Abstract
To provide a novel direction for the diagnosis of gastric cancer (GC). The differentially expressed blood microRNAs (miRNAs) in gastric carcinoma were screened through SangerBox using the datasets GSE113486, GSE112264, and GSE113740. The miRNA-target genes prediction, conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses were performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.8. STRING analysis was further investigated with Cytoscape. The correlations among the expression levels of key miRNAs and prognosis/diagnostic value in GC patients were determined by survival prognosis and Receiver Operating Characteristic (ROC) curve analysis. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR) was employed to detect the different expression levels of key miRNAs in human blood samples. In the process, the influence of Helicobacter pylori (H. pylori) infection on expression levels of miRNAs was analyzed with Gene Expression Omnibus 2R (GEO2R) in dataset GSE108307. To evaluate these findings, the expression level of Cytotoxin-associated gene A (CagA), along with clinical markers used in the help of GC pathologic diagnosis, were measured and compared with the key miRNAs obtained in this study. The bioinformatics analysis identified five crucial blood miRNAs, including hsa-miR-124-3p, hsa-miR-125a-3p, hsa-miR-29b-3p, hsa-miR-4276, and hsa-miR-575. The detection in human blood samples combined with cross-analysis involving H. pylori infection, the expression levels of CagA and clinical markers underscored the significance and effectiveness of these specific miRNAs in early diagnosis and monitoring of gastric carcinoma. This study identified five potential blood miRNA biomarkers for GC through bioinformatics analysis coupled with detection in human blood samples, thus providing new possibilities for important biomarkers related to diagnosis and prognosis of GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a type of malignant tumour that originates from the epithelium of the gastric mucosa. Each year, over 1,000,000 new patients are diagnosed with GC every year, resulting in approximately 769,000 people deaths in 20211,2. GC has been imposing significant mental and financial burdens on individuals and society as a whole. Although there have been advancements in early diagnosis and treatment of GC, predictive biomarkers are necessary to establish precision medicine as the cornerstone for achieving efficient, reliable, and measurable outcomes in the management of GC.
Biomarkers are objectively measured and evaluated characteristics that serve as indicators of normal biologic process, pathogenic processes, or the pharmacological response to therapeutic interventions3. Current molecular markers associated with diagnosis, prognosis, and prediction of therapeutic responses in GC include tissue-based biomarkers such as metastasis-related genes [eg. human epidermal growth factor receptor 2 (Her-2) and E-cadherin], comprehensive analysis genes, epigenetic alterations, as well as genetic polymorphism4. Blood is a commonly used test material due to its easy to obtain, detectability, and minimal invasiveness. Additionally, considering the significant influence of the tumour microenvironment on carcinogenesis, invasion, and metastasis, blood components may also impact tumorigenesis. Therefore, circulating tumour cells, circulating cell-free DNA, non-coding RNAs (e.g. microRNAs, long noncoding RNAs and circular RNAs) and exosomes have been demonstrated to be specifically expressed in blood during gastric carcinogenesis5,6,7. The clinical utilization of blood antigens has facilitated the diagnosis and selection of chemical drugs, encompassing carcinoemacmbryonic antigen (CEA), alpha-fetoprotein (AFP), cancer antigen 19 − 9 (CA19-9), cancer antigen 72 − 4 (CA72-4), and cancer antigen 125 (CA125). Specific biomarkers have also been examined for diagnosis and treatment of GC, such as dysregulation of genes and non-coding RNAs found in gastric washes/gastric juice8; volatile markers detected in exhaled breath9; p-cresol and 4-hydroxybenzoic acid present in urine samples10; H. pylori key virulence factor CagA overexpressed in tissue samples11. However, most tissue-based biomarkers of GC carry the risk of a sampling error due to intratumoral heterogeneity, and adequate tissue sampling is of paramount importance. The biomarkers in blood, gastric washes/gastric juice, exhaled breath, or urine samples are still not clear and definite. The noninvasive blood biomarkers for early diagnosis and/or screening of high-risk population for GC may be a good choice.
