Abstract
This study explores the production and characterization of chromium reductase from Bacillus paramycoides S48, focusing on its ability to effectively reduce toxic hexavalent chromium to less harmful chromium. The strain exhibited 65% reduction in Cr(VI) within 96 h at 30 °C. Clear morphological and functional group shifts on strain S48 cell surface treated with metal were noted using analytical tools i.e. SEM–EDX and FT-IR. The physico-chemical conditions such as temperature, pH and nutritional factors were optimized for better chromate reductase yield using Placket-Burman and Central Composite design software. The purified chromate reductase, obtained through size exclusion column chromatography, demonstrated a specific activity of 1416.5 U/mg, and 6.6-fold increase in purity, with a molecular mass of approximately 35 kDa. The enzyme exhibited stability at temperature 30–40 °C and pH 5.0–8.0. Furthermore, the purified chromium reductase achieved, 80% reduction of toxic Cr(VI) at temperature 35 °C after 96 h. The BparChR gene was successfully cloned into the pET-28a vector, expressed in E. coli BL21, and purified through Ni-Affinity ion exchange chromatography. The recombinant BparChR protein displayed a specific activity of 1680 U/mg, and a purification fold of 5.73 times. The BparChR exhibited a remarkable 90% reduction in chromium after 96 h, surpassing the efficacy of whole-cell and native chromium reductase. This study concludes that B. paramycoides S48, holds promise for the cost-effective and environmentally friendly detoxification of chromium in contaminated industrial effluents.
Similar content being viewed by others
Introduction
Heavy metals (HMs) are toxic environmental pollutants and are present as natural constituents of the earth’s surface1. High concentration of heavy metals has significant and harmful effects on humans, plants, aquatic creatures and microorganisms. The uptake of elevated levels of heavy metals by plants, followed by their subsequent buildup in human tissues via bioaccumulation in the food chain, presents a risk to both human well-being and the natural surroundings. Metals such as copper serve as a co-factor for oxidative stress-associated enzymes and important for the photosynthesis2. Whereas some toxic heavy metals such as Pb, As, Cr, and Hg have a detrimental effect on kidneys and central nervous system (CNS) resulting in mental disorders, headache, weakness, anemia, abdominal cramps, and diarrhea3.
The adverse effects of industrial emissions on the environment are evident, causing widespread harm to agricultural land and water bodies on a global scale. Consequently, this has raised substantial concerns as a significant issue4. Chromium (Cr), a frequently encountered heavy metal contaminant, where the ground water is polluted5. Chromium is a firm, steel-gray metal present in the form of chromite ore in the earth’s mantle. It occurs in different oxidation states (from Cr3+ to Cr6+) and hexavalent form of the chromium are the most stable6,7.
The toxicity of chromium in eukaryotes and prokaryotes is linked to its fast diffusion of chromium through the bacterial membrane and produces free radicals through reduction of chromium. These free radicals cause alteration of DNA and other lethal effects8. The level of chromium in drinking water is increasing and it’s above the permissible limit of 0.015 mg/L (EPA-US). Therefore, it is urgently required to reduce, remove and recover chromium from industrial effluents for safety of human health and environmental protection9. Hexavalent chromium (CrVI)) is regarded as the most detrimental form of chromium due to its strong oxidation potential, increased solubility in water, rapid penetration through natural layers10,11. Heavy metals have been eliminated using a variety of conventional physicochemical methods from contaminated sites but, these approaches are incompetent as a result of high energy and power supplies, produce unwanted by-products and unsuccessful to eradicate all-metal pollution12. Biological treatments of chromium using indigenous and non-toxic bacteria are recognized as a rapid, cost-effective, environmentally friendly, chemical-free, and energy-efficient technique. Among microbes, bacteria are particularly effective in converting toxic hexavalent chromium into a non-toxic form. Their efficiency adds to their economic advantage, making them an inexpensive and eco-friendly option13,14. Utilizing enzymes isolated from microbial cells holds greater value compared to employing the whole cell. While microbial biotransformation yields potentially hazardous by-products harmful to the environment, enzymatic biotransformation ensures environmental safety by avoiding the formation of lethal substances. Many studies have shown that different strains isolated from the contaminated environment have greater efficacy towards metals and are highly considerable in bioremediation processes i.e. biosorption, bioaccumulation, biotransformation, and enzymatic bioreduction can be effectively immobilized or convert the toxic form of metal into nontoxic form15. Molecular techniques can improve the removal of heavy metals, therefore genetically modified microorganisms can be used in locations with high levels of heavy metal contamination16. Furthermore, through genetically modified approaches, bacteria can enhance the production of metal-binding proteins and enzymes crucial for the removal of heavy metals. This results in a 33% increase in the efficiency of heavy metal removal17.
In this current study, bioreduction of chromium by Bacillus paramycoides S48 (NH24A2), was assessed using different analytical methods such as atomic absorption spectroscopy, Fourier-transform infrared spectroscopy (FTIR) and Scanning electron microscopy (SEM). The physico-chemical parameters were optimized for production of chromate reductase using Placket Burman and Central Composite design. Furthermore, the enzyme was purified and characterized. The gene that codes for chromium reductase from strain S48 was successfully cloned and expressed and its chromium reduction efficiency was compared with that of wild type.
Material and method
Materials
Nutrient broth (NB) and nutrient agar (NA), 1,5 diphenylcarbazide (DPC), potassium di chromate (K2Cr2O7), nicotinamide adenine dinucleotide hydrogen (NADH) and phosphoric acid were purchased from Sigma Aldrich. DNA/Plasmid isolation and purification kits, Taq polymerase, T4 DNA ligase, NcoI and HindIII restriction enzymes, pGEM-T Easy vector and pET-28a (+) vector were obtained from Promega (Madison, WI, USA) and Novagen respectively. E. coli strains JM101 and BL21 (DE3) (NEB) were used as a host for gene cloning and expression of protein respectively. LB broth medium supplemented with kanamycin or ampicillin (50 μg/mL) was used as a selective medium for cultivation of recombinant strain. All additional reagents and chemicals used in the research were of the highest quality that was commercially available.
