Abstract
Inflammatory bowel disease (IBD) is a chronic condition characterized by intestinal inflammation and dysbiosis, with limited treatment options and significant challenges in long-term management. This study investigated the therapeutic potential of novel strains belonging to Bifidobacterium longum and Limosilactobacillus species, in a dextran sodium sulfate (DSS)–induced mouse model of colitis. In this study, our primary objective was to determine whether ingestion of these strains alleviates colitis symptoms and, if so, to elucidate how they restored gut microbial balance and modulated microbial metabolic functions. In most probiotic-treated groups, colitis disease activity index scores were significantly improved and colon length was preserved, with strains CBA7106 and BBH exhibiting efficacy comparable to that of the Lactobacillus rhamnosus GG (used as a positive control). Moreover, histological analyses confirmed reduced inflammation and enhanced mucosal integrity. Microbial diversity assessments demonstrated a marked restoration of gut microbial composition, highlighted by increased abundances of beneficial taxa such as Lactobacillus and Akkermansia. Metabolomic profiling identified key anti-inflammatory metabolites, including 6-hydroxycaproic acid, indole-3-lactic acid, and choline, which were significantly elevated in the probiotic-treated groups. Notably, functional analyses using PICRUSt2 revealed a sustained decrease in the siderophore biosynthesis pathway (ko01053), suggesting that these candidate probiotic strains may inhibit siderophore production—a pivotal mechanism by which pathogenic bacteria aggravate intestinal inflammation. Taken together, these findings indicate that the candidate probiotic strains CBA7106 and BBH effectively mitigate DSS-induced colitis by modulating the gut microbiota, promoting the production of anti-inflammatory metabolites, and suppressing siderophore biosynthesis. This study provides valuable insights into the development of targeted probiotic therapies for IBD, underscoring their potential as a complementary approach to restoring intestinal health and reducing inflammation. Further clinical studies are warranted to validate these observations in human populations.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD), which includes Crohn’s disease and ulcerative colitis (UC), is a chronic inflammatory disorder of the gastrointestinal tract that substantially impairs the quality of life of patients1. It is characterized by severe symptoms, such as abdominal pain, diarrhea, intestinal bleeding, and weight loss, along with systemic manifestations, such as fatigue and malnutrition2. Despite advancements in therapeutic strategies, the existing treatments, including corticosteroids, immunomodulators, biologics, and small-molecule inhibitors, are limited by their suboptimal efficacy, high relapse rates, and adverse side effects3. Consequently, there is an urgent need to explore alternative or complementary therapies to address these challenges and improve the long-term management of IBD4.
Gut microbiota plays a central role in maintaining intestinal homeostasis by regulating immune responses, supporting epithelial integrity, and facilitating nutrient metabolism. However, in IBD, substantial dysbiosis of the intestinal microbiota occurs, which is characterized by an increased abundance of pro-inflammatory microbial taxa, such as Enterobacteriaceae, and a reduction in beneficial microbes, including Lactobacillus and Bifidobacterium. Dysbiosis contributes to epithelial barrier dysfunction, promotes inflammatory immune responses, and alters microbial metabolic output, all of which exacerbate intestinal inflammation5. Therefore, targeting dysbiosis and restoring microbial homeostasis have emerged as promising strategies for mitigating IBD symptoms and progression. Probiotics, live microorganisms that confer health benefits when administered in adequate amounts, have garnered considerable attention as potential therapeutic agents for IBD treatment. These microorganisms modulate the composition of the gut microbiota, enhance epithelial barrier integrity, and regulate immune responses. Lactobacillus rhamnosus GG (LGG) has demonstrated efficacy in experimental colitis models by reducing pro-inflammatory cytokine levels, enhancing the production of anti-inflammatory cytokines, such as IL-10, and promoting epithelial regeneration6. Similarly, Bifidobacterium bifidum alleviates DSS-induced colitis in mice by modulating gut microbiota composition and reducing mucosal inflammation7. These studies highlight the potential of probiotics in addressing both microbial and immune dysregulation in IBD.
Probiotics, such as Lactobacillus plantarum and Bifidobacterium longum, reduce siderophore biosynthesis, thereby limiting the proliferation of pathogenic bacteria and mitigating inflammation8. VSL#3, a multi-strain probiotic containing Lactobacillus and Bifidobacterium, has shown efficacy in inducing remission in patients with UC by reducing inflammatory cytokines and restoring the gut microbial balance9. The yeast-based probiotic Saccharomyces boulardii suppresses the expression of inflammatory cytokines, such as TNF-α, strengthens epithelial cell defense mechanisms, and reduces mucosal inflammation10. Polysaccharide A produced by Bacteroides fragilis modulates immune responses, suppresses inflammation, and restores mucosal homeostasis in experimental models of IBD11. The nonpathogenic Escherichia coli strain Nissle 1917 (EcN) promotes the production of antimicrobial peptides, enhances the function of the intestinal epithelial barrier, and reduces inflammatory responses in patients with IBD12. Furthermore, Clostridium butyricum produces short-chain fatty acids, particularly butyric acid, which exert potent anti-inflammatory effects, suppress inflammation, and facilitate mucosal repair in colitis13.
As previously noted, multiple studies have demonstrated that various probiotics exert anti-inflammatory effects via distinct mechanisms. These probiotics have shown anti-inflammatory and mucosal regenerative activities against IBD, suggesting their potential as key therapeutic targets. Considering the limitations of existing treatment modalities, the identification and validation of additional yet-to-be-characterized probiotics are warranted, as newly discovered strains may display effects comparable to or exceeding those previously reported. In this study, we evaluated the therapeutic potential of B. longum and Limosilactobacillus species as candidate probiotics for IBD management.
The primary objective of this study was to determine whether novel strains (CBA7104, CBA7106, CBA7108, and BBH) obtained from healthy human fecal samples exerted anticolitic effects in a DSS-induced colitis mouse model. In addition, we aimed to characterize any shifts in the gut microbial community and identify the metabolites produced by these strains that exhibit therapeutic potential in ameliorating colitis.
