Abstract
To delve into the role and molecular mechanism of lysosome-associated membrane protein 3 (LAMP3) in tongue squamous cell carcinoma (TSCC). Differential expression of LAMP3 in TSCC from GEO microarray was analyzed, and pathway enrichment analysis of LAMP3 was performed utilizing GSEA. LAMP3 expression was detected by western blot, immumohistochemical staining and qRT-PCR. LAMP3 knockdown plasmid was constructed for a variety of biological function assays to verify the involvement of LAMP3 in TSCC. The upstream transcription factors and binding sites of LAMP3 were bioinformatically predicted. Dual luciferase reporter gene assay was applied to check the presence of c-Myc binding to the promoter region of LAMP3 and the regulation of its transcription. Further, a xenograft tumor model was developed to corroborate the impact of LAMP3 on tumor growth in vivo. LAMP3 expression was enhanced in TSCC tissues and cells. LAMP3 knockdown attenuated TSCC cell proliferation, DNA replication and metastatic capacity while induced glucose metabolism reprogramming in vitro. Meanwhile, LAMP3 depletion contributed to the delay of tumor progression in vivo. c-Myc was found bind to the LAMP3 promoter region to positively modulate LAMP3 transcriptional expression. LAMP3 regulated by c-Myc enhanced TSCC cell proliferation, DNA replication capacity while induced glucose metabolism reprogramming, suggesting a potential target for clinical trials in TSCC.
Similar content being viewed by others
Introduction
Head and neck neoplasms occur as the 6th most common malignancy, the majority of which are oral squamous cell carcinomas1. Tongue squamous cell carcinoma (TSCC), which is characterized primarily by high malignancy, advanced metastasis as well as poor prognosis, originates from the squamous epithelial cells of the tongue complex underlying the buccal mucosa and accounts for approximately 1/3 to 1/2 of oral squamous cell carcinomas. Current clinical treatment relies primarily on surgical excision with adjunctive radiation and chemotherapy2. Unfortunately, the five-year overall survival has been limited to 33–40% for TSCC patients due to the abundance of lymphatic and blood vessels around the tongue, the rapid growth rate of TSCC, the tendency of lymph node and distant metastasis in the early stage, as well as the absence of effective molecular markers3. As a consequence, the identification of potential emerging molecular biomarkers to guide the initial diagnosis and follow-up management of TSCC patients is of critical importance.
Lysosome-associated membrane protein 3 (LAMP3), a member of the lysosomal membrane-associated glycosylated protein family, is a high glycosylation single transmembrane protein. LAMP3 has previously been reported as a driver of oncogenesis associated with various malignancies. High LAMP3 expression was an independent prognostic biomarker for oral squamous cell carcinoma4. Individuals with elevated LAMP3 or TP53 expression experienced unfavorable overall survival, in addition, epithelial LAMP3 and TP53 expression provided independent prognostic markers for laryngeal squamous cell carcinoma5. LAMP3 downregulation enhanced the therapeutic effectiveness of radiation exposure in laryngeal squamous cell carcinoma through the modulation of LAMP3/LAMC2/TNC signaling pathway6. RPL21 has been demonstrated to stimulate TFEB transcription for the enhancement of LAMP3, which in turn facilitated invasion and metastasis by the activation of FAK/paxillin/ERK pathway for the guidance of adhesive plaques development via LAMP3-dependent mechanisms in colorectal cancer pathogenesis7. Currently, the involvement of LAMP3 in TSCC is unclear.
Oncogene Myc, one of the hot spots in molecular biology research in recent years, has been proven to contribute significantly to the occurrence and development of malignant tumors8. Moreover, c-Myc, as an essential member of MYC family, not only plays an indispensable role in normal cellular proliferation processes such as transcription9, glycolysis10, differentiation11, and apoptosis12, but also its overexpression has been implicated in tumor stemness13 and immune escape14. c-Myc was discovered to suppress LAPTM5 expression in B lymphoma cells through transcriptional and post-transcriptional modifications15. c-Myc functioned as the transcription factor that bound to hsa_circ_0001479 parental gene promoter in order to drive the production of hsa_circ_0001479, and in addition, hsa_circ_0001479 accelerated gastric cancer development and metastasis along with the mediation of CD8 + T-cell immune escape14. Perilipin 3 knockdown suppressed immune resistance through c-Myc regulation of PD-L1 transcription in lung adenocarcinoma16. Angelicin suppressed oral squamous cell carcinoma cell growth and enhanced apoptosis through the negative regulation of the DUSP6-mediated c-MYC signaling pathway17. c-Myc was demonstrated to directly bind to the SOD2 promoter and facilitate the migration, invasion as well as stemness of TSCC18.
Therefore, in this work, we observed the influence of LAMP3 on the malignant biological behavior of TSCC cells and tumor growth in vivo by regulating its expression in order to investigate the specific regulation mechanisms of c-Myc and LAMP3.