Methods
Bacterial strains
One of previously reported bacterium, Bacillus paramycoides S48 (NH24A2) NCBI accession number ON338038, was selected for the current study due to its tolerance to high concentration of chromium. The culture was maintained on nutrient agar under optimum growth conditions for further studies.
Bioreduction assay for chromium removal
A 250 mL Erlenmeyer flask with 100 mL of nutritional broth supplemented with 300 ppm of K2Cr2O7 was incubated with a 24 h fresh culture of B. paramycoides S48, while a negative control without chromium was also run in separate at 150 rpm for 72 h. 1 mL sample was collected after every 24 h incubation and centrifuged at 14,000 rpm for 5 min at 4 °C in order to collect supernatant for analysis of Cr(VI) reduction by 1,5-diphenylcarbazide method18. The reaction mixture was prepared, it consists of 400 μL cell free supernatant, 10 mL of distilled water, 1 mL Diphenyl carbazide (DPC) (0.25 g DPC in 100 mL acetone) and one drop of phosphoric acid. The reaction mixture was incubated for 10 min. and then absorbance was measured at 540 nm (Spectrophotometer, Agilent Technologies, G6860A, Malaysia). The K2Cr2O7 standard curve was plotted for measurement of chromium reduction. Batch bio-reduction experiment was run under optimized conditions (pH 7.0 and temperature 35 °C) at varying Cr(VI) concentration (0–400 mg/L) and contact time (24–120 h), abiotic control without inoculum was run in separate. The sample was taken after every 24 h and cell free supernatant was analyzed for Cr(VI) reduction by 1,5-diphenylcarbazide method18.
Preparation of cell biomass for chromium absorption
B. paramycoides S48 was cultured in nutrient broth supplemented with 300 ppm of K2Cr2O7 for 72 h. Bacterial cells were recovered by centrifugation at 8000 rpm for 10 min and washed gently with sterilized deionized water three times. Cells were dried in a hot oven at 55 °C for 48 h. 200 mg of dried biomass was treated with 5 mL of 30% nitric acid and kept at 150 °C for 5 h to completely evaporate nitric acid. The dried biomass was dissolved in 3 mL of 30% hydrogen peroxide and placed at 150 °C for 2 h to oxidize any remaining organic matter. The dried sample was again suspended in 1 mL of 30% nitric acid with 20 mL deionized water and finally filtered using 0.20 Millipore syringe filter. The chromium adsorption efficiency of strain S48 was evaluated using inductively coupled plasma mass spectrometry (ICP-MS).
Chromate reductase quantitative assay
The chromate reductase assay was performed by the method as previously described with minor modifications19. The reaction mixture containing 0.2 mL (0.2 mM) K2Cr2O7, 0.2 mL (0.2 M) phosphate buffer, pH (7.0), 0.2 mL (0.2 mM) NADH solution and 0.4 mL enzyme solution was prepared in a volume of 1 mL and was incubated at 37 ˚C for 30 min. Following the incubation, 0.5 mL of 20% trichloroacetic acid (TCA) was added to stop the reaction. Finally, 2 mL of DPC (0.5%) was added and the absorbance was measured spectrophotometrically at 540 nm. A control was run in separate by adding distilled water rather than enzyme solution. The enzyme unit was defined as the rate of chromate synthesis, measured in μmol/mL per min. The total protein in crude sample was determined by Lowry’s method20.
Optimization of physico-chemical parameter for maximum chromate reductase production
The physico-chemical parameters were statistically optimized by Placket-Burman design (PBD) and central composition design (CCD) using Design Expert 10.1 software, as described in the supplementary materials (S1 experimental design). The model (F) was statistically analyzed using ANOVA. PBD optimized a total of 11 components, including Na2HPO4, MgSO4.7H2O, KH2PO4, CaCl2, NaCl, sucrose, K2Cr2O7, Na2HPO4, K2HPO4, and (NH4)2SO4. Chromium reductase was subsequently produced in bulk quantity using the optimized medium.
Bulk production of chromate reductase under optimized culture conditions
The chromium reductase enzyme was produced by growing 1% fresh culture of B. paramycoides S48 was inoculated in a 500 mL production medium in an 2L Erlenmeyer Conical flask. The composition of enzyme production medium was [g/L; KH2PO4, 1.0; Na2HPO4, 4.0; NaH2PO4, 3.0; CaCl2, 0.1; NaCl, 0.6; MgSO4·7H2O, 5.0; Sucrose, 15.0; K2Cr2O7, 0.002; K2HPO4, 3.0; Yeast extract, 8.0 and (NH4)2SO4, 2.5]. The flask was incubated at 35 °C in a shaking incubator with 150 rpm for 72 h. The supernatant was collected and processed for extraction and purification of enzyme from it.
Bioreduction of hexavalent chromium by purified chromate reductase
An effluent was prepared by adding 500 mg/L of K2Cr2O7 in a sterile distilled water and treated with varying concentration of crude and purified enzyme in terms of percentage (1–10 v/v). The mixture was incubated at 35 °C for 72 h and concentration of residual chromium was measured after every 24 h using DPC method. Moreover, chromate reductase gene was successfully cloned and expressed in E. coli host in order to achieve the highest chromium reduction efficiency.
Cloning and sequencing of chromium reductase gene (ChR gene)
The primers for chromate reductase (ChR) were designed on complete sequence of B. subtilis FMN chromium reductase obtained from GenBank. The following primers having NcoI and HindIII restriction sites were used for gene amplification: Forward primer (5ʹ-CCATGGGCATATTAGTGATCAGC-3ʹ) and reverse primer (ChR-R 5ʹ- CCATGGGCATATTAGTGATCAGC-3ʹ). The gene was subjected to amplification by using the following conditions: 95 °C initial heating, 3 min of denaturation at 94 °C, 58 °C annealing, and 30 cycles of elongation at 72 °C for 60 min and 40 s. The amplified PCR product was cloned into pGEM-T vector and then transformed into E. coli JM101 competent cells. The plasmid (pGEM-T-ChR) was extracted from transformed cells using GeneJET plasmid Miniprep Kit (Thermo Scientific) and digested with EcoRI to confirm cloning. The pGEM-T-ChR clone was double digested with NcoI and HindIII, then ligated in pET28a expression vector using T4 Ligase enzyme and finally transformed into expression host E. coli BL21 (DE3). The plasmid from expression host was sequenced and analyzed using Taq Dye Deoxy Terminator Cycle Sequencing Kit and Applied Biosystems (Macrogen, Korea, Model370A automatic sequencer), respectively.