Materials and methods
Isolation, culture, and whole-genome sequencing of potential probiotic bacterial strains
Bacterial isolation and culture
The candidate probiotic strains used in this study were isolated from healthy Korean human fecal samples as part of a microbiota project aimed at identifying beneficial strains. The three Lactobacillus strains (CBA7104, CBA7106, CBA7108) and the Bifidobacterium longum (BBH) strain were chosen based on their consistent presence in healthy subjects and their superior anti-inflammatory properties demonstrated through preliminary in vitro screening, including antimicrobial activity, maintenance of epithelial integrity, and anti-inflammatory potential. Four bacterial strains CBA7104: Lacticaseibacillus paracasei (isolated from healthy Korean fecal samples), CBA7106: Limosilactobacillus fermentum (isolated from healthy Korean fecal samples), CBA7108: Lactobacillus gasseri (isolated from healthy Korean fecal samples), BBH: Bifidobacterium longum (isolated from healthy Korean fecal samples), including Lactobacillus rhamnosus (positive control) were isolated from the feces of healthy individuals based on their performance in antimicrobial and cell culture assays. The strains were revived from − 80 °C glycerol stocks onto De Man-Rogosa-Sharpe (MRS) agar plates, a selective medium for lactic acid bacteria, and incubated at 37 °C for 48 h. Single colonies were then cultured in MRS broth at 37 °C for 24 h.
Genomic DNA extraction
The culture was centrifuged at 12,000 × g for 3 min to remove the supernatant, and the pellet was washed three times with 1 mL of NaCl-EDTA. The pellets from two tubes were combined into one. The cell pellet was resuspended in 100 µL of NaCl-EDTA and 100 µL of lysozyme (10 mg/mL), and 1 µL of RNase (20 mg/mL) was added. The mixture was incubated at 37 °C with periodic shaking for 1 h. Next, 229 µL of NaCl-EDTA, 50 µL of 10% SDS, and 20 µL of Proteinase K were added, mixed thoroughly, and incubated at 55 °C for 1 h. Subsequently, 200 µL of cold protein precipitation solution was added, vortexed, and the mixture was placed on ice for 5 min before centrifugation at 12,000 × g, 4 °C, for 3 min. The supernatant was transferred to a clean 1.5 mL tube, centrifuged again (12,000 × g, 4 °C, 3 min), and the supernatant was moved to another clean 1.5 mL tube. DNA was precipitated with 600 µL of cold isopropanol and centrifuged at 12,000 × g, 4 °C, for 3 min to remove the supernatant. The pellet was washed with 600 µL of fresh 70% ethanol, and the supernatant was discarded. The tube was left to air-dry. Finally, the pellet was dissolved in 50 µL of H2O, incubated at 55 °C for 5 min, and stored at − 20 °C. The DNA samples were quantified using Qubit, yielding concentrations of 25–50 ng/µL, with a minimum volume of 20 µL, and used for whole-genome sequencing (WGS).
Library preparation and whole-genome sequencing
Genomic DNA was processed on a next-generation sequencing platform following protocols and kits from Pacific Biosciences library preparation. The libraries were multiplexed on 1 × SMRTcell 8 M using a Sequel IIe. After sequencing HiFi long reads, the demultiplexing was performed with lima, and the HiFi reads were quality-controlled with FastQC. Assembly was conducted using version 130.0.0207600 of the SMRTlink pb_microbial_analysis pipeline. Where required, the polishing, circularization, and origin rotation of the assembly were also carried out using the SMRTlink pipeline.
Genome comparison and annotation
The assemblies were compared using Pyani (version 0.2.13.1), revealing that three strains belonged to the same species with an average nucleotide identity (ANI) > 95%. A phylogenetic tree was constructed using the neighbor-joining (NJ) algorithm with default parameters in MEGA 11.0 software, based on the matrix of percent identity. Close relatives were identified by Basic Local Alignment Search Tool (BLAST) searches in the National Center for Biotechnology Information (NCBI) GenBank public database. Finally, the genomes were annotated using Prokka (v. 1.13)14.
Analysis of candidate microbiome-derived metabolites in vitro
Bacterial cultures were initiated by inoculating 100 μL of stock bacterial culture into 5 mL of Lactobacilli MRS broth (Difco™ Laboratories, Detroit, MI, USA) and incubating at 37 °C with agitation at 1000 rpm for 1 h; Subsequently, 500 μL of the resulting pre-culture was transferred to 20 mL of MRS medium and incubated for 24 h under the same conditions. For each strain (e.g., BBH, CBA7104, CBA7106, and CBA7108), eight 20 mL cultures were prepared (totaling 160 mL per strain). To perform liquid–liquid extraction, 100 μL of each culture was mixed with extraction solvent (MeOH:IPA:DW = 3:3:2), vortexed for 1 min, sonicated for 10 min, then centrifuged at 13,200 rpm for 10 min at 4 °C, after which the upper layer was transferred to a new tube and concentrated using a speed vacuum. For freeze-drying, cultures were centrifuged at 4000 rpm for 10 min at 4 °C (#1736R, LABOGENE, Lynge, Denmark), and the supernatant was discarded. The bacterial pellet was then lyophilized in an OPERON freeze-dryer (#FDCF-12012, Gimpo, Korea) at − 90 °C under vacuum for 24 h. The freeze-dried pellet was weighed using an OPTIKA analytical scale (#B214Ai, Ponteranica, Italy), and 0.01 g of the powder was resuspended in 1 mL of distilled water, after which the optical density (A600) was measured with a NanoDrop One (Thermo Fischer Scientific, Waltham, MA, USA) to estimate cell concentration. For the LC-Orbitrap MS (#90,207,135, ThermoFisher Scientific, Waltham, MA, USA)–based untargeted metabolomic profiling, water containing 0.1% formic acid (Solvent A) and acetonitrile containing 0.1% formic acid (Solvent B) were used. Chromatographic separation was carried out on a Waters BEH C18 column (flow rate: 0.3 mL/min) with a total runtime of 15 min, and mass spectral data were acquired in both positive and negative ionization modes. Metabolite identification was performed using Compound Discoverer 3.2 software; to ensure data quality, duplicate compounds within each ionization mode were removed, and only those compounds commonly detected (i.e., overlapping) in both modes were designated as final candidate metabolites (A total of 73 samples, comprising all culture supernatants, controls, and replicates, were analyzed in this manner).