Materials and methods
Tissue collection
The work was approved by Medical Ethics Committee of Nanjing Medical University and study was performed in accordance with relevant guidelines. TSCC samples and adjacent tissues at least 1 cm away from the tumor were obtained from 10 patients undergoing oral and maxillofacial surgery in The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University. All patients had not received radiotherapy or chemotherapy prior to surgery. Informed consent forms were signed by all patients.
Cell culture and transfection
TSCC cells (H357, SCC-9, SCC-7 as well as CAL-27; ATCC, USA) were cultivated and maintained in RPMI-1640 medium supplemented with 10% FBS (Gibco, USA) at a temperature of 37 °C with 5% CO2. Human oral keratinocytes (HOK; ScienCell) were cultured under identical conditions as TSCC cells except for the medium used, keratinocyte growth medium (KGM Gold, Lonza). Small hairpin RNA specifically targeting LAMP3 (sh-LAMP3), shRNA negative control (sh-NC); c-Myc overexpression plasmid pcDNA3.1-c-Myc (Oe-c-Myc), and empty pcDNA3.1 plasmid (Oe-NC) were obtained from GenePharma (Shanghai, China). Cells were inoculated into 6-well plates and cultured overnight at 37 °C under 5% CO2. Transfection was subsequently performed utilizing Lipofectamine 2000. Following transfection, cells expressing sh-LAMP3 were screened with purinomycin and cells overexpressing c-Myc were screened with neomycin. The growth medium was then refreshed to continue the incubation for another 48 h. Cells were harvested and transfection efficiency was determined utilizing quantitative real time fluorescence quantification PCR (qRT-PCR) and western blot.
Cell proliferation assays
CCK-8 and colony forming assays were utilized to evaluate the proliferation potential of TSCC cells. For CCK-8 assay, harvested transfected CAL-27 and SCC-7 cells underwent pipetting into 96-well plates followed by incubation for the periods of 0, 1, 2 or 3 d. Following the addition of 20 μL CCK-8 reagent (5 mg/mL; Beyotime, China) at above time points, the cells were maintained at 37 °C for a further 2 h. The absorbances were read at the wavelength of 450 nm utilizing a multi-well spectrophotometer. For the performance of colony forming assay, cells underwent pipetting into six-well plates and kept in a cell incubator for 10–14 d. The culture was terminated at the appearance of cell colonies, which were fixed utilizing methanol followed by the application of 0.5% crystal violet for cell visualization. The colonies containing up to 50 cells were number-counted utilizing Image J software to evaluate cell colony formation ability after they were photographed under the microscope.
EdU assay
The EdU assay was utilized to evaluate the DNA replication capacity. The harvested cells were pipetted into 96-well plates for overnight cultivation. Thereafter, with 100 µL of 50 µM EdU medium filled into each well, the cells were kept at 37 °C with 5% CO2 for a period of 2 h. Following two washes utilizing PBS, an addition of 4% paraformaldehyde (50 µL) was applied for fixation for an additional 30 min at room temperature. Glycine at a concentration of 2 mg/mL (50 µL per well) was given prior to the incubation with a shaker for the next 5 min, followed by 100 µL of 0.5% TritonX-100 solution for another 10 min. Following repeat washing steps, 100 µL of Apollo staining solution was pipetted and incubated for 30 min at room temperature without exposure to light. 100 µL of Hoechst33342 reagent was applied followed by incubation in the dark for an additional 30 min. Afterwards, cells were visualized and photographed under a fluorescence microscope, and the percentage of cells in the replicative phase was calculated.
Transwell assay
Transwell assay was utilized to characterize the migration and invasion abilities of TSCC cells. For migration assay, cells of each group were resuspended with cell culture medium without FBS, and the cell suspension was inoculated into the upper Transwell chamber (Corning, NY), while the lower chamber was supplied with 600 μL of complete medium with 10% FBS as chemotactic agent. At the completion of the 24-h incubation, cells were fixated utilizing methanol followed by the stain with 0.1% crystal violet, dried, and then monitored through a microscope (Olympus) for visualization and photography. To carry out the invasion assay, we pre-diluted Matrigel gel with FBS-free cell culture medium at a ratio of 1:8, spread it evenly into the upper Transwell chamber, then kept at the temperature of 37 °C for a period of 4 h. Cells were inoculated onto the solidified gel, with the addition of 600 μL of complete medium with 20% FBS into the lower chamber. The further procedure was the same as for the migration assay.
Cell adhesion assay
30 μL of matrix gel was added to each well, allowed to dry for 1 h, and then gently washed twice with 37 °C pre-warmed PBS solution. Bovine serum albumin diluted in PBS was applied following a 37 °C, 5% CO2 incubation with a time of 1 h to block nonspecific sites. The harvested cells were added to 96-well plates and kept under incubation for a period of 2 h. Following gentle wash steps with PBS, medium with 2% fetal bovine serum was added, and 20 μL of tetrazolium blue salt at 5 mg/mL was added to each well, with a 4-h incubation. The culture solution was aspirated, with the addition of 150 μL DMSO per well, after which the plates were allowed to grow at room temperature for a period of 10 min and shaken on a micro-oscillator for a further 15 min. Absorbance was recorded at a wavelength of 570 nm utilizing a Multiskan FC automatic enzyme labeling instrument. The relative number of adherent cells was calculated utilizing the formula: (average OD value of treated cells)/(average OD value of control cells)19.