ChR gene expression in E. coli BL 21 host cell
E. coli BL21 (DE3) containing pET28a-ChR vector was cultured overnight at 37 °C in 10 mL of LB medium containing kanamycin (50 ug/mL). 2 mL of an overnight grown culture was added to 300 mL freshly prepared LB medium and placed in a shaking incubator at 37 °C for 2 h until the optical density (OD) reached 0.6 at 600 nm. 0.5 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the medium and incubated again at 20 °C for 16 h to induce protein expression. The cells were harvested after centrifugation (13,000 rpm for 10 min) and re-suspended in 0.2 M potassium phosphate buffer (pH 7.5). The cells were sonicated in ultrasonic homogenizer (Sartorius Labsonic M, 80% amplitude, 0.6 cycles for 5 min) to release intracellularly produced chromate reductase. After sonication, the cells were centrifuged at 13,000 rpm for 15 min in order to remove the debris and clear supernatant containing chromate reductase enzyme was collected.
Purification of native and recombinant chromate reductase
The cell free supernatant obtained in the previous section was subjected to ammonium sulfate precipitation by adding solid ammonium sulfate to it with swirled continuously at 4 °C until the solution was 80% saturated. Then dialysis was performed to remove ammonium sulfate from enzyme. The enzyme chromate reductase was purified through gel permeation chromatography technique21. Approximately 4 mL of crude enzyme was loaded to the Sephadex G-100 gel column. Potassium phosphate buffer (pH 7.0) was passed through the column with a flow rate of 3 mL/15 min. A total of 30 fractions were collected and absorbance was noted at 280 nm to determine total protein concentration in each fraction. The protein quantification and enzyme assay were performed for all fractions. Fractions with high chromate reductase activity were collected and stored at 4 °C for further study.
Similarly, the chromate reductase from recombinant BL21 was purified using 3 steps. ammonium sulfate precipitation, dialysis, and flow through DEAE Sepharose column. The molecular size of chromate reductase was determined by SDS-PAGE. The sample was run on 12% SDS-PAGE gel followed by staining with Coomassie brilliant blue R-250 dye to visualize the protein bands22.
Bioinformatics analysis
The BLASTp and BLASTn tools (http://www.ncbi.nlm.nih.gov/BLAST/) were used to analyze nucleotides and determine the corresponding amino acids, respectively. The CLUSTALW tool (http://www.ebi.ac.uk/clustalW) was used to perform multiple sequence alignment analysis for the chr gene. A dendogram was generated using MEGA 6.0 program and phylogenetic analysis was performed for chromate reductase.
Characterization of native and recombinant chromate reductase (BparChR) from B. paramycoides S48
Effect of temperature and pH on enzyme activity and stability
The effect of temperature and pH on the activity of both native and recombinant purified chromate reductase enzyme (BparChR) was measured at various temperatures ranging from 30–60 °C and pH 3.0–9.0 for 30–120 min. Different buffers i.e. 100 mM acetate buffer (pH 3.0–5.0), phosphate buffer for (pH 6.0–8.0), and Glycine–NaOH buffer (pH 9.0–11.0) was used. The effect of temperature and pH was determined in terms of relative activity using standardized assay.
Effect of metal ions on enzyme activity
The effect of various divalent cations i.e., Ca2+, Cu2+, Mg2+, Zn2+, Hg2+, Ba2+, Fe2+, Ni2+, K2+, Cd2+, Co2+, and Na2+ on activity of purified chromate reductase was determined using two different metals concentration (2 mM and 10 mM) for 30 min. The standard chromium reductase test was used to calculate residual activity.
Effect of surfactant and organic solvents on enzyme activity
The effect of various surfactants such as Tween 80, Tween 60, Tween 20, SDS, CTAB, and Triton X-100 and organic solvents such as methanol, acetonitrile, acetone, propanol, butanol, ethanol, formaldehyde, and ethyl acetate, on enzyme activity was analyzed after incubation for 30 min. The standard enzyme assay was used to calculate residual activity.
Determination of kinetic parameters
The Michaelis–Menten constant (Km) and maximum velocity (Vmax) for purified chromate reductase activity was determined at 40 °C and pH 7.0 in a reaction mixture with various concentrations of K2Cr2O7 as a substrate (0.1–1 mM). The data was plotted in the Lineweaver–Burk software in order to determine the Vmax and Km value of kinetic constant.
Bioreduction of hexavalent chromium by recombinant BparChR from B. paramycoides S48
A K2Cr2O7 solution was prepared after mixing 100–500 mg/L of K2Cr2O7 in sterile distilled water in 500 mL Erlenmeyer flask. 10 mg/mL of BparChR enzyme was added to the flask and then incubated at 35 °C for 72 h. The residual chromium concentration was measured after every 24 h using DPC method as previously described.
Statistical analysis
Analyzing the variance (ANOVA) allowed us to quantitatively determine the PBD model and CCD for optimizing media components. To assess the interplay between different nutritional factors, response surface plots of the projected model were used.
Results
Characterization of Bacterial strain S48
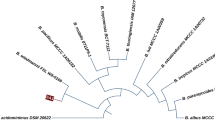
The bacterial strain S48 was previously identified through 16S rRNA gene sequencing. The ITS sequence analysis of strain S48 showed its close homology (99%) to Bacillus paramycoides. The sequence was submitted to NCBI with accession number ON338038 (Fig. 1).