MicroRNA(miRNA) is a class of non-coding single-stranded RNA molecules of approximately 22 nucleotides in length encoded by endogenous gene12. It plays a crucial role in the post-transcriptional regulation of gene expression in plants and animals13. Through base pairing with complementary sequences within mRNA molecules, miRNAs effectively silence their expression. This dysregulation of negative genes expression control is closely associated with tumorigenesis. In recent years, manipulation of miRNA clusters expression has been explored for cancer diagnosis, treatment, and prognosis14. MiRNAs have pivotal functions in gastric carcinogenesis, including oncogenic miRNAs (oncomiRNAs) such as miR-17, miR-130, and miR-181; tumour suppressor miRNAs like miR-27b, miR-29 family, miR-34, miR-124, miR-128, miR-218, miR-429, and miR-49715. MiRNAs can also modulate sensitivity to chemotherapeutic agents and alter the function of cancer-related signal pathways such as phosphatase and tensin homolog deleted on chromosome ten/phosphatidylinositol 3-kinase/protein kinase B (PTEN/PI3K/Akt)16,17. Notably, abnormal levels of miRNAs can discriminate cancer patients from healthy individuals, which have been detected not only in tissues but also in various body fluids. Therefore, miRNAs have emerged as promising biomarkers for cancer management including diagnosis, clinical staging, prediction of tumour behaviour, assessment of treatment response, and patient survival. For example, the significance of miR-21 and miR-222 lies in their distinct expression patterns in serum samples which emphasizes their utility as non-invasive biomarkers18. Henceforth investigating the characteristics of miRNAs in blood may hold immense promise for improving tumour prediction and diagnose19. The utilization of miRNAs as highly tissue-specific biomarkers has led to their incorporation into a diverse range of diagnostic tests, offering great potential for clinical diagnosis and determination of metastatic origin20.Therefore, investigating differentially expressed miRNAs in the blood of GC patients holds promise for identifying novel tumour markers and guiding future research directions. However, current studies on differentially expressed miRNAs in GC patients’ blood are not without limitations. In fact, comprehensive profiling using high-throughput sequencing is necessary to fully elucidate the dysregulation of blood miRNAs during gastric carcinogenesis.
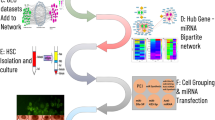
In recent years, the utilization of tumour databases, sequencing technology, and bioinformatics analysis has become prevalent in cancer research at the molecular level21,22. These advancements provide novel ideas and tools to identify differentially expressed miRNAs from patients’ blood samples and explore their functional pathways that potentially impact gastric carcinoma. Therefore, this study searched for the relevant datasets from the GEO database23. Subsequently, the bioinformatics tools were employed to analyze the acquired data to identify key miRNAs that were implicated in biological mechanisms through functional enrichment and protein network construction. The results were detected in human blood samples and further analyzed for their prognosis and diagnostic value in patients with GC. These identified crucial blood miRNAs and their corresponding target genes have the potential to serve as biomarkers for early screening and diagnosis of GC. CagA has been identified to be involved in tumorigenesis-correlated signal pathways. Therefore, the significance and effectiveness of the key miRNAs was also assessed through relevance analysis with different Pathological Tumour-Node-Metastasis (pTNM) stages, H. pylori infection and CagA expression by comparison them with clinical markers. The flow chart illustrating the methods employed in this study can be seen in Fig. 1.
Materials and methods
Data selection
GEO (https://www.ncbi.nlm.nih.gov/geo/) is developed by the National Center for Biotechnology Information (NCBI) and serves as a repository for high-throughput gene expression data submitted by scientific researchers worldwide. The database was queried using the keywords “gastric cancer”, “blood”, “human”, and “miRNA”. Subsequently, GSE113486, GSE112264, and GSE113740 were identified as the most appropriate datasets containing 115 cases of gastric cancer samples and 151 cases of normal samples. Additionally, GSE108307 was utilized to elucidate miRNAs influenced by H. pylori infection.