Cell cycle assay
Cells in the logarithmic growth phase after transfection were washed with precooled PBS, then resuspended with the addition of precooled 70% ethanol followed by overnight fixation at a temperature of 4 °C. Next, after another wash utilizing PBS, the cells were resuspended with the addition of 500 μL propidium iodide (PI) and further incubated for 30 min at 37 °C under light protection. Finally, the cells were passed through a 300-mesh filter and the cell cycle distribution was analyzed utilizing flow cytometry.
Activity of the glycolytic and gluconeogenic pathways
The activity of the gluconeogenic pathway was quantified through the determination of glycolytic products, including pyruvate, lactate, citrate and malate. Glycolytic products and glucose levels were measured utilizing several kits provided by Sigma, USA. The catalogue of kits was MAK071-1KT for pyruvate, MAK064 for lactate, MAK333 for citrate, MAK067 for malate and CBA086 for glucose, respectively according to the instructions. Gene expression levels related to gluconeogenesis and aerobic glycolysis were detected utilizing qRT-PCR.
Immunofluorescence staining
Cells were inoculated into 24-well plates pre-positioned with cell crawlers for overnight incubation prior to three washes with PBS. Subsequently, 4% paraformaldehyde was sequentially added and fixed for a period of 2 h at room temperature. 0.5% Triton X-100 was applied for permeabilization at room temperature for an additional 20 min, following which 10% goat serum was allowed to block for a further 30 min at room temperature. Afterwards, primary antibody was added and incubated overnight at 4 °C, and FITC fluorescent secondary antibody was added and incubated away from light for 1 h. Finally, DAPI stain was given with the incubation in the dark for 5 min, followed by observation of the cells under a fluorescence microscope and image acquisition.
Dual luciferase reporter assay
The HumanTFDB served as a prediction tool for the predicted binding between c-Myc and the LAMP3 promoter sequence. The promoter sequences of wild-type or mutant LAMP3 were inserted into a luciferase reporter gene plasmid (pGl3) to construct a wild-type vector (LAMP3-WT) or a mutant vector (LAMP3-MUT). The synthesized plasmids were simultaneously transfected into 293 T cells with Oe-NC or Oe-c-Myc utilizing Lipofectamine 2000. After 48-h transfection, the luciferase activity of the reporter gene was assessed according to the instructions of the Dual Luciferase Kit (Promega, USA).
qRT-PCR
Total RNA was obtained utilizing the Trizol method, and cDNA was synthesized utilizing the Prime Script RT Master Mix reagent kit (Takara, Japan). Then, qPCR assays were performed using SYBR Green Universal Master Mix (Thermo Fisher Scientific, USA) with a PCR LightCycler480 (Roche, Switzerland). β-actin was used as the internal control, and the reaction settings were that 95 °C for 30 s, 95 °C for 5 s, and 60 °C for 30 s, with 40 amplification cycles in total. The relative mRNA expression levels of target genes were quantified by 2−△△Ct method utilizing β-actin as an internal reference. Primers were as follows: LAMP3: F: 5’-TGAAAACAACCGATGTCCAA-3’ and R: 5’-TCAGACGAGCACTCATCCAC-3’; c-Myc: F: 5’-GCCTCAGAGTGCATCGAC-3’ and R: 5’-TCCACAGAAACAACATCG-3’; β-actin: F: 5’- CATGTACGTTGCTATCCAGGC-3’, R: 5’- CTCCTTAATGTCACGCACGAT-3’.
Western blot
Total proteins were obtained by means of RIPA lysate, and its concentration was determined with the BCA protein assay kit. Following the separation of proteins through SDS-PAGE, the proteins were transferred to PVDF membrane. The membrane was incubated with 5% nonfat milk for a period of 2 h, then exposed to the primary antibodies including LAMP3 (ab315108, 1/1000, Abcam, UK), γ-H2AX (ab229914, 1/1000, Abcam, UK), Ku70 (ab108604, 1/10,000, Abcam, UK), E-cadherin (ab314063, 1/1000, Abcam, UK), N-cadherin (ab76011, 1/5000, Abcam, UK), Snail (ab229701, 1/1000, Abcam, UK), c-Myc (ab32072, 1/1000, Abcam, UK) and β-actin (ab5694, 1/2000, Abcam, UK) for overnight incubation at a temperature of 4 °C, followed by exposure to the HRP-labeled secondary antibody for an additional 1 h. Protein bands were visualized with ECL detection reagent and semi-quantified by analyzing the gray values of the bands utilizing ImageJ software (version 2.0.0).