Bioreduction of hexavalent chromium by Bacillus paramycoides S48
Chromium (VI) reduction capability of strain S48 was determined at varying concentrations of K2Cr2O7 ranging from 25–500 mg/L. About 90% chromium reduction was observed at 25–50 mg/L after 48 h. The rate of reduction was gradually decreased with an increase in K2Cr2O7 concentration (100–500 mg/L), (Fig. 2A).
Bioreduction of chromium and their SEM images of B. paramycoides S48 after Cr reduction (A) Chromium reduction by strain S48 with different initial concentration (25- 500 mg/L) (B) S48 without Cr treatment and EDX Spectrum of control containing no detectable Cr Peak (C) S48 treated with Cr and EDX Spectrum of S48 containing detectable Cr Peak.
Analysis of bioreduction of hexavalent chromium by Bacillus paramycoides S48
FT-IR spectroscopy analysis of biomass
Strain S48 cells were subjected to FTIR spectroscopy for chemical changes in its structure after incubation with chromium. FTIR spectrum for bacterial cells incubated with and without chromium (treated & untreated) showed potential difference in their functional groups (Supplementary materials figure S1A and B). The peaks in the range of 3500–3280 cm−1 showed –OH group of glucose and –NH stretching of protein. The peak 2923.80 cm−1 wavenumber was moved to 2929.89 cm−1 after chromium adsorption depicting stretching of CH3 groups. Similarly, other peaks at 1633.96 cm−1, 1525.08 cm−1, 1394.59 cm−1, 1039.74 cm−1 and 610.88 cm−1 shifted to 1626.81 cm−1, 1516.40 cm−1, 1395.08 cm−1, 1047.01 cm−1 and 610.40 cm−1, respectively, that corresponds to the presence of stretching at C=O, amid bond, COO, phosphate groups and deformation of S=O and –C–C– stretching. In control, two peaks were obtained at 574.67 and 554.95 cm−1 which completely disappeared in Cr (VI) treated sample, furthermore a new peak at 539.47 cm−1 appeared in treated sample which was not seen in control. The FTIR analysis clearly indicates the effect of chromium metal on different functional groups after attachment to cell wall of strain S48.
SEM–EDX analysis of B. paramycoides S48
The changes in morphology of B. paramycoides S48 after incubation with chromium, were visualized through SEM. The SEM analysis of metal untreated biomass showed elongated, rod shaped, and uniform sized bacterial cells with smooth surface, however EDX analysis showed no peak of chromium (Fig. 2B), whereas, cells exposed to Cr(VI) were observed with marked changes in its appearance such as increase in width, reduce in length, with rough appearance and clumping of cells and EDX analysis showed a peak at a binding energy of 5.4, which attributed to intracellular absorption of Cr by B. paramycoides S48 (Fig. 2C).
Optimization of physico-chemical parameter for chromate reductase production
Optimization of physical conditions
The effect of several physical conditions on chromate reductase production from B. paramycoides S48 was evaluated by determining its specific activity. The optimal conditions for maximum enzyme activity was observed at 35 °C, pH 7.0, after 48 h of incubation.
Optimization of the nutritional factors using Plackett–Burman design (PBD) and central composite design (CCD)
The factors optimized through PBD showed a significant effect on production of chromate reductase. Maximum specific activity 305.386 U/mg was achieved in run number 08 (Supplementary materials Table S1). Pareto chart from PBD explains the effect of individual factor on production of chromate reductase. 04 out of 11 factors were found to have significant effect on enzyme production including B: KH2PO4, G; K2Cr2O7, F; Sucrose, and C; MgSO4.7H2O (Fig. 3A). The model equation for specific activity (U/mg) can be written as:
Showing the significant factors effect on enzyme production using Pareto chart and a three-dimensional response surface plot. (A) The effect of different factors generated by PBD on the rate of enzyme production (B) Response surface plots showing the combined effects KH2PO4 and K2Cr2O7; (C) Response surface plots showing the combined effects KH2PO4; and sucrose.
The Model F-value of 12.11 implies that the model is significant. Value of “Prob > F” less than 0.05 indicates model terms are significant. In this case B, C, F, G, are significant model terms (Supplementary materials Table S2).
The major components acquired by PBD were screened using a two-level CCD. Thirty runs were conducted to examine the impact of the four dietary components included in the model on the activity of chromate reductase (Supplementary materials Table S3). The acceptability of model was evaluated using ANOVA (Supplementary materials Table S4). Among the quadratic and linear terms, only interactive term AB and AC were found to be significant model terms, whereas A represents KH2PO4, and B represent K2Cr2O7 while, C represent sucrose. Response surface plot (AB) shows that increase in K2Cr2O7 and a decrease in KH2PO4 enhanced enzyme activity (Fig. 3B). The response plot (AC) shows that chromate reductase activity was increased by decreasing the concentration of KH2PO4 and increasing sucrose concentration (Fig. 3C). The second polynomial equation was obtained after multiple regression analysis data. Final Equation in terms of coded factors:
The Model F-value of 8.64 implies that the model is significant.Values of “Prob > F” less than 0.05 indicate model terms are significant (Supplementary materials Table S4). In this case, C, AB, AC, B2, C2 are significant model terms.
Purification of chromate reductase from B. paramycoides S48
Gel filtration chromatography using Sephadex G-100 was used to purify chromate reductase (Fig. 4A). The purification steps for chromate reductase are shown in Table 1. The molecular weight of chromate reductase was found to be approximately 35 kDa using standard denaturing marker protein (Fig. 4B).
Purification and characterization profile of chromium reductase (A) Total protein and specific activity profile of chromate reductase from B. paramycoides S48 in fractions from column chromatography (B) SDS-PAGE of chromate reductase from B. paramycoides S48 after purification by column chromatography. Lane 1, (Color Prestained Protein Standard) 11–245 kDa; Lane 2, Chromate reductase of 35 kDa (C) Effect of temperature on the activity and stability of chromate reductase from B. paramycoides S48 (D) Effect of pH on the activity and stability of chromate reductase from B. paramycoides S48.