The initial screening of differentially expressed MiRNAs and their predicted target genes
SangerBox, an R language-based biometric analysis platform (http://sangerbox.com), was utilized for the differential analysis of sample files using the GEO convenience converter and DECenter. Subsequently, the network tool library in SangerBox was employed to perform robust rank aggregation (RRA) analysis. As a result, 20 miRNAs (10 up-regulated and 10 down-regulated) exhibiting significant changes in the GC group compared to normal samples were identified as research targets. Targetscan (http://www.targetscan.org/), miRDB (http://mirdb.org/), and miRtarBase (https://mirtarbase.cuhk.edu.cn) were utilized as online prediction websites for identifying potential target genes of these miRNAs.
Functional enrichment of differentially expressed genes
The DAVID 6.8 database (https://david.ncifcrf.gov/) is widely utilized in the field of bioinformatics to provide comprehensive and systematic information for large-scale gene or protein lists. In this study, it was employed to perform GO and KEGG24,25,26 enrichment analyses on differentially expressed genes and their associated signalling pathways, aiming to gain insights into the underlying molecular mechanisms involved in gastric carcinoma. Statistical significance was determined based on a P-value < 0.05 and a minimum gene count of ≥ 2.
PPI network
PPI network is composed of individual proteins connected through their interactions. Studying PPI network can facilitate understanding of functional links among proteins and identification of core regulators. STRING (https://string-db.org) is a protein interaction database that offers exhaustive specimens and comprehensive interaction information. After obtaining the PPI network consisting of 20 differentially expressed miRNAs and their target genes, the significant sub-network and its corresponding genes were analyzed using Cystoscape’s MCODE plug-in clustering function module (version 3.6.1). The minimum confidence score required for an interaction was set at medium level (0.900).
Selection and survival curve analysis of the key MiRNAs and relative target genes
The sub-network nodes of the PPI network were clustered to sort the most important genes that were negatively regulated by key miRNAs. The frequency of a miRNA’s occurrence was used to determine its significance in the study. ONCOLNC and GEPIA tools were utilized for interactive exploration of survival dependencies, and correlation analysis between key nodal genes and the of GC patient survival rate. The survival curves of the key miRNAs and the target genes were examined by the Kapler-Meier Plotter online tools (https://kmplot.com/analysis/) to determine the influences of their expression levels on GC patient prognosis with statistical significance was defined as P<0.05.
The human blood specimens and qRT-PCR determination
The human peripheral blood specimens were collected from Xiajin County People’s Hospital, Dezhou, and Shandong University Affiliated Hospitals, Jinan, Shandong Province. The participants were allocated into two groups: an experimental group (n = 49) and a control group (n = 37). The experimental group comprised newly diagnosed GC patients with definitive histopathological confirmation who had no prior antitumor therapy and absence of severe comorbidities or complications. Demographic characteristics (age and gender), clinicopathological parameters (pTNM stage), and immunohistochemical marker profiles including Ki67, E-cadherin, CK19, Her-2, AFP, CEA, CA19-9, CA125, and CA72-4 were extracted from standardized histopathology reports. The control group consisted of age- and gender-matched healthy volunteers recruited through randomized selection. Exclusion criteria for controls included personal history of malignancy or precancerous lesions, chronic systemic diseases affecting physiological function or metabolic homeostasis, and occupational exposure to known carcinogens. This research has been approved by the Ethics Committee of Shandong University School of Basic Medical Sciences (No.: ECSBMSSDU2021-1–097).