In vivo animal study
All experimental protocols were approved by Medical Ethics Committee of Nanjing Medical University. This study is performed in accordance with relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines. Nude mice were purchased from the experimental animal center of Nanjing Medical University. Each group had 6 mice. Mice were injected intraperitoneally with fluorescein substrate and allowed to react with luciferase for 10 min. Following isoflurane anesthesia, nude mice were imaged in vivo using a small animal live imaging instrument and photographed for documentation. Finally, after the sacrifice, the tumor weight and volume following the completion of removal.
Hematoxylin and eosin (H&E) staining
The histopathology examination of TSCC tissues was performed using H&E staining. The TSCC tissues samples were put in 10% formaldehyde solution, dehydrated in ethanol gradient, embedded in paraffin, and cut down into slices of 4 μm. After deparaffinage, the samples were stained using hematoxylin and eosin. Then the slices were mounted and observed under a light microscope (Leica Microsystems, Wetzlar, Germany).
Immunohistochemical staining
After pre-treatment with heat mediated antigen retrieval for 20 min in a microwave oven, the endogenous peroxidase activity of the Sects. (4-μm thick) was blocked by incubating the tissues in 3% H2O2 for 30 min. The sections were treated with primary antibody of LAMP3 (1/2000, Abcam, UK) and Ki-67 (1/1000, Abcam, UK) for 15 min at room temperature and subsequently treated with corresponding secondary antibody (1/1000, Abcam, UK) at room temperature for 30 min. Next, the sections were developing using Diaminobenzidine (DAB) solution (Beyotime, China), counterstained by Hematoxylin Staining Solution (Beyotime, China), and then mounted with DPX, and observed under a microscope (IX71; Olympus, Tokyo, Japan).
Bioinformatics analysis
Microarray data of TSCC were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) to obtain the GSE9844, GSE13601, and GSE34105 gene microarray packages for the analysis of the differences in the levels of LAMP3 mRNA between TSCC tissues and corresponding paracancerous tissues. These 3 microarrays were merged and the data were batch corrected utilizing the sva package in R. KEGG enrichment analysis of LAMP3 mRNA was performed utilizing GSEA v4.1.0 software.
Statistical analysis
Data were displayed in the form of mean ± SD, a and GraphPad Prism 8 was utilized for statistical analysis and graphing. Comparisons between two groups were obtained utilizing the independent samples t-test, while comparisons among multiple groups were accomplished utilizing one-way ANOVA analysis. Differences were regarded as statistically significant at a P of less than 0.05.
Results
LAMP3 was overexpressed in TSCC
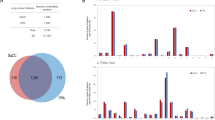
To evaluate the difference in LAMP3 expression in TSCC and normal tissues, mRNA sequencing data were analyzed utilizing the GEO database, containing 26 TSCC tissues as well as 12 normal tissues in GSE9844; 31 TSCC tissues as well as 26 normal tissues in GSE13601; or 62 TSCC tissues as well as normal tissues. It was discovered that LAMP3 was upregulated in TSCC tissues in comparison to normal tissues (Fig. 1A). The strong expression of LAMP3 in TSCC samples compared to paracancerous tissues was further verified by the examination of our TSCC and paracancerous samples (Fig. 1B, C). In addition, we detected the LAMP3 expression level in HOK and TSCC cells (H357, SCC-9, SCC-7, and CAL-27) utilizing qRT-PCR, with the finding of a markedly elevated LAMP3 expression level in TSCC cells in contrast to HOK cells (Fig. 1D). We subsequently performed GSEA-KEGG enrichment analysis on these three tissue microarray datasets and found that LAMP3 was significantly enriched in DNA replication, cell adhesion molecules, glycolysis and gluconeogenesis pathways (Fig. 1E). The above results demonstrated that LAMP3 was strongly hyper-expressed in TSCC and possibly implicated in DNA replication, cell adhesion molecules, glycolysis and gluconeogenesis.
LAMP3 was highly expressed in TSCC. (A) GEO microarray analysis of LAMP3 expression in tissues of TSCC patients. (B) Western blot detection of LAMP3 expression in tissues of TSCC patients. (C) Immunohistochemical staining detection of LAMP3 expression in tissues of TSCC patients. (D) qRT-PCR and western blot to detect the expression level of LAMP3 in TSCC cells. (E) LAMP3 was analyzed for KEGG enrichment in the GSEA database. *p < 0.05; ***p < 0.001.