Verification of the colitis-improving effect of a novel probiotic strain in a dextran sulfate sodium-induced colitis mouse model
Animals
Eight-week-old male C57BL/6 mice (n = 42) were purchased from Koatech (Pyeongtaek, Korea). The mice were maintained under standard laboratory conditions (25 ± 2 °C, 50 ± 10% humidity, and a 12 h/12 h light/dark cycle). The mice were acclimatized to laboratory conditions for a week. After a week of acclimatization to the environment, forty mice equally divided into five groups. The experimental process was shown in Fig. 1. In brief, colitis was induced in mice by administering drinking water containing 2% DSS (#160,110, MP Biomedicals Ltd., Santa Ana, CA, USA) for five consecutive days. A one-week washout period was then implemented to allow partial recovery and prevent mortality associated with prolonged DSS exposure. Following this recovery period, candidate probiotic strains were administered orally to each mouse. In the Sham (n = 6) group, no interventions were administered from the start to the end of the experiment, and only PBS was provided orally. In the DSS-CON (n = 6) group, following colitis induction, PBS was likewise administered solely by the oral route. After the completion of the washout period, which lasted for 7 days following DSS administration, the probiotic-treated groups (DSS-LGG, DSS-BBH, DSS-CBA7104, DSS-CBA7106, DSS-CBA7108; n = 6 for each) received 10⁹ CFU/day of their respective strains, mixed into fresh drinking water for oral administration. This approach was designed to minimize the risk of toxic reactions during the acute inflammatory phase induced by DSS. Probiotic administration was conducted once daily from day 12 to day 17, and sacrifice was performed on day 17. After completion of the intervention experiments, the mice were euthanized under isoflurane inhalation anesthesia. Following blood collection, the abdominal cavity was opened, and the entire intestinal tract (from the stomach to the anus) was carefully excised without damage. All methods were performed in accordance with the relevant guidelines and regulations. Also, the study is reported in accordance with ARRIVE guidelines. The total length of each excised intestine was measured, and the samples were subsequently fixed in 10% formalin for future histological analysis.
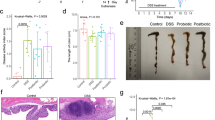
Schematic representation of the experimental design for DSS-induced colitis and probiotic administration in mice. Acute colitis was induced in mice by administering dextran sulfate sodium (DSS) in drinking water for 5 days (days 0–5), followed by a 7-day washout and recovery period (days 5–12). Candidate probiotic strains were orally administered during the recovery phase (days 12–17), and all animals were sacrificed on day 17. The experimental groups included: Sham (no DSS or probiotic), DSS control (DSS-CON), positive control (Lactobacillus rhamnosus GG; DSS-LGG), Bifidobacterium longum (DSS-BBH), Lacticaseibacillus paracasei (DSS-CBA7104), Limosilactobacillus fermentum (DSS-CBA7106), and Lactobacillus gasseri (DSS-CBA7108).
Fecal sample collection, evaluation of the disease activity index
Throughout the experimental period, daily body weight measurements were taken for each mouse, fecal samples were collected and inspected, and the anal region was examined. All measurements and sample collections were conducted after transferring each mouse to a 100 mL clean plastic beaker, and once fecal collection was complete, the anal region was assessed. Before proceeding to the next mouse, the beaker was thoroughly disinfected with 70% ethanol and allowed to dry. The disease activity index (DAI) was assessed by scoring the extent of daily body weight loss (0: none; 1: 1–5%; 2: 6–10%; 3: 11–15%; 4: > 15%), stool consistency (0: normal; 2: loose stool; 4: diarrhea), and the presence of occult or gross blood (0: negative; 2: fecal occult blood test positive; 4: gross bleeding).
Histological analysis
For histological evaluation, the mid-region of the colon was excised, fixed in 10% neutral-buffered formalin, and embedded in paraffin. Paraffin-embedded samples were sectioned at 5 μm thickness using a microtome, and sections were mounted on glass slides. The sections were deparaffinized and rehydrated in a graded ethanol series, then stained with hematoxylin for 30 s, rinsed twice in distilled water, rinsed briefly in 95% ethanol, and counterstained with eosin for 15 s. Following dehydration in ascending concentrations of ethanol (95% and 100%) and clearance in xylene, sections were mounted with Permount™ Mounting Medium (#SP15-100, Thermo Fischer Scientific, USA) and cover slipped. Images of H&E-stained sections were acquired at 100 × magnification using the EVOS M7000 Imaging System (Invitrogen, Carlsbad, CA, USA). Histological scoring was performed independently by two blinded investigators, and their scores were averaged. Three parameters were assessed: (i) Severity of inflammation (0–3), (ii) Depth of injury (0–3), and (iii) crypt damage (0–4). Each parameter’s score was multiplied by a factor corresponding to the percentage of tissue involvement (× 1 for < 25%, × 2 for 26–50%, × 3 for 51–75%, and × 4 for > 75%). The weighted values were summed to generate a total histological score15.