Total RNA was extracted using Invitrogen TRIzol (Thermo Fisher SCIENTIFIC, Code: 15596018). For miRNA quantification, the synthesis of cDNA was performed using Evo M-MLV RT Kit for qPCR (Accurate Biology, Code: AG11707). The qPCR primers of miR-575, miR-125a-3p, miR-124-3p, miR-4276 and miR-29b-3p were designed and synthesized with the Bulge-Loop miRNA qRT-PCR Primer sets (RIBOBIO, Code: MQPSCM001). Real-time PCR was carried on with SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology, Code: AG11701) on a Bio-Rad CFX-96 real-time system. U6 was detected as the internal control. CagA expression was determined with the primers forward: 5’-CAAGTCCGTGGGCATCATGT-3’ and reverse: 5’-GAGGACACTCGGTCTCTAGC-3’. All qRT-PCR reactions were conducted in triplicates, and relative quantification was calculated by the 2-ΔΔCt method (95% confidence interval) with calibration to the corresponding control. The expression data were analyzed with a Student’s t-test. The P < 0.05 was considered significant difference in statistics.
ROC analysis
The ROC curves were constructed using the expression data of key miRNAs through SPSS 26.0 software, with area under the ROC curve (AUC) values serving as diagnostic indicators. The P < 0.05 was considered to be statistically significant.
Statistical analysis
All experiments were performed in triplicate. Quantitative data were analyzed using GraphPad Prism 8.0.2 (GraphPad Software) and SPSS 26 (IBM) for statistical computations and graphical representations, with final figure preparation conducted in Adobe Photoshop 2023. Statistical comparisons were carried out using Student’s t-test and one-way ANOVA, with results expressed as mean ± standard deviation (SD). Statistical significance was defined as P < 0.05 for all analyses.
Results
The blood MiRNAs differentially expressed in gastric carcinoma
The SangerBox was utilized for the analysis of GSE113486, GSE112264, and GSE113740 datasets to screen and identify aberrantly expressed miRNAs in the blood samples of GC patients. The results revealed that in GSE113486, there were 2086 miRNAs exhibiting statistically significant differences between the GC group and normal controls. The top 10 significantly up-regulated and down-regulated miRNAs in GSE113486 were presented in Fig. 2A. The top 10 most significantly up-regulated and down-regulated miRNAs in GSE112264 were shown in Fig. 2B. The top 10 differentially expressed miRNAs in GSE113740 were displayed in Fig. 2C. A more detailed analysis of the GEO database was shown in Figure S1. The RRA algorithm was utilized to identify the top 10 up-regulated miRNAs and the top 10 down-regulated miRNAs from the three datasets (Fig. 2D).
The differentially expressed blood miRNAs in gastric carcinoma. (A) The top 10 up-regulated and down-regulated miRNAs in GSE113486. (B) The top 10 up-regulated and down-regulated miRNAs in GSE112264. (C) The top 10 up-regulated and down-regulated miRNAs in GSE113740. (D) The highest-scoring up-regulated and down-regulated miRNAs identified using RRA algorithm. The selected key miRNAs were shown in red frames.
The identification of target genes directly silenced by the most differentially expressed miRNAs and the subsequent enrichment analysis of GO and KEG
By integrating and comparing predictions from Targetscan, Mirtarbase and miRDB, a total of 691 over-expressed genes and 1074 down-regulated ones were determined as potential targets of the selected 20 miRNAs. GO and KEGG enrichment analyses using DAVID revealed the molecular functions (MFs), cellular components (CCs), biological processes (BPs), and signalling pathways associated with these target genes. GO enrichment results for Up-regulated genes suggested that key BPs included autophagosomes maturation, regulation of mitochondrial membrane potential, and folic acid metabolic process. The main MFs identified were protein binding, metal ion binding, and RNA polymerase II core promoter proximal region sequence-specific DNA binding. The CCs observed included cytosol, cytoplasm, and nucleus (Fig. 3 A). On the contrary, the down-regulated genes (targets of up-regulated miRNAs) were primarily involved in BPs such as regulation of transcription from RNA polymerase II promoter, platelet-derived growth factor receptor signalling pathway, and DNA-templated positive regulation of transcription. The main MFs encompassed protein binding, chromatin binding, transcriptional activator activity, and RNA polymerase II core promoter proximal region sequence-specific binding. The CCs comprised nucleoplasm, nucleus and focal adhesion (Fig. 3B). The signalling pathways implicated in the regulation of down-expressed genes included focal adhesion, prolactin signalling pathway, PI3K-Akt signalling pathway, and focal adhesion (Fig. 3 C). KEGG analysis indicated that endocytosis, proteoglycans in cancer, and glycosylphosphatidylinositol-anchor biosynthesis were among the main signalling pathways involved in overexpressed genes (targets of down-regulated miRNAs)-correlated mechanism (Fig. 3D).