LAMP3 knockdown suppressed TSCC cell proliferation and DNA replication
To investigate the possible bio-function of LAMP3 in TSCC, various in vitro assays were undertaken. CAL-27 and SCC-7 cells with the higher LAMP-3 expression were selected for subsequent cell function tests. Firstly, LAMP3 silencing was applied to CAL-27 and SCC-7 cells, which showed that LAMP3 was markedly suppressed compared with the sh-NC group, especially sh-LAMP3#2 (Fig. 2A and Supplementary Fig. S1A). Therefore, we selected LAMP3#2 for the subsequent study. CCK-8 (Fig. 2B and Supplementary Fig. S1B) and colony formation assay (Fig. 2C and Supplementary Fig. S1C) results demonstrated that LAMP3 knockdown greatly impaired the proliferation ability of CAL-27 and SCC-7 cells compared to the sh-NC group. The influence of LAMP3 on DNA replication was monitored utilizing EdU assay. EdU-positive cells were markedly diminished following LAMP3 knockdown with respect to the control group (Fig. 2D and Supplementary Fig. S1D), indicating that LAMP3 down-regulation had a strong suppressive effect on DNA replication. Furthermore, we monitored the cell cycle and noticed that LAMP3 knockdown enhanced the percentage of G1/G2 and lowered the percentage of S phase in the CAL-27 and SCC-7 cell cycle (Fig. 2E and Supplementary Fig. S1E). Besides, LAMP3 downregulation increased the expression level of DNA double-strand break marker 53BP1 (Fig. 2F and Supplementary Fig. S1F) as well as the expression level of DNA damage-associated protein γ-H2AX, while decreased the expression level of DNA repair-associated protein Ku70 (Fig. 2G and Supplementary Fig. S1G). Collectively, these observations revealed that LAMP3 knockdown impaired the proliferation as well as DNA replication of TSCC cells.
Interference with LAMP3 inhibited proliferation and DNA replication of CAL-27 cells. (A) qRT-PCR and western blot to detect the expression level of LAMP3. (B) CCK-8 assay for cell viability. (C) Colony forming assay to detect cell proliferation. (D) EdU assay to detect cellular DNA replication. (E) Flow cytometry to detect cell cycle. (F) Immunofluorescence to detect the level of DNA double-strand break marker 53BP1. (G) Western blot to detect the expression of DNA damage repair-related proteins. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Knockdown of LAMP3 inhibited TSCC cell metastasis, adhesion and metabolic reprogramming
To determine the impact of LAMP3 on TSCC cell metastasis, transwell assay was employed, which revealed that the invasion capability of CAL-27 and SCC-7 cells with sh-LAMP3 was greatly reduced as compared to sh-NC (Fig. 3A, B and Supplementary Fig. S2A, B). Since Epithelial-mesenchymal transition (EMT) is an important process by which tumor cells acquire high invasive ability, we further examined the expression of EMT biomarkers in the cells. As shown in Fig. 3C and Supplementary Fig. S2C, the protein expression level of E-cadherin was markedly enhanced in CAL-27 and SCC-7 cells with sh-LAMP3 by comparison with sh-NC, whereas that of N-cadherin and Snail was substantially decreased. LAMP3 downregulation remarkably diminished cell adhesion to the extracellular matrix as determined with the cell adhesion assay (Fig. 3D and Supplementary Fig. S2D). Moreover, LAMP3 knockdown also markedly enhanced gluconeogenic gene (PEPCK1, FBP1, and PFK) mRNA levels and reduced aerobic glycolysis gene (PKM2, LDHA, and GLUT1) mRNA levels (Fig. 4A). LAMP3 downregulation blocked the production of pyruvate, lactate, citrate and malate in CAL-27 and SCC-7 cells (Fig. 4B, C), whereas glucose production was stimulated (Fig. 4D). These results suggested that LAMP3 knockdown suppressed TSCC cell metastasis and adhesion, blocked glycolysis, and stimulated gluconeogenesis.
Interference with LAMP3 inhibited CAL-27 cell metastasis and adhesion. (A, B) Transwell assay to detect cell invasion and migration ability. (C) Western blot to detect the expression of EMT-related proteins. (D) Cell adhesion assay to assess cell adhesion to extracellular matrix. p < 0.001; ****p < 0.0001.
Interference with LAMP3 inhibited metabolic reprogramming in TSCC cells. (A) qRT-PCR was performed to detect the mRNA levels of genes related to gluconeogenesis and aerobic glycolysis. (B) Corresponding kits were used to detect the levels of pyruvate, citrate and malate in cells. (C) L-lactic acid kit detects the level of lactic acid in cell supernatants. (D) Glucose detection kit to detect glucose levels in cell supernatants. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
c-Myc drove the transcription of LAMP3
To further clarify the mechanism of LAMP3 in TSCC cells, we predicted the upstream transcription factors of LAMP3 utilizing the HumanTFDB database, which found that c-Myc had a potential binding site for LAMP3 with a high score (Fig. 5A). We investigated the mRNA as well as protein expression levels of c-Myc in HOK, CAL-27 and SCC-7 cells, and indicated that they were remarkably upregulated in the latter (Fig. 5B). We constructed a c-Myc overexpression plasmid and transfected it into CAL-27 and SCC-7 cells. qRT-PCR as well as western blot were utilized to confirm the transfection efficiency, which indicated that the overexpression plasmid greatly enhanced c-Myc expression level in CAL-27 and SCC-7 cells (Fig. 5C). The result of dual luciferase reporter gene assay demonstrated that Oe-c-Myc had a marked upregulation of LAMP3 promoter transcriptional activity in 293 T cells with LAMP3 wild-type promoter. While in 293 T cells with LAMP3 mutant promoter, the LAMP3 promoter transcriptional activity was not impacted by Oe-c-Myc (Fig. 5D). This implied that c-Myc as a transcription factor could directly bind to the LAMP3 promoter region. To verify the role of c-Myc in the transcriptional regulation of LAMP3, LAMP3 expression level in c-Myc hyperexpressing CAL-27 and SCC-7 cells was examined utilizing qRT-PCR and western blot. As a result, c-Myc overexpression markedly enhanced LAMP3 expression level (Fig. 5E). The results indicated that LAMP3 expression was tightly modulated through c-Myc transcription factor.