Microbial genomic DNA extraction from mouse fecal microbiota
All experimental groups, including the CBA7104, CBA7108 group, underwent the same DSS induction protocol. However, since the DAI score of the CBA7108 group remained relatively low, a separate gut microbiota analysis for this group was not conducted. Instead, microbiota analysis was focused on fecal samples from mice treated with strains expected to exhibit similar efficacy to the positive control. Fecal samples were collected from mice on day 0, day 5, and day 17, grouped accordingly. Fresh fecal samples from each group were collected, snap-frozen in liquid nitrogen, and stored at—20 °C. Total microbial metagenomic DNA from each sample was extracted using the QIAamp DNA Microbiome Kit (#51,704, Qiagen, Hilden, Germany), and the experiments were conducted following the protocol provided with the kit. The quality of the extracted genomic DNA was assessed using a bioanalyzer (Agilent 2100, Agilent Technologies, Inc., Santa Clara, CA, USA) and stored at—20 °C until further analysis. The Illumina platform targeted the V3—V4 hypervariable regions of the bacterial 16S rRNA gene. PCR amplification of the target region began immediately after mDNA extraction. The 16S V3—V4 amplicons were amplified using KAPA HiFi HotStart ReadyMix (#09,420,398,001, Roche, Penzberg, Germany). For this purpose, a pair of universal primers targeting the V3—V4 regions recommended by Illumina were used. The primer sequences were as follows: 16S 341F forward primer: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′; 16S 806R reverse primer: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′. After PCR amplification, all PCR products were purified using AMPure XP beads (#A63881, Beckman Coulter, California, USA). Additional PCR amplification was then performed to add multiplexing indices and Illumina sequencing adapters using the Nextera XT Index Kit (#FC-131–2004, Illumina, San Diego, CA, USA). The final PCR products were purified again using AMPure XP beads. After constructing the amplicon library, 16S metagenomic sequencing was carried out using the paired-end 2 × 300 bp Illumina MiSeq protocol (Illumina MiSeq, San Diego, CA, USA).
Bioinformatic analysis
The gut metagenomics data obtained through Illumina MiSeq was analyzed using Quantitative Insights into Microbial Ecology version 2 (QIIME 2)16 pipeline plugin (2022.08). Before implementing DADA2 (Divisive Amplicon Denoising Algorithm 2)17 in QIIME2, the Figaro was used to get the optimal options by based on sequence quality. Amplicon sequence variants (ASVs) were obtained by Denoising step in DADA2 algorithm. The ASVs sequences were subjected to bacterial classification using a Naïve Bayes classifier based on V3-V4 hypervariable reads extracted from the SILVA 138v 99% rRNA database to improve accuracy and then several features annotated with Archaea, Eukaryotes, Mitochondria, or Chloroplasts were removed. Focused bacterial features were subjected to phylogeny tree construction (align-to-tree-mafft-fasttree plugin) and taxonomy composition. This features were rarefied, and used to analysis of α-diversity (Observed features, Chao1 index, Shannon’s index, Simpson’s index, Pielou’s evenness) and β-diversity (Bray–Curtis, unweighted UniFrac).
KEGG orthologs and pathways prediction
Functional attributes of the identified microbial communities were predicted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) pipeline, version 2.5.218. With the previously obtained ASVs dataset as an input, PICRUSt2 was used to infer the metagenomic functional content. Predicted functional genes were categorized using the Kyoto Encyclopedia of Genes and Genome (KEGG)19,20,21 ontology and pathway analysis. KEGG is a database designed to facilitate the understanding of the basic biological system from genomic, chemical, and molecular data.
Statistical analysis
All statistical analyses related to animal experiments were performed using GraphPad Prism 9 software (GraphPad Software Inc., San Diego, CA, USA). One-way ANOVA and Tukey post hoc test were performed to determine differences between groups. The statistical significance level was set at p < 0.05.
Results and discussion
Genomic characteristics of candidate probiotic strains
We performed WGS to analyze the potential properties of the strains, CBA7104, CBA7106, CBA7108, and BBH, which were isolated from the feces of healthy participants of the intestinal microbiota dataset of 745 healthy Korean individuals from the Korea Food Research Institute (accession no. PRJEB33905). CBA7104 was identified as Lacticaseibacillus paracasei, CBA7106 as Limosilactobacillus fermentum, CBA7108 as Lactobacillus gasseri, and BBH as a strain of B. longum (Table 1, Supplementary Fig. S1). The size of each bacterial chromosome was similar to the previously reported ranges for CBA7104 (L. paracasei, 3 Mbp), CBA7106 (L. fermentum, 2 Mbp), CBA7108 (L. gasseri, 1.99 Mbp), and BBH (B. longum, 2.5 Mbp). Additionally, the CBA7104, CBA7106, CBA7108, and BBH isolates contained 2918, 1972, 1871, and 2482 coding sequences with GC content of 46.2%, 51%, 35%, and 43%, respectively. The numbers of tRNA (CBA7104: 60, CBA7106: 58, CBA7108: 46, and BBH: 47), rRNA (CBA7104: 15, CBA7106: 15, CBA7108:12, and BBH: 12), and tmRNA (CBA7104: 1, CBA7106: 1, CBA7108: 1, and BBH: 1) genes were also determined for each isolate (Table 1, Supplementary Fig. S1). CBA7104 was assigned to L. paracasei, which is known as L. paracasei strain_L1 from the complete whole genome 85 of the RefSeq of L. paracasei. In addition, CBA7106 was assigned to two L. fermentum strains, EFEL6800 and GR1008, from whole genome 21 of the RefSeq of L. fermentum strains. This was assigned to L. fermentum. The strain was assigned L. gasseri from whole genome 63 of RefSeq the of L. gasseri strains, known as L. gasseri strain_Lg1266. Finally, BBH strains were assigned to well-documented B. longum strains, JCL127 from whole genome 91 of the RefSeq of B, longum isolates (Supplementary Table S1–S5 and Supplementary Fig. S2)16.
The candidate strains alleviate the overall symptoms of colitis in mice
To evaluate the effects of the novel probiotic strains on gut health, a comparative analysis was conducted involving the sham, DSS-CON, DSS-LGG, DSS-BBH, CBA7104, CBA7106, and CBA7108 groups (Fig. 1). The probiotic-treated groups received candidate probiotic strains after the washout period, which lasted for 7 days following the DSS administration phase. This approach was designed to reduce the risk of toxic reactions during the acute inflammatory phase. The probiotics were administered once daily from day 12 to day 17, after which the animals were sacrificed. All groups exhibited a gradual decrease in the DAI scores from day 9 (Fig. 2A, B). Notably, the CBA7108 (75%) and DSS-BBH (56%) groups showed a significant reduction compared with the DSS-CBA7104 group (35%) and DSS-CON (33%) groups. Upon macroscopic inspection, the probiotic-treated groups exhibited colon lengths comparable to or longer than those observed in the DSS-CON group (Fig. 2C). Statistical analysis revealed that the DSS-CON group had shorter colon length compared with that of the sham group (P < 0.01). In contrast, the DSS-LGG, DSS-BBH, and DSS-CBA7106 groups had significantly longer colons compared with that of the DSS-CON group (p < 0.05) (Fig. 2D).