The analysis of proteins interaction network and identification of key blood MiRNAs
Furthermore, the 20 most differently expressed blood miRNAs and their corresponding target genes were analyzed in the STRING database (Fig. 3E). The results of PPI network analysis were imported into Cytoscape software for further investigation. A total of 38 nodes were identified, indicating a close relationship among the top 20 differently expressed blood miRNAs. These miRNAs were found to be involved in multiple signalling pathways, suggesting their potential roles in tumorigenesis. Additionally, the up-regulated gene-encoded proteins network consisted of 413 nodes with 143 edges, an average node degree of 0.692, and an average local clustering coefficient of 0.253. The enrichment P value was less than 0.0287 (Figure S2A). Similarly, the down-regulated gene-encoded proteins network comprised of 1323 nodes with 1021 edges, an average node degree of 1.54, and an average local clustering coefficient of 0.319. The enrichment P value was less than 2.62e-10 (Figure S2B). These findings indicate the data is reliable and can be utilized for further analysis.
The identification of target genes directly regulated by the key blood miRNAs from the three datasets by bioinformatics analysis. (A) The principal BPs, MFs, and CCs of down-regulated miRNAs and corresponding up-regulated target genes. (B) The principal BPs, MFs, and CCs of up-regulated miRNAs and corresponding down-regulated target genes. (C) Signalling pathway enrichment results of up-regulated miRNAs and corresponding down-regulated target genes in KEGG (The permission to use KEGG software has been obtained from Kanehisa Laboratories). (D) Signalling pathway enrichment results of down-regulated miRNAs and the corresponding up-regulated target genes in KEGG. (E) Network of target gene-encoded proteins corresponding to all 20 most significantly differentially expressed blood miRNAs.
The Cytoscape software was utilized to determine the number of occurrences of the blood miRNAs on the key nodes, thereby assessing the significance of these miRNAs (Table S1). The findings indicated that hsa-miR-124-3p and hsa-miR-29b-3p were involved in up-regulated cases, while hsa-miR-125a-3p, hsa-miR-4276, and hsa-miR-575 were associated with down-regulated cases (Fig. 2, in red frames). These miRNAs could potentially serve as crucial biomarkers for early diagnosis and treatment of gastric carcinoma. Furthermore, the sub-network genes obtained from the PPI network analysis represented potential targets for these key miRNAs and were subjected to survival analysis. The results revealed 27 blood miRNA-targeted genes that exhibited an association with GC (Table S2). According to online data, similar specific miRNA expression patterns were not found in colon adenocarcinoma (COAD), liver hepatocellular carcinoma (LIHC) and lung adenocarcinoma (LUAD), which were also derived from epithelial cells, confirming the specificity of this key miRNAs expression model in gastric carcinogenesis (Figure S3).
The key blood MiRNAs were identified through their expression levels detection in human blood samples
The demographic information (including age, gender, and pTNM) of the patients who provided blood samples and their correlation with the expression of key miRNAs was presented in Table 1.
The results of qRT-PCR, which determined the expression levels of selected key miRNAs in human blood samples, were depicted in Fig. 4A-E. Furthermore, the expression levels of tissue-based markers and blood antigens to help diagnosis and treatment of GC and five key miRNAs across different pTNM stages from GC patients’ samples and online databases were also analyzed, which were shown in Fig. 4 F, S4, S5, and Tables 2 and 3. From these findings, it was preliminarily observed that there was a significant up-regulation of hsa-miR-124-3p and hsa-miR-29b-3p, as well as a down-regulation of hsa-miR-575, hsa-miR-125a-3p and hsa-miR-4276, whose expression levels in human blood specimens were consistent with bioinformatics predictions and online data. Moreover, it was determined that the patients’ age and gender did not exert a significant influence on the expression of key miRNAs (P > 0.05). However, there were significant differences in the expression levels of key miRNAs among different stages of pTNM (*P < 0.05) individually, and these expression trends were partially dependent on the pathological stages (from I to III); whereas the expression trends of each clinical marker did not exhibit substantial variation or correlation with the stages. Further testing and verification using larger cohorts is necessary to confirm these results.