c-Myc promoted LAMP3 transcription. (A) HumanTFDB predicted transcription factor c-Myc and LAMP3 promoter binding site. (B) qRT-PCR and western blot detection of c-Myc expression in HOK, CAL-27 and SCC-7 cells. (C) qRT-PCR and western blot to detect the expression level of c-Myc. (D) Luciferase reporter gene assay to detect promoter activity. (E) qRT-PCR and western blot to detect the expression level of LAMP3 after c-Myc overexpression. **p < 0.01; ***p < 0.001; ****p < 0.0001.
LAMP3 knockdown impaired TSCC growth in vivo
To further assess the role of LAMP3 in vivo, we established a subcutaneous tumor model of TSCC in nude mice by injecting luciferase-labeled CAL-27 cells, the schematic diagram was displayed in Fig. 6A. Following isoflurane anesthesia, nude mice were imaged in vivo. As shown in Fig. 6B, after LAMP3 silencing, the photon intensity of the transplanted tumors was substantially diminished compared with that in the controls, indicating a reduced tumor growth rate in nude mice. Moreover, the volume and weight of subcutaneous tumors in mice with LAMP3 knockdown considerably lowered (Fig. 6C–E). Moreover, the results of H&E staining in TSCC tissues demonstrated that tumor tissues in sh-NC group showed obvious necrosis, whereas silencing of LAMP3 improved the necrosis in tumor (Fig. 6F). In addition, Ki-67 expression level was clearly suppressed in the transplanted tumors with ectopic expression of LAMP3 compared to the sh-NC group (Fig. 6G). The results implied that LAMP3 could potentially suppress tumor growth in vivo.
Interference with LAMP3 inhibited TSCC growth in vivo. (A) Schematic diagram of in vivo study. (B) Small animal live imaging technique to monitor the size of luciferase-labeled transplanted tumors. (C) Representative images of xenograft tumors. Changes in tumor (D) weight and (E) volume. (F) The histopathological characteristics of the tumors were examined by H&E staining. (G) Expression levels of Ki-67 in tumor tissues detected by IHC. ****p < 0.0001.
Discussion
In our present work, LAMP3 was demonstrated to be elevated in TSCC tumor tissues based on microarray analysis of GSE9844, GSE13601, and GSE34105, and was further confirmed that LAMP3 expression level was markedly enhanced in TSCC tissues and cells, indicating that LAMP3 was strongly overexpressed in TSCC. Furthermore, the transcriptional regulation relationship between c-Myc and LAMP3 was supported by bioinformatic prediction and dual luciferase assay. Other studies have reported that elevated LAMP3 expression might serve as a prognostic marker independently in oral squamous cell carcinoma4 and laryngeal squamous cell carcinoma5. Diminished expression of LAMP3 enhanced the response to irradiation in laryngeal squamous cell carcinoma by the regulation of the LAMP3/LAMC2/TNC signaling pathway6. RPL21 stimulated TFEB transcription and enhanced LAMP3 expression, which facilitated invasion and metastasis by the modulation of FAK/paxillin/ERK signaling pathway through LAMP3-dependent mechanisms in colorectal cancer development7. LAMP3 deficiency improved PKA-mediated phosphorylation of VASP and suppressed invasion and metastasis of esophageal squamous cell carcinoma20. LAMP3 stimulated osteosarcoma invasion and metastasis by the regulation of SPP1 downstream signaling21. In our study, we revealed that LAMP3 deregulation diminished the proliferative activity, DNA replication, invasion and migration ability of TSCC cells. EMT has been a hot spot in the study of the invasion, metastasis and the enhancement of tumor cell stemness genesis mechanism of malignant tumorigenesis. Studies have indicated that EMT is implicated in the invasion and metastasis of various malignant tumors22. When malignant tumor cells underwent EMT, their gene expression profiles changed accordingly, with the expression of epithelial cell marker protein such as E-cadherin downregulated and the expression of mesenchymal cell marker proteins such as vimentin and Snail upregulated23. We also noticed that LAMP3 silencing led to a reduction in the occurrence of EMT in TSCC cells.