Characterization of chromate reductase
Effect of temperature and pH on chromate reductase activity and stability
The chromate reductase from B. paramycoides S48 showed maximum activity at temperature ranges 30–40 °C and retained 100% of its activity for 120 min at 35 °C (Fig. 4C). The enzyme showed maximum activity at pH 6.0–8.0 and retained 100% of its activity at pH 7.0 for 120 min (Fig. 4D).
Effect of metal ions
The effect of different concentrations (2 mM and 10 mM) of mono and divalent metals on chromate reductase activity was analyzed. Metals such as Ca2+, Ba2+, Fe2+, K2+, and Mg2+ have shown positive effect on chromate reductase activity with the increase in metal concentration in terms of increase in its residual activity. A slight decrease in residual activity was observed in the presence of Cu2+, Co2+ and Na2+ at both concentrations. A strong inhibitory effect was observed in the presence of Hg2+ and Cd2+ at both 2 mM and 10 mM concentrations (Table 2).
Effect of surfactants and organic solvents
Various surfactants in two different surfactants concentrations (0.5% and 1.0%) were used to analyze their effect on activity of purified chromate reductase. Among surfactants: Tween 20, 60 and 80 enhanced enzyme activity with increase in its concentration, while SDS CTAB and triton X-100 inhibited enzyme activity (Supplementary materials Table S5). Similarly, the effect of organic solvents on enzyme activity was determined by incubating it in organic solvent for 120 min. Acetonitrile, ethyl acetate and n-Hexane suppressed enzyme activity, while, methanol, propanol, chloroform and DMSO increased the activity up to 120% with an increase in incubation time (Supplementary materials Table S6).
Determination of kinetic parameters
The Michaelis Menten constant (Km) and rate of reaction (Vmax) values of purified chromate reductase from B. paramycoides S48 were calculated by Lineweaver and Burk’s plot. The enzyme was incubated with varying concentrations potassium dichromate (1–10 µM). The Km and Vmax of enzyme were found to be 2.33 µM and 222.22 µmol mg−1 min−1, respectively (Supplementary materials Fig. S2).
Bioreduction of hexavalent chromium by purified chromate reductase
Potassium dichromate (100 mg/L) was treated with 0.1, 1, and 5 mg/mL of both crude and purified enzymes. Crude enzyme showed 20, 45, and 75% chromate reduction with 0.1, 1 and 5 mg/mL after 120 h, whereas purified chromate reductase showed 42, 64, and 87% chromate reduction with 0.1, 1 and 5 mg/mL after 120 h, respectively (Fig. 5).
Gene cloning and expression of chromate reductase
The chromium reducing gene (BparChR) from strain S48 was amplified using S48 genomic DNA as a template in the presence of newly designed primers. Approximately 0.6 kb size of chromate reductase encoding gene from B. paramycoides S48 was amplified (Fig. 6A). The recombinant pGEMT- BparChR plasmid was sequenced by Macrogen, Netherland. Phylogenetic relationship of BparChR from B. paramycoides S48 with other chromate reductase showed highest identity with chromate reductase of B. paramycoides sp OV166 (Fig. 6C). 0.5 mM IPTG was used to induce recombinant chromate reductase, and SDS-PAGE was used to verify expression. The expressed band of chromate reductase was clearly visible on SDS-PAGE (Fig. 6B).
Cloning and expression of BparChR gene from B. paramycoides S48 (A) PCR amplification of BparChR gene; Lane 1, DNA ladder (NEB, B7025); Lane 2, BparChR amplicon approximately 0.6 kb (B) Recombinant chromate reductase expression by SDS-PAGE; Lane 1. Protein marker (11–245 kDa, Lane 2,3 Induced protein of E. coli BL 21 (DE3) with pET-28a, Lane 4: Induced protein of E. coli BL 21 (DE3) with pET28BparChR. (C) Phylogenetic relationship of B. paramycoides S48 chromate reductase with other chromate reductase available in NCBI database, Neighbor-joining tree showed maximum identity with chromate reductase of B. paramycoides sp OV166.
Purification of recombinant chromate reductase
The recombinant chromate reductase was purified by ion-exchange chromatography using DEAE Sepharose (with elution buffer NaCl 0.9 M concentration along with 100 mM phosphate buffer). The purified band of recombinant chromate reductase was clearly visible on SDS-PAGE (Supplementary materials figure S3). Table 3 showed the steps of purification of chromate reductase with total yield and purification fold. of recombinant enzyme.
Characterization of BparChR enzyme
Effect of temperature and pH on activity and stability
The optimum BparChR enzyme activity was achieved at 35 °C. The BparChR enzyme retained 94% of its activity at 35 °C for 120 min (Fig. 7A). The optimum activity of recombinant enzyme was observed at pH 7.0. The enzyme retained 100% of its activity at pH 6.0–7.0 for 120 min. (Fig. 7B).
Temperature and pH profile of purified recombinant enzyme and treatment of chromium by recombinant chromate reductase from B. paramycoides S48 (A) Effect of temperature on activity and stability (B) Effect of pH on activity and stability (C) Aqueous solution containing different concentration of Cr (100–500 mg/L) treated with recombinant chromate reductase (10 mg/mL).
Bioreduction of hexavalent chromium by purified recombinant chromate reductase
The deionized water containing different concentrations of chromium (25–500 mg/L) was treated with purified recombinant enzyme (10 mg/mL) at 35 °C for 96 h. The recombinant BparChR reduced 100% of chromate between 25–100 mg/L after 24 h, while the higher concentrations (200–500 mg/L) were completely reduced after 96 h. (Fig. 7C).
Discussion
The researchers are focusing on biological methods utilizing microorganisms for the removal of toxic metals present in industrial effluents worldwide, as a response to the increase in number of industries in recent years. Chromium (Cr), a significant heavy metal pollutant predominantly found in wastewater, raises environmental concerns. When microorganisms encounter metal ions in their environment, they adopt various strategies: developing resistance to toxic levels, utilizing trace amounts for metabolic activities, detoxifying excessive metal species, or tolerating metal ions up to a threshold limit. Consequently, microbial species like bacteria, fungi, and yeasts engage in the uptake of Cr through various biological processes i.e. bioaccumulation, biosorption and chromium efflux. An abundance of microorganisms naturally inhabits water and soil that receive industrial effluents. These microbes have evolved a range of mechanisms to defend themselves against the harmful effects of heavy metals23.
