).
Tian, X. et al. Identification and characterization of microRNAs in white and brown alpaca skin. BMC Genom. 13, 555. https://doi.org/10.1186/1471-2164-13-555 (2012).
Li, Z. W. et al. GNAI1 and GNAI3 reduce colitis-associated tumorigenesis in mice by blocking IL6 signaling and down-regulating expression of GNAI2. Gastroenterology 156, 2297–2312. https://doi.org/10.1053/j.gastro.2019.02.040 (2019).
Johansson, P. et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 7, 4624–4631. https://doi.org/10.18632/oncotarget.6614 (2016).
Muir, A. M. et al. Variants in GNAI1 cause a syndrome associated with variable features including developmental delay, seizures, and hypotonia. Genet. Sci. 23, 881–887. https://doi.org/10.1038/s41436-020-01076-8 (2021).
Liu, H. Y. Screening and validation of differentially expressed genes based on the study of seasonal changes of coat color in white arctic foxes. Master thesis (Hebei Normal University of Science and Technology, 2022).
Li, J. L. The research on the expression of coat color-related miRNAs and their target genes in the goats of different breeds and ages. Master thesis (Southwest University, 2020).
Kervarrec, T. et al. GRM1 gene fusions as an alternative molecular driver in blue nevi and related melanomas. Mod. Pathol. 36, 100264. https://doi.org/10.1016/j.modpat.2023.100264 (2023).
van de Nes, J. A. P. et al. Activating CYSLTR2 and PLCB4 mutations in primary leptomeningeal melanocytic tumors. J. Invest. Dermatology. 137, 2033–2035. https://doi.org/10.1016/j.jid.2017.04.022 (2017).
Hou, C. et al. Mutations of GNAQ, GNA11, SF3B1, EIF1AX, PLCB4 and CYSLTR in unveal melanoma in Chinese patients. Ophthalmic Res. 63, 358–368. https://doi.org/10.1159/000502888 (2020).
Itoh, T., Fukatani, K., Nakashima, A. & Suzuki, K. MicroRNA-141-3p and microRNA-200a-3p regulate α-melanocyte stimulating hormone-stimulated melanogenesis by directly targeting microphthalmia-associated transcription factor. Sci. Rep. 10, 2149. https://doi.org/10.1038/s41598-020-58911-w (2020).
Cai, W. et al. Bio responsive self-assembly of Au-miRNAs for targeted cancer theranostics. EBioMedicine 54, 102740. https://doi.org/10.1016/j.ebiom.2020.102740 (2020).
Chen, X. et al. Transcriptomic mapping uncovers purkinje neuron plasticity driving learning. Nature 605, 722–727. https://doi.org/10.1038/s41586-022-04711-3 (2022).
Funding
This study was funded by the Education Department of Jilin Province, China (Grant No. JJKH20220399 KJ).
Author information
Authors and Affiliations
Contributions
J.L. and Q.Z. conceived and designed the study. J.L., Q.Z., and X.S. conducted the literature review. J.L., B.Z., R.L., and X.L. performed the experiments. J.L., R.L., and B.Z. conducted the statistical analysis. J.L., Q.Z., and X.S. wrote the manuscript. All authors contributed to the revision of the manuscript and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, J., Zhang, B., Liu, R. et al. miRNA-200a suppresses GNAI1 and PLCB4 to modulate skin pigmentation in cashmere goats. Sci Rep 15, 17456 (2025). https://doi.org/10.1038/s41598-025-01956-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01956-6