Moreover, we discovered that LAMP3 might also affect gluconeogenesis and glycolysis processes via GSEA-KEGG enrichment analysis. Cancer cells drove tumor growth, metastasis, apoptosis and other processes through the aerobic glycolysis pathway, and targeting the glycolysis pathway might offer a novel strategy for cancer treatment24,25. GLUT1 has an essential function in glucose uptake in cancer cells, and some studies have demonstrated that GLUT1 had a positive correlation with poor prognosis of patients with gastric cancer26. LDHA was described as a critical regulator of lactic acid production, and the suppression of LDHA activity provided a central link in the blockade of aerobic glycolysis27. PKM2 is the enzyme which is key to gluconeogenesis in cancer cells and can promote cell proliferation and tumorigenesis by non-metabolic means28. Gluconeogenesis occupies an extensive role in glucose metabolism, catalyzed by a number of enzymes to convert substances such as lactate, glycerol, and gluconeogenic amino acids into glucose and glycogen. PEPCK1, FBP1 and PFK are the essential rate-limiting enzymes in gluconeogenesis. FBP1 was identified to exert tumor suppression effects through the prevention of glycolysis during lung cancer progression29. PEPCK1 suppressed tumorigenesis through the enhancement of gluconeogenesis, ATP depletion and cell growth arrest, and the suppression of glycolysis30,31. PFK catalyzes the first irreversible reaction of glycolysis to promote glucose depletion, which may further accelerate apoptosis32,33. To investigate the involvement of LAMP3 in glycolysis and gluconeogenesis in TSCC, we silenced LAMP3 and demonstrated that modulation of LAMP3 gene expression severely impacted glucose uptake and the levels of genes related to lactate, glycolysis and gluconeogenesis in CAL-27 and SCC-7 cells. These findings indicated that LAMP3 might contribute to the progression of TSCC through the induction of glucose metabolism reprogramming.
c-Myc is generally overexpressed in human cancer tissues. Li et al.34 reported that c-Myc-dependent FBP1 downregulation suppressed proliferation and invasion, reduced aerobic glycolysis, and sensitized ovarian cancer cells to cisplatin-induced apoptosis through the regulation of STAT3. Liu et al.18 demonstrated that c-Myc stimulated SOD2 transcription, which further enhanced TSCC migration, invasion, and stemness. Our findings suggested the possibility of direct binding of c-Myc to the LAMP3 promoter and the enhancement of LAMP3 expression level.
However, our study has some limitations. First, the detailed mechanism by which c-Myc regulates LAMP3 transcription remains to be further elucidated. In addition, whether increase in LAMP3 expression is associated with resistance towards chemotherapeutic drugs or radiation remains unclear. We would consider these limitations in further studies.
To summarize, the present study confirmed that LAMP3 regulated with c-Myc facilitated TSCC cell proliferation, DNA replication and metastatic capabilities, and regulated glucose metabolism reprogramming, which was expected to provide a potentially viable target for clinical trials in TSCC.
Data availability statement
The data are available from the corresponding author upon reasonable request.
References
Chamoli, A. et al. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral Oncol 121, 105451 (2021).
Schoenfeld, J. D. et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in untreated oral cavity squamous cell carcinoma: A phase 2 open-label randomized clinical trial. JAMA Oncol 6(10), 1563–1570 (2020).
Naruse, T. et al. Immunohistochemical study of PD-1/PD-L1 axis expression in oral tongue squamous cell carcinomas: Effect of neoadjuvant chemotherapy on local recurrence. Pathol Oncol Res 26(2), 735–742 (2020).
Lu, J. et al. Clinical significance and prognostic value of the expression of LAMP3 in oral squamous cell carcinoma. Dis Markers 2017, 1218254 (2017).
Qiu, X. et al. LAMP3 and TP53 overexpression predicts poor outcome in laryngeal squamous cell carcinoma. Int J Clin Exp Pathol 8(5), 5519–5527 (2015).
Wu, H. et al. Efficacy of radiation exposure in laryngeal squamous cell carcinoma is mediated by the LAMP3/LAMC2/tenascin-C pathway. Exp Biol Med (Maywood) 244(13), 1070–1080 (2019).
Zhu, J. et al. RPL21 interacts with LAMP3 to promote colorectal cancer invasion and metastasis by regulating focal adhesion formation. Cell Mol Biol Lett 28(1), 31 (2023).
Matsumoto, M. et al. Control of the MYC-eIF4E axis plus mTOR inhibitor treatment in small cell lung cancer. BMC Cancer 15, 241 (2015).
Zhao, Y., Wang, Y., Miao, Z., Liu, Y. & Yang, Q. c-Myc protects hepatocellular carcinoma cell from ferroptosis induced by glutamine deprivation via upregulating GOT1 and Nrf2. Mol Biol Rep 50(8), 6627–6641 (2023).
Gauthier, T. et al. TGF-β uncouples glycolysis and inflammation in macrophages and controls survival during sepsis. Sci Signal 16(797), eade0385 (2023).
Li, S., Yu, J., Zhang, J., Li, X. & Yu, J. LSD1 interacting with HSP90 promotes skin wound healing by inducing metabolic reprogramming of hair follicle stem cells through the c-MYC/LDHA axis. Faseb J 37(7), e23031 (2023).