An elemental analysis of both sludge and soil samples, revealing an elevated concentration of chromium (Cr) at approximately 2648.644, surpassing the standard values set by the US-EPA. The levels of Cr, measured at 40.06 ± 0.21, were notably high, with the water comprehensive pollution index reaching levels 1000 times higher than the critical values established by Chen et al.24. The primary screening of Cr resistant strains was performed and Bacillus paramycoides S48 from Korangi sludge was found to be the best among Cr resistant strains. Industrial effluents had bacterial populations ranging from 200 to 300 colonies per 100 mL, depending on the sample site25.
B. paramycoides S48 exhibited highest Cr (1500 mg/L) tolerance level at temperature 35 °C and pH 7.026. In a previous study the minimum inhibitory concentration (MIC) for heavy metal was found in the range from 50 to 1900 µg/mL27. A Cr(VI) reducing bacterium Pseudomonas dechromaticans was first time reported in 1970s from an anaerobic environment that reduce Cr(VI) into Cr(III)28. In our study, the Cr(VI) reduction rate was observed as 60% at 500 mg/L initial concentration at 35 °C and pH 7.0 after 96 h. This was significantly higher in comparison to a previous report where Pseudomonas sp. MAI4 reduced 150 µg/mL of Cr(VI) after 120 h. The bacterial growth and Cr(VI) reducing effect of bacteria were negatively affected by increasing temperature due to inhibition of physiological activity as a result of damage to bacterial macromolecules (DNA and proteins) as well as outer membranous structures23.
The FTIR spectrum of B. paramycoides S48 after incubation with Cr(VI) showed remarkable difference in the cellular functional groups. These results correspond to the attachment of metal to the functional group present in the cell wall of bacterium. The peaks at position 2929 cm−1 and 1633 cm−1 represent stretching vibrations of C–H bond and vibration of C=O group, respectively. The stretching vibration of C=O and C–H groups corresponds to Cr(VI) binding to membranous functional groups29. The scanning electron micrographs of B. paramycoides S48 after incubation with chromium showed clear morphological changes in comparison with control. The surface of strain S48 with Cr(VI) appeared distorted, dense and attached to each other in comparison to control. According to the previous reports, several bacterial strains, such as Pseudomonas sp. B50D, Acinetobacter sp. B9 and Bacillus sp. strain PZ-1 were observed with similar morphological changes in the presence of toxic heavy metals30,31.
Maximum production of chromate reductase by B. paramycoides S48 was achieved at temperature 35 °C and pH 7.0. The nutritional parameters for maximum enzyme production were statistically optimized using Plackett–Burman and central composite designs, and the total yield was increased up to 545.993 U/mg. Sucrose and MgSO4.7H2O exhibited a positive impact on maximizing the production of the chromium reductase. These findings suggest that sucrose induces enzyme production, while the salts (MgSO4·7H2O) provide the necessary nitrogen for bacterial growth and enzyme synthesis. The reciprocal interaction analysis indicated that increase in concentration of K2Cr2O7 and sucrose enhanced the enzyme activity enzyme. This is further supported by a study on inducible chromate reductase from Bacillus methylotrophicus with initial activity of 212.84 U/mg after 48 h in a medium supplemented with 0.25 mM chromate32. Very few studies are available where statistical design has been used for optimization of chromate reductase production and reduction of Cr(VI).
Gel filtration column chromatography was used to purify chromate reductase to homogeneity, and its molecular size was estimated to be 35 kDa. A chromate reductase enzyme from a novel bacterium, Ochrobactrum sp. strain Cr-B4, with molecular mass approximately 31.53 kDa, was partially characterized33.
The activity of chromium reductase was reported to be stable throughout a broad temperature range (30 to 45 °C) and pH range (6.0 to 8.0), with its optimum values seen at 40 °C and pH 7.0. Additionally, the enzyme activity was also enhanced in presence of various metals except for Hg2+ , which indicated the presence of critical vicinal sulfhydryl groups34. Chromate reductase from Rhodopseudomonas palustris KU003 was strongly inhibited by metals such as Cd2+, Zn2+ and Cu2+35. The purified chromate reductase retained 100% activity in the presence of various surfactants, while strong inhibition was seen in the presence of CTAB and, SDS. The limitation in the activity is due to non-ionic surfactants reacting with enzyme hydrophobic region and lead to modification in 3-D structure36. Organic solvents such as acetonitrile, n-hexane and ethyl acetate decreased the enzyme activity with increase in incubation time35. The chromate reductase exhibited promising low Km and high Vmax values which indicated a high viability of the reaction system in the presence of K2Cr2O7 as a substrate.
The chromate reductase (ChR) gene from B. paramycoides S48 was cloned and expressed in the E. coli BL21 host. The native chromate reductase activity was lower than the recombinant one that may be attributed to high levels of protein expression. The recombinant enzyme from E. coli BL21 was functionally active and able to eliminate harmful Cr (VI). Several researchers have reported their success stories on cloning and expression of ChR genes in E. coli host37. The cloned chromate reductase (ChR gene) from B. paramycoides S48 of 0.6 kb was comparable to the gene studied in other Bacillus species24. The ChrR gene has mostly been identified in Gram-positive bacteria such as Bacillus atrophaeus Rhodococcus erythropolis and Arthrobacter aurescens with a potential role in bioconversion of toxic Cr(VI) into non-toxic Cr(III)10,38,39. Numerous Cr(VI) reductases have been identified, including ChrT from Serratia sp. S240, NemA from E. coli41, ChrR from Pseudomonas42 and NfrA from Bacillus subtilis43. However, only ChrR has been elucidated at the molecular level, and its spatial structure has been confirmed. While the exact molecular mechanisms used by various bacterial strains for Cr(VI) bioremediation remain largely unexplored44.