Differences in disease activity index scores and colon length in DSS-induced colitis mice following probiotic administration. (A) Disease activity index score. (B) Colon length measurements at the endpoint of the experiment. (C) Body weight changes in DSS-induced colitis mice treated with candidate probiotic strains. Statistical comparisons were made using two-way ANOVA followed by Tukey’s multiple comparisons test. p < 0.05, p < 0.01, p < 0.001, p < 0.0001; ns = not significant. (D) Statistical comparison of colon length. All data presented as the mean (M) ± standard deviation (S.D.). ‡p < .01, vs. Sham; *p < .05, vs. DSS-CON.
Histological scores were evaluated based on the observation of H&E-stained colon tissue. Compared with the sham group, the DSS-CON group exhibited markedly more severe histological damage (P < 0.001). However, among the probiotic-treated groups, DSS-BBH (P < 0.0001), DSS-CBA7104 (P < 0.001), and DSS-CBA7106 (P < 0.0001) showed a significantly lower degree of damage compared with that of DSS-LGG. Although all DSS-treated groups displayed significantly greater damage than the sham group, significant markers are omitted from the figure for clarity. These findings suggest that supplementation with probiotics during the recovery period following DSS treatment and washout may accelerate the restoration of intestinal injury. Notably, DSS-BBH, DSS-CBA7104, and DSS-CBA7106 demonstrated high therapeutic efficacy (Fig. 3A, B). These findings suggest that probiotics can effectively alleviate inflammation and protect the intestinal tissues in DSS-induced colitis models. The pronounced effects observed in the BBH group, which contained B. longum and Limosilactobacillus strains, were particularly noteworthy. This aligns with the existing research. A study reported that Bifidobacterium bifidum BGN4 paraprobiotics reduced inflammatory responses and improved the gut microbial balance in DSS-induced colitis mouse models21. Additionally, research using lactic acid bacteria isolated from kimchi has demonstrated improvements in DSS-induced colitis models, including the inhibition of weight loss, reduction in DAI scores, and restoration of colon length22. These studies indicate that probiotics can modulate the gut microbiota balance and suppress inflammatory responses, thereby alleviating colitis symptoms. B. longum and Limosilactobacillus strains are recognized as beneficial microbes in the gut environment, with potential preventive and therapeutic effects against IBDs.
Histological evaluation of colonic damage and corresponding histology scores in DSS-induced colitis mice. (A) Representative hematoxylin and eosin (H&E) stained colon sections from each group: Sham, DSS-CON, DSS-LGG, DSS-CBA7104, DSS-CBA7106, DSS-CBA7108, and DSS-BBH. DSS treatment caused substantial epithelial disruption, crypt loss, and inflammatory cell infiltration, which were partially restored in probiotic-treated groups. Scale bars = 100 μm. (B) Quantitative histology scores based on epithelial integrity, crypt architecture, and inflammatory cell infiltration. Data are presented as mean ± SEM (n = 6 per group). Statistical significance was assessed using one-way ANOVA followed by Tukey’s post hoc test. p < 0.05, p < 0.01, p < 0.001.*p < .0001, vs. Sham; ***p < .001, ****p < .0001, vs. DSS-CON.
Metabolomic characterization of candidate strains
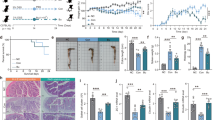
To identify the basic metabolic features of the isolated strains, metabolomic analysis was performed using in vitro cultivation of each strain. The strains were cultured in MRS medium and a comparative analysis was conducted based on the metabolites secreted, excluding the basic components of the MRS medium. The metabolite patterns of each probiotic strain were confirmed. Compared with the MRS medium, increases in dipeptides, amino acids, and pyridines were commonly observed in the CBA7104, CBA7106, BBH, and LGG groups, whereas increases in indoles were characteristically observed in CBA7104, CBA7106, and BBH (Supplementary Fig. S3, Supplementary Table S9–S11). In the comparative analysis with MRS medium, a total of 47 metabolites were commonly identified across all three strains. Notably, strain 7106 exhibited 47 unique metabolites. Furthermore, 13 metabolites were shared between strains 7106 and BBH, 9 metabolites were common between strains 7106 and LGG, and 6 metabolites were identified as common between strains BBH and LGG (Fig. 4A). The hierarchical clustering analysis revealed that the distance between the LGG and BBH samples was the shortest, confirming the similarity between the LGG and BBH groups (Fig. 4B). For the CBA7106 samples, while the distance to the LGG group was smaller than that of the MRS (negative control) and CBA7108 groups, distinctive compounds showed characteristic differences. In both the CBA7106 and BBH groups, 47 compounds were significantly increased or decreased compared to those in the MRS medium (negative control), with 44 compounds exhibiting an increasing pattern and three compounds exhibiting a decreasing pattern. Notably, the CBA7106 group showed a significantly higher quantity of metabolites than the BBH and LGG groups (Fig. 4B, Supplementary Table S8). Among the 47 compounds that significantly increased or decreased in the CBA7106, BBH, and LGG groups compared with the MRS medium (negative control), three compounds 6-hydroxycaproic acid (14.94 and 1.98), indole-3-lactic acid (5.86 and 1.43), and choline (1.0 and 2.05), showed a common pattern of significant increase in both the CBA7106 and BBH groups compared with those in the LGG group (Table 2). Notably, 6-hydroxycaproic acid acts as an intermediate in fatty acid metabolism, and contributes to anti-inflammatory and tissue recovery processes. In DSS models, 6-hydroxycaproic acid has been reported to suppress the secretion of pro-inflammatory cytokines, such as TNF-α and IL-6, while increasing the production of the anti-inflammatory cytokine, IL-10.