Hui, M. et al. PAK2/beta-catenin/c-Myc/PKM2 signal transduction suppresses ovarian granulosa cell apoptosis in polycystic ovary syndrome. Biochem Biophys Res Commun 677, 54–62 (2023).
Yoshida, G. J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J Exp Clin Cancer Res 37(1), 173 (2018).
Zang, J. et al. Hsa_circ_0001479 accelerates tumorigenesis of gastric cancer and mediates immune escape. Int Immunopharmacol 124(Pt A), 110887 (2023).
Zhang Y, Zhang X, Zhang Y, Xu H, Wei Z, Wang X, Li Y, Guo J, Wu F, Fang X et al. c-Myc inhibits LAPTM5 expression in B-cell lymphomas. Ann. Hematol. 2023.
Men, X. & Zhu, W. Silencing of perilipin 3 inhibits lung adenocarcinoma cell immune resistance by regulating the transcription of PD-L1 through c-Myc. Immunol Invest 52(7), 815–831 (2023).
Liu N, Li C, Shang Q, Qi J, Li Q, Deng J, Dan H, Xie L, Chen Q. Angelicin inhibits cell growth and promotes apoptosis in oral squamous cell carcinoma by negatively regulating DUSP6/cMYC signaling pathway. Exp. Cell Res. 2023:113793.
Liu, Z. et al. SOD2 is a C-myc target gene that promotes the migration and invasion of tongue squamous cell carcinoma involving cancer stem-like cells. Int J Biochem Cell Biol 60, 139–146 (2015).
Zhang, Q. et al. C1632 inhibits ovarian cancer cell growth and migration by inhibiting LIN28 B/let-7/FAK signaling pathway and FAK phosphorylation. Eur J Pharmacol 956, 175935 (2023).
Huang, F. et al. Depletion of LAMP3 enhances PKA-mediated VASP phosphorylation to suppress invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett 479, 100–111 (2020).
Li, Y., Du, W., Han, J. & Ge, J. LAMP3 promotes the invasion of osteosarcoma cells via SPP1 signaling. Mol Med Rep 16(5), 5947–5953 (2017).
Saitoh, M. Involvement of partial EMT in cancer progression. J Biochem 164(4), 257–264 (2018).
Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules 2016;21(7).
Abbaszadeh, Z., Çeşmeli, S. & Biray Avcı, Ç. Crucial players in glycolysis: Cancer progress. Gene 726, 144158 (2020).
Ganapathy-Kanniappan, S. & Geschwind, J. F. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 12, 152 (2013).
Zhou, D. et al. Glucose transporter-1 cooperating with AKT signaling promote gastric cancer progression. Cancer Manag Res 12, 4151–4160 (2020).
Liu, X. et al. LDHA and LDHB overexpression promoted the Warburg effect in malignantly transformed GES-1 cells induced by N-nitroso compounds. Food Chem Toxicol 180, 114007 (2023).
Dayton, T. L., Jacks, T. & Vander Heiden, M. G. PKM2, cancer metabolism, and the road ahead. EMBO Rep 17(12), 1721–1730 (2016).
Cong, J. et al. Dysfunction of natural killer cells by FBP1-induced inhibition of glycolysis during lung cancer progression. Cell Metab 28(2), 243-255.e245 (2018).
Bian, X. L. et al. Nur77 suppresses hepatocellular carcinoma via switching glucose metabolism toward gluconeogenesis through attenuating phosphoenolpyruvate carboxykinase sumoylation. Nat Commun 8, 14420 (2017).
Chen, Z., Tang, W. J., Zhou, Y. H., Chen, Z. M. & Liu, K. Andrographolide inhibits non-small cell lung cancer cell proliferation through the activation of the mitochondrial apoptosis pathway and by reprogramming host glucose metabolism. Ann Transl Med 9(22), 1701 (2021).
Vaz, C. V. et al. Androgens enhance the glycolytic metabolism and lactate export in prostate cancer cells by modulating the expression of GLUT1, GLUT3, PFK, LDH and MCT4 genes. J Cancer Res Clin Oncol 142(1), 5–16 (2016).
Xu, R. H. et al. Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res 65(2), 613–621 (2005).
Li, H. et al. FBP1 regulates proliferation, metastasis, and chemoresistance by participating in C-MYC/STAT3 signaling axis in ovarian cancer. Oncogene 40(40), 5938–5949 (2021).
Author information
Authors and Affiliations
Contributions
Jing Feng: Methodology, Software, Data Curation, Writing- Original Draft; Wei He: Resources, Validation, Visualization; Jin Fang: Supervision, Conceptualization, Writing- Reviewing and Editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study is performed in accordance with relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, J., He, W. & Fang, J. c-Myc-dependent LAMP3 regulates the proliferation, metastasis and metabolic reprogramming of tongue squamous cell carcinoma. Sci Rep 15, 19179 (2025). https://doi.org/10.1038/s41598-025-02172-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-02172-y