The recombinant BparChR was found stable at temperature range 30–54 °C with optimum activity at 35 °C and pH 7.0. The highest activity at neutral pH may offer sustainability of the BparChR for bioremediation of industrial effluents. Zhou45 reported ChrT-engineered bacterium that was capable of removing Cr(VI) at 37 °C and pH 7.0. Sometime unsuitable environmental conditions affect the growth and survival of host bacteria, as a result removal of Cr(VI) may be affected by changes in environmental conditions.
In our study, the enzyme was active in the presence of metal ions except Hg2+ that inhibited BparChR activity. The purified recombinant BparChR showed 98% chromate reduction efficieny in a sample containing 100–500 mg/L of K2Cr2O7 after 96 h. The main issue in bioremediation technology is the presence of other pollutants that affect the process of Cr(VI) removal. The high activity of chromate reductase in the presence of metal ions is crucial for efficient wastewater bioremediation. Recombinant BparChR is the most potent protein for removal of toxic Cr(VI) from effluent and thus has the most potential and efficient method for Cr treatment in industrial effluents before release into water.
Conclusion
It is concluded that chromium resistant B. paramycoides S48 isolated from metals contaminated sludge, could bioremediate chromium at ambient temperature. Primary screening showed that strain S48 exhibited the highest tolerance to Cr (1500 mg/L). The chromium reductase activity from strain S48 in chromium contaminated effluent demonstrates potential role in chromium removal. The purified enzyme was found stable to various environmental factors, especially metals ions which could efficiently remove chromium at mesophilic temperature. The chromate reduction of yield and efficiency were further enhanced to approximately 98% after cloning and overexpression of chromate reductase from B. paramycoides S48. Considering its reduction ability, strain S48 and recombinant BparChR against hexavalent chromium could be a potential candidate in bioremediation of heavy metals.
Data availability
All data supporting the findings of this study are available within the paper and its supplementary information.
References
Vardhan, K. H., Kumar, P. S. & Panda, R. C. A review on heavy metal pollution, toxicity and remedial measures: Current trends and future perspectives. J. Mol. Liq. 290, 111197 (2019).
Chrysargyris, A., Papakyriakou, E., Petropoulos, S. A. & Tzortzakis, N. The combined and single effect of salinity and copper stress on growth and quality of Mentha spicata plants. J. Hazard. Mater. 368, 584–593 (2019).
Rana, M. N., Tangpong, J. & Rahman, Md. M. Toxicodynamics of Lead, Cadmium, Mercury and Arsenic- induced kidney toxicity and treatment strategy: A mini review. Toxicol. Rep. 5, 704–713 (2018).
Ricciardi, L., D’Odorico, P., Galli, N., Chiarelli, D. D. & Rulli, M. C. Hydrological implications of large-scale afforestation in tropical biomes for climate change mitigation. Philos. Trans. R. Soc. B Biol. Sci. 377, 20210391 (2022).
Yang, X. et al. Mechanism and enhancement of Cr(VI) contaminated groundwater remediation by molasses. Sci. Total Environ. 780, 146580 (2021).
Coetzee, J. J., Bansal, N. & Chirwa, E. M. N. Chromium in environment, its toxic effect from chromite-mining and ferrochrome industries, and its possible bioremediation. Expo. Health 12, 51–62 (2020).
Qian, J. et al. Direct Cr (VI) bio-reduction with organics as electron donor by anaerobic sludge. Chem. Eng. J. 309, 330–338 (2017).
Jobby, R., Jha, P., Yadav, A. K. & Desai, N. Biosorption and biotransformation of hexavalent chromium [Cr(VI)]: A comprehensive review. Chemosphere 207, 255–266 (2018).
Li, S. et al. New method for efficient removal of Cr(VI) by recoverable magnetic nitrogen-doped carbon aerogel microspheres: Kinetics and mechanism. J. Mol. Liq. 360, 119564 (2022).
Baldiris, R., Acosta-Tapia, N., Montes, A., Hernández, J. & Vivas-Reyes, R. Reduction of hexavalent chromium and detection of chromate reductase (ChrR) in Stenotrophomonas maltophilia. Molecules 23, 406 (2018).
Zheng, N. et al. Rust triggers rapid reduction of Cr6+ by red phosphorus: The importance of electronic transfer medium of Fe3+. Chemosphere 303, 134971 (2022).
Singh, A. & Prasad, S. M. Remediation of heavy metal contaminated ecosystem: an overview on technology advancement. Int. J. Environ. Sci. Technol. 12, 353–366 (2015).
Trellu, C. et al. Removal of hydrophobic organic pollutants from soil washing/flushing solutions: A critical review. J. Hazard. Mater. 306, 149–174 (2016).
Verma, S. & Kuila, A. Bioremediation of heavy metals by microbial process. Environ. Technol. Innov. 14, 100369 (2019).
Jacob, J. M. et al. Biological approaches to tackle heavy metal pollution: A survey of literature. J. Environ. Manag. 217, 56–70 (2018).
Naik, M. M. & Dubey, S. K. Lead resistant bacteria: Lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol. Environ. Saf. 98, 1–7 (2013).
Yin, K., Wang, Q., Lv, M. & Chen, L. Microorganism remediation strategies towards heavy metals. Chem. Eng. J. 360, 1553–1563 (2019).
Fulladosa, E., Desjardin, V., Murat, J.-C., Gourdon, R. & Villaescusa, I. Cr(VI) reduction into Cr(III) as a mechanism to explain the low sensitivity of Vibrio fischeri bioassay to detect chromium pollution. Chemosphere 65, 644–650 (2006).
Thatoi, H., Das, S., Mishra, J., Rath, B. P. & Das, N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: A review. J. Environ. Manag. 146, 383–399 (2014).
Lowry, O. H. [17] Micromethods for the assay of enzymes. In Methods in Enzymology Vol. 4 366–381 (Academic Press, 1957).