Additionally, indole-3-lactic acid, a tryptophan-derived metabolite produced by Lactobacillus species, plays a crucial role in maintaining the gut microbial balance and immune regulation. Specifically, indole-3-lactic acid activates the aryl hydrocarbon receptor, which helps to maintain immune homeostasis in the intestinal mucosa and suppresses the inflammatory responses23. Finally, choline has been reported to inhibit the NF-κB signaling pathway, thereby reducing the secretion of pro-inflammatory cytokines24. Therefore, the probiotic strains CBA7106, BBH, and LGG produced metabolites with anti-inflammatory properties. This study focused on their impact on gut health through changes in the gut microbiota.
Evaluation of probiotic effects on gut health: microbiome analysis
We compared changes in the gut microbiota based on fecal samples collected from different groups and treatments with candidate strains in a colitis-induced mouse model. To assess the effects of DSS treatment and probiotic administration on the gut microbiome, microbiome analysis was performed using fecal samples collected from all experimental groups at designated time points. In the DSS-CON group, microbiome analysis was conducted using fecal samples obtained at three time points: day 0 (before DSS treatment), day 5 (during treatment), and day 17 (post-treatment), with three biological replicates collected at each time point. Similarly, in the Sham group, fecal samples were collected at day 0, day 5, and day 17, with three replicates per time point. For the probiotic-treated groups (DSS-CBA7106, DSS-LGG, DSS-BBH), microbiome analysis was performed using fecal samples collected on day 17 after probiotic administration, with three replicates per group (Supplementary Table S6). The analysis primarily focused on comparing α diversity and β diversity among the groups. Alpha diversity, a metric that measures microbial species diversity within a single sample, showed a decreasing trend following DSS treatment. However, this reduction was not significant (Fig. 5A). Beta diversity, which measures differences in microbial composition between samples, was evaluated using the Bray–Curtis statistical method. The results revealed significant differences between the groups. Notably, the DSS-treated group (day 5) exhibited substantial changes in the gut microbial community structure compared with that of the sham group. Additionally, groups treated with control and candidate probiotic strains (LGG, BBH, and CBA7106) demonstrated statistically significant differences in gut microbial diversity (Fig. 5B, Table 3). As shown in Fig. 6A the microbial composition of each group revealed the characteristics of microbes influenced by inflammation. At the family level, the abundance of Enterococcaceae increased in all the groups after DSS treatment. Although a decreasing trend was observed in the DSS-CON group on day 17, the DSS-LGG, BBH, and DSS-CBA7106 groups showed a continuous increase. At the genus level, Enterococcus also showed a significant increase in the DSS-treated groups (days 0 and 5), as well as in the DSS-LGG, BBH, and DSS-CBA7106-treated groups.
Gut microbial diversity in a DSS-induced colitis mouse model following administration of a candidate probiotic strains. (A) Alpha diversity; black bar = Sham, orange bar = DSS-CON, brown bar = 5 days after DSS-CON; red bar = 17 days after DSS-CON, blue bar = DSS-LGG, light blue bar = DSS-BBH, and green = DSS-CBA7106. From left, observed features, Shannon index, and Pielou’s evenness index are shown respectively. (B) Beta diversity. (C) Bar plot showing significant differences for each group based on the 5th day after DSS treatment.
The genus-level relative abundances and heatmap of LefSe analysis results comparing probiotic-treated groups following administration of novel probiotic strains in a DSS-induced colitis mouse model. The Fig. 6A shows the relative abundances of microbial genera according to the DSS administration schedule, while Fig. 6B presents a heatmap of LefSe analysis results comparing the groups. The red and blue colors represent the comparison groups, respectively.
These results align with previous findings that Enterococcus increases in IBD and colitis models25. DSS treatment damages the intestinal epithelial barrier, allowing pathogenic bacteria to gain access to the mucosa. Enterococcus thrives in this environment, adheres to the mucosa, and exacerbates the inflammatory responses26. Furthermore, nutrient availability in the gut changes under inflammatory conditions. Enterococcus efficiently exploits these resources, outcompeting other microbes and negatively impacting the growth of beneficial bacteria27.
The LEfSe analysis revealed distinct changes in gut microbiota composition between the Sham and DSS control groups at different time points (0, 5, and 17 days). Compared to the DSS-CON.0 day group, the Sham group showed significantly higher levels of Roseburia and Candidatus Arthromitus. As the inflammatory condition progressed to DSS-CON.5 day, the Lachnospiraceae NK4A136 group became more prevalent. By DSS-CON.17 day, Turicibacter and members of the Erysipelotrichaceae family were significantly enriched, indicating a shift towards these taxa as inflammation persisted. Within the DSS group itself, time series comparisons showed that Clostridia and Lachnospiraceae NK4A136 group increased notably by day 5, while Turicibacter and Helicobacter became dominant by day 17. These findings demonstrate that DSS treatment leads to distinct microbial shifts as the inflammation progresses over time. Further analysis focusing on the probiotic-treated groups revealed similar dynamic changes. At the initial time point (DSS-CON.0 day), the analysis showed that the 7106 group exhibited a significantly higher abundance of Peptostreptococcaceae and Romboutsia, while the BBH group demonstrated an increased presence of Enterococcaceae, Akkermansia muciniphila, and Verrucomicrobiota. In contrast, the DSS-CON.0 day group itself was dominated by the Lachnospiraceae family compared to the LGG group (Fig. 6B, Supplementary Figs. S3, S4 Supplementary Tables S7, S8).
Next, we used PICRUSt2 to analyze and predict the functional characteristics of the microbiota in the groups treated with candidate probiotic strains. Notably, among the various KEGG Orthology pathways, a consistent decrease was observed in the ko01053 (Fig. 7A, B) pathway, which is associated with the biosynthesis of siderophore group non-ribosomal peptides. Notably, our results indicated that Lactobacillus and Akkermansia genera showed a significant increase in the DSS-LGG (P = 0.024) and DSS-BBH (P = 0.033) groups compared with those in the other groups, except for the DSS-CBA7106 group (P = 0.042). Lactobacillus is generally considered a beneficial bacterium of the gut microbiota28. They can thrive in inflammatory environments and gain a competitive advantage in conditions where harmful bacteria are reduced, such as after DSS treatment29. The administration of probiotics likely facilitated the proliferation of Lactobacillus. Moreover, lactic acid, a metabolic byproduct of Lactobacillus, contributes to mucosal protection and pH regulation in the gut, further promoting its growth30. L. plantarum reduces intestinal inflammation and promotes mucosal recovery in DSS models31. Similarly, certain Lactobacillus strains, including L. rhamnosus GG (often employed as a positive control), benefits DSS-induced colitis models by enhancing beneficial Lactobacillus populations and alleviating inflammatory responses, thereby contributing to the restoration of microbial balance. In particular, clinical data suggest that Lactobacillus may help improve UC symptoms, although additional large-scale studies are necessary to confirm its efficacy in Crohn’s disease32. Akkermansia muciniphila is known for its mucin-degrading capability, facilitating mucosal regeneration, and strengthening the intestinal barrier33,34. Although there are relatively few studies that directly demonstrate an expansion of A. muciniphila precisely during the inflammation recovery phase in DSS-induced colitis models, as the damaged mucosa is repaired, A. muciniphila can proliferate and accelerate the restoration of the mucus layer35,36.
Furthermore, recent studies have suggested that Bifidobacterium and Lactobacillus strains promote the growth of Akkermansia, contributing to mitigation of inflammation37. This suggests that probiotic treatments alter microbial function, specifically by reducing siderophore biosynthesis. IBD and colitis models have shown that siderophores produced by pathogenic bacteria are key mechanisms that exacerbate gut inflammation38. Siderophores are high-affinity iron-chelating molecules that are produced by microorganisms that acquire iron (Fe) in limited environments. Pathogenic bacteria use siderophores to efficiently acquire iron from their host environment39. In IBD and colitis models, siderophore production by pathogenic bacteria plays a major role in exacerbating gut inflammation40. Our results suggest that DSS-induced changes in the gut environment (e.g., changes in oxygen levels and pH) may affect siderophore production.
Genome analysis revealed key genetic features of CBA7106 and CBA7104 strains, including the absence of virulence genes and transmissible antibiotic resistance, ensuring their safety. Functional annotations indicated that these strains were equipped with genes related to carbohydrate and amino acid metabolism, suggesting their adaptability and ability to positively influence the gut microbial function. Evaluation of the genetic and functional stability of the probiotic candidates identified in our study is of paramount importance. Furthermore, a previously published special focus review emphasized that genetic and functional stability assessments similar to ours are essential to minimize potential adverse effects41.
The reduction in DAI scores and restoration of colon length in the CBA7106 and BBH groups underscore their anti-inflammatory potential. Tissue analysis further confirmed the improvements in mucosal integrity and reduced inflammation. Similar outcomes have been reported for B. bifidum and Lactobacillus plantarum in DSS models, where probiotics modulated inflammatory responses and enhanced epithelial barrier function17,42. These effects are attributed to the ability of probiotics to regulate cytokine secretion. Metabolites, such as 6-hydroxycaproic acid and indole-3-lactic acid, may promote an anti-inflammatory state.
The increase in beneficial microbes, such as Lactobacillus and A. muciniphila, in the probiotic-treated groups highlights their role in restoring microbial balance. Lactobacillus contributes to mucosal protection through lactic acid production, which regulates gut pH and supports epithelial repair43. A. muciniphila is recognized for its role in strengthening the mucus layer and enhancing barrier integrity, particularly in inflammatory environments36,37. Furthermore, α and β diversity analyses indicated significant differences in microbial composition after probiotic administration, suggesting that these strains could mitigate DSS-induced dysbiosis. One of the novel findings of this study was the consistent decrease in the ko01053 pathway, which is associated with the siderophore group non-ribosomal peptide biosynthesis. Siderophores are critical for pathogenic bacteria to sequester iron in the host environment, thereby exacerbating inflammation in conditions, such as IBD44. Therefore, our newly identified probiotic strains may suppress siderophore biosynthesis, thereby indirectly inhibiting the growth of pathogenic bacteria and alleviating inflammation.
The murine DSS colitis model, which was effective in preliminary studies, cannot fully replicate human IBD. Further clinical trials are required to validate these findings in humans. Although metabolites, such as 6-hydroxycaproic acid, indole-3-lactic acid, and choline, have been implicated in anti-inflammatory effects, their specific mechanisms require further elucidation. This study primarily focused on short-term outcomes. Long-term studies are required to assess the sustainability and safety of probiotic interventions, explore the synergistic effects of these probiotics with existing IBD therapies, investigate the impact of these probiotics on iron metabolism and siderophore activity in human models, and evaluate personalized probiotic formulations based on individual gut microbiota profiles.
The findings of this study highlight the potential of probiotics, particularly CBA7106 and BBH, as adjunct therapies for IBD. Their ability to modulate the gut microbiota, produce anti-inflammatory metabolites, and inhibit siderophore biosynthesis presents a multifaceted approach to improving gut health. By integrating genetic, microbial, and metabolic analyses, this study provided a comprehensive framework for understanding the influence of probiotics on gut health. These insights will pave the way for the development of targeted probiotic therapies tailored to specific inflammatory and dysbiotic conditions. Our experimental design primarily focused on evaluating probiotic effects during the recovery phase after DSS-induced acute colitis. While the washout period allowed partial recovery, we acknowledge that the lack of fecal sample collection during this phase limits our understanding of longitudinal microbiota changes, including potential autoimmune modulation. Future studies will address this limitation by incorporating fecal sampling during the washout period. This study has the limitation of not directly verifying the effects of the metabolites of the candidate strains on the improvement of intestinal inflammation. However, it was observed that the metabolites of the new candidate strains included substances associated with inflammation mitigation, and additional studies are needed to elucidate the relationship between these findings and genomically predicted results in clinical settings. These findings provide a solid foundation for the use of probiotics in the treatment of IBD.
Data availability
The probiotics novel strain WGS sequences data that supported the findings of this study are publicly available at the NIH National Center for Biotechnology Information Sequence Read Archive (SRA) with BioProject ID PRJNA390215, PRJNA1221993, PRJNA1223017.