Determann, H. Gel Chromatography Gel Filtration · Gel Permeation · Molecular Sieves: A Laboratory Handbook (Springer, 2012).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Wani, P. A. et al. Prospective of chromium (VI) reduction under in vitro and in vivo conditions and stimulation of antioxidant defense of cowpea under the exposure of Cr (VI). Appl. Soil. Ecol. 132, 187–193 (2018).
Chen, L., Hoff, J. & S,. A two-stage wood chip-based biofilter system to mitigate odors from a deep-pit swine building. Appl. Eng. Agric. 28, 893–901 (2012).
Mustapha, M. U. & Halimoon, N. Screening and isolation of heavy metal tolerant bacteria in industrial effluent. Procedia Environ. Sci. 30, 33–37 (2015).
Kalsoom, et al. Isolation and screening of chromium resistant bacteria from industrial waste for bioremediation purposes. Braz. J. Biol. 83, e242536 (2021).
Marzan, L. W., Hossain, M., Mina, S. A., Akter, Y. & Chowdhury, A. M. M. A. Isolation and biochemical characterization of heavy-metal resistant bacteria from tannery effluent in Chittagong city, Bangladesh: Bioremediation viewpoint. Egypt. J. Aquat. Res. 43, 65–74 (2017).
Romanenko, V. I. & Korenkov, V. N. Bacterial reduction of ions. Inform Byul In-ta Biol Vnutr Vod Akad Nauk SSR 25, (1975).
Srinath, T., Verma, T., Ramteke, P. W. & Garg, S. K. Chromium (VI) biosorption and bioaccumulation by chromate resistant bacteria. Chemosphere 48, 427–435 (2002).
Bhattacharya, A. & Gupta, A. Evaluation of Acinetobacter sp. B9 for Cr (VI) resistance and detoxification with potential application in bioremediation of heavy-metals-rich industrial wastewater. Environ. Sci. Pollut. Res. 20, 6628–6637 (2013).
Giovanella, P., Cabral, L., Bento, F. M., Gianello, C. & Camargo, F. A. O. Mercury (II) removal by resistant bacterial isolates and mercuric (II) reductase activity in a new strain of Pseudomonas sp. B50A. New Biotechnol. 33, 216–223 (2016).
Sandana Mala, J. G., Sujatha, D. & Rose, C. Inducible chromate reductase exhibiting extracellular activity in Bacillus methylotrophicus for chromium bioremediation. Microbiol. Res. 170, 235–241 (2015).
Hora, A. & Shetty, V. K. Partial purification and characterization of chromate reductase of a novel Ochrobactrum sp. strain Cr-B4. Prep. Biochem. Biotechnol. 45, 769–784 (2015).
Harboul, K., El Aabedy, A., Hammani, K. & El-Karkouri, A. Reduction of hexavalent chromium using Bacillus safensis isolated from an abandoned mine. Environ. Technol. 1–17 (2023).
Rajyalaxmi, K., Merugu, R., Girisham, S. & Reddy, S. M. Chromate reduction by purple non sulphur phototrophic bacterium Rhodobacter sp. GSKRLMBKU–03 isolated from pond water. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 89, 259–265 (2019).
Gulcin, İ, Buyukokuroglu, M. E. & Kufrevioglu, O. I. Metal chelating and hydrogen peroxide scavenging effects of melatonin. J. Pineal Res. 34, 278–281 (2003).
Wei, W. et al. Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ. Sci. Technol. 48, 3363–3371 (2014).
Patra, R. C., Malik, S., Beer, M., Megharaj, M. & Naidu, R. Molecular characterization of chromium (VI) reducing potential in Gram positive bacteria isolated from contaminated sites. Soil Biol. Biochem. 42, 1857–1863 (2010).
Chaney, R. L. & Baklanov, I. A. Phytoremediation and Phytomining: Status and Promise. In Advances in Botanical Research Vol. 83 (eds Cuypers, A. & Vangronsveld, J.) 189–221 (Academic Press, 2017).
Gu, R. et al. Chromium metabolism characteristics of coexpression of ChrA and ChrT gene. Ecotoxicol. Environ. Saf. 204, 111060 (2020).
Robins, K. J., Hooks, D. O., Rehm, B. H. A. & Ackerley, D. F. Escherichia coli NemA is an efficient chromate reductase that can be biologically immobilized to provide a cell free system for remediation of hexavalent chromium. PLoS ONE 8, e59200 (2013).
Park, C. H., Keyhan, M., Wielinga, B., Fendorf, S. & Matin, A. Purification to Homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl. Environ. Microbiol. 66, 1788–1795 (2000).
Moch, C., Schrögel, O. & Allmansberger, R. Transcription of the nfrA-ywcH operon from Bacillus subtilis is specifically induced in response to heat. J. Bacteriol. 182, 4384–4393 (2000).
Eswaramoorthy, S. et al. Crystal structure of ChrR—A quinone reductase with the capacity to reduce chromate. PLoS ONE 7, e36017 (2012).
Zhou, S. et al. Reducing capacity and enzyme activity of chromate reductase in a ChrT-engineered strain. Exp. Ther. Med. 14, 2361–2366 (2017).
Funding
The current study was funded by HEC under grant No. 50043034 and Government of Turkiye Burslari Scholarship Grant No. 20PK038055.
Author information
Authors and Affiliations
Contributions
KW, SC; AOB, AAS: Preparation of the overall research plan as well as protocols for various experiments; KW, SUD, EC: Performed experimental work in lab as per the pre-designed research plan; EC, FH, SK, MB: Facilitated in interpretation of various analysis and statistical optimization experiments in the current research project; SK, SC, AOB, AAS: Guided the student to perform cloning and expression experiment KW, SUD: Facilitated in statistical analysis of the results KW, SUD, SC, AOB, AAS: Write up of the manuscript; FH, SK, MB, AAS: Proof reading of the overall manuscript for English comprehension and typing mistakes.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kalsoom, K., Din, S.U., Ceylan, E. et al. Cloning and expression of chromate reductase from Bacillus paramycoides S48 for chromium remediation. Sci Rep 15, 18796 (2025). https://doi.org/10.1038/s41598-025-03412-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03412-x