References
Rogler, G., Singh, A., Kavanaugh, A. & Rubin, D. T. Extraintestinal manifestations of inflammatory bowel disease: Current concepts, treatment, and implications for disease management. Gastroenterology 161, 1118–1132 (2021).
Larsen, S., Bendtzen, K. & Nielsen, O. H. Extraintestinal manifestations of inflammatory bowel disease: Epidemiology, diagnosis, and management. Ann. Med. 42, 97–114 (2010).
Neurath, M. F. Current and emerging therapeutic targets for IBD. Nat. Rev. Gastroenterol. Hepatol. 14, 269–278 (2017).
Ananthakrishnan, A. N. et al. Environmental triggers in IBD: A review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 15, 39–49 (2018).
Ni, J., Wu, G. D., Albenberg, L. & Tomov, V. T. Gut microbiota and IBD: Causation or correlation?. Nat. Rev. Gastroenterol. Hepatol. 14, 573–584 (2017).
Gao, J. et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 8, 13 (2018).
Cui, Q. et al. Bifidobacterium bifidum ameliorates DSS-induced colitis in mice by regulating AHR/NRF2/NLRP3 inflammasome pathways through indole-3-lactic acid production. J. Agric. Food Chem. 71, 1970–1981 (2023).
Cassat, J. E. & Skaar, E. P. Iron in infection and immunity. Cell Host Microbe 13, 509–519 (2013).
Sood, A. et al. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin. Gastroenterol. Hepatol. 7, 1202–1209 (2009).
Guslandi, M., Mezzi, G., Sorghi, M. & Testoni, P. A. Saccharomyces boulardii in maintenance treatment of Crohn’s disease. Dig. Dis. Sci. 45, 1462–1464 (2000).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Kruis, W. et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53, 1617–1623 (2004).
Stoeva, M. K. et al. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 13, 1–28 (2021).
Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Jung, Y. K., Lee, S., Yoo, J. I. & Baek, K. W. The protective effect of IL-12/23 neutralizing antibody in sarcopenia associated with dextran sulfate sodium-induced experimental colitis. J. Cachexia Sarcopenia Muscle 14, 1096–1106 (2023).
Bolyen, E. et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ 2018, 1–53 (2018).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Douglas, G. M. et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688 (2020).
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011).
Lee, S. Y. et al. Bifidobacterium bifidum BGN4 paraprobiotic supplementation alleviates experimental colitis by maintaining gut barrier and suppressing nuclear factor kappa B activation signaling molecules. J. Med. Food 25, 146–157 (2022).
Lee, S. Y. et al. Anti-inflammatory effect of lactic acid bacteria isolated from kimchi on acid-induced acute colitis in model mice. Toxicol. Environ. Health Sci. 9, 279–283 (2017).
Zelante, T. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013).
Parrish, W. R. et al. Modulation of TNF release by choline requires alpha7 subunit nicotinic acetylcholine receptor-mediated signaling. Mol. Med. 14, 567–574 (2008).
Mohamed, S. S. et al. Effect of the standard herbal preparation, STW5, treatment on dysbiosis induced by dextran sodium sulfate in experimental colitis. BMC Complement Med Ther. 21, 168 (2021).
He, W., Ni, W. & Zhao, J. Enterococcus faecium alleviates gut barrier injury in C57BL/6 mice with dextran sulfate sodium-induced ulcerative colitis. Gastroenterol. Res. Pract. 2021, 2683465 (2021).
Lebreton, F., Willems, R. J. L. & Gilmore, M. S., (2014) in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (ed. Gilmore, M. S., Clewell, D. B., Ike, Y. & Shankar, N.).
Hill, C. et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014).
Khan, I. et al. Lactobacillus plantarum strains attenuated DSS-induced colitis in mice by modulating the gut microbiota and immune response. Int. Microbiol. 25, 587–603 (2022).
Salminen, S. & Salminen, E. Lactulose, lactic acid bacteria, intestinal microecology and mucosal protection. Scand. J. Gastroenterol. Suppl. 222, 45–48 (1997).
Huang, R. et al. Lactobacillus and intestinal diseases: mechanisms of action and clinical applications. Microbiol. Res. 260, 127019 (2022).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA 110, 9066–9071 (2013).
Derrien, M., Vaughan, E. E., Plugge, C. M. & de Vos, W. M. Akkermansia muciniphila . nov, sp. Nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 (2004).
Reunanen, J. et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl. Environ. Microbiol. 81, 3655–3662 (2015).
Chelakkot, C. et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 50, e450 (2018).
Deriu, E. et al. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 (2013).
Miethke, M. & Marahiel, M. A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451 (2007).
Gerner, R. R. et al. Siderophore immunization restricted colonization of adherent-invasive Escherichia coli and ameliorated experimental colitis. mBio 13, e0218422 (2022).
Sanders, M. E. et al. Safety assessment of probiotics for human use. Gut Microbes 1, 164–185 (2010).
Ren, R. et al. Therapeutic effect of Lactobacillus plantarum JS19 on mice with dextran sulfate sodium induced acute and chronic ulcerative colitis. J. Sci. Food Agric. 103, 4143–4156 (2023).
Wu, H. et al. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes 11, 997–1014 (2020).
Khasheii, B., Mahmoodi, P. & Mohammadzadeh, A. Siderophores: Importance in bacterial pathogenesis and applications in medicine and industry. Microbiol. Res. 250, 126790 (2021).
Author information
Authors and Affiliations
Contributions
KA and KWB contributed to conception and design of the study. YO, YL and JC participated in the main experiments. KA, KY and YJA organized the data. KWB and KA performed the statistical analysis. KA and KWB wrote the first draft of the manuscript. KA and KWB wrote sections of the manuscript. YSK, EI, EJS, YSP, DHL, WL DYL and KH revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Ethical approval
All animal experiments were approved by the Institutional Animal Care and Use Committee of Gyeongsang National University (GNU-220522-M0053).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ahn, K., Baek, KW., Yun, K. et al. The effects of candidate probiotic strains on the gut environment in dextran sulfate sodium-induced colitis mouse. Sci Rep 15, 21103 (2025). https://doi.org/10.1038/s41598-025-03860-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03860-5