The expression of key miRNAs in human blood specimens by RT-qPCR detection and the correlation with the stages. (A) The expression of hsa-miR-124-3p increased in the blood samples of GC patients (P < 0.01 vs. normal). (B) The blood expression of hsa-miR-125a-3p decreased in the blood samples of GC patients (P < 0.01 vs. normal). (C) The expression of hsa-miR-29b-3p increased in the blood samples of GC patients (P < 0.05 vs. normal). (D) The expression of hsa-miR-4276 decreased in the blood samples of GC patients (P < 0.01 vs. normal). (E) The expression of hsa-miR-575 decreased in the blood samples of GC patients (P < 0.01 vs. normal). (F) The expression levels of key miRNAs in different stages of pTNM (P < 0.05 vs. Stage I).
The diagnostic value of key blood MiRNAs in patients with GC
The survival curves indicating the correlation between the expression of key miRNAs and patient prognosis could be found in Figure S6. Additionally, Figure S7 displayed the survival curves for key miRNAs’ target genes and corresponding patient prognosis. The expression levels of hsa-miR-124-3p, hsa-miR-29b-3p, and hsa-miR-575 were closely associated with GC patient prognosis (P < 0.05). However, regarding hsa-miR-125a-3p and hsa-miR-4276, their expressions in human blood specimens were consistent with bioinformatics predictions but inconsistent with GC patient prognosis. Furthermore, although the prognosis analysis indicated that there was no significant association between hsa-miR-4276 expression and patient survival rate (P > 0.05), lower expression levels of hsa-miR-4276 tended to decreased patient survival rate. The prognosis analysis also revealed that most target genes’ expressions were related to patients’ likelihoods for survival.
In order to further elucidate the diagnostic value of key blood miRNAs in gastric carcinoma, ROC analysis was performed based on their expression levels. The results demonstrated that miR-124-3p, miR-125-3p and miR-29b-3p held potential for diagnosis, with area under the ROC curve (AUC) values of 0.952, 0.812 and 0.824 respectively. Combining miR-124-3p, miR-125-3p, and miR-29b-3p yielded an improved ROC value of 0.975. Despite relatively lower ROC values (0.556 and 0.41) for hsa-miR-4276 and hsa-miR-575 respectively, the model composed of these key miRNAs exhibited excellent discriminatory ability between patients with and without GC (Fig. 5A-F).
The key miRNAs could have diagnostic value in GC determined by ROC analysis (A) The value of hsa-miR-124-3p AUC was 0.952, P < 0.001. (B) The value of hsa-miR-125a-3p AUC was 0.812, P < 0.001. (C) The value of miR-29b-3p AUC was 0.824, P < 0.001. (D) The value of hsa-miR-4276 AUC was 0.556, P > 0.05. (E) The value of hsa-miR-575 AUC was 0.41, P >0.05. (F) The value of (hsa-miR-124-3p + hsa-miR-125a-3p + miR-29b-3p) AUC was 0.975, P < 0.001.
The model was further validated in GSE108307, which involved the miRNAs induced by H. pylori infection. Cross-validation demonstrated the presence of up-regulated hsa-miR-29b-3p and down-regulated hsa-miR-4276 in the dataset (Figure S8). Furthermore, CagA expression was assessed in human blood specimens using qRT-PCR. The results indicated a significant overexpression of CagA in GC patients compared to normal individuals (Fig. 6 A). The expression levels of five key miRNAs and clinical markers were detected in CagA-positive samples from GC patients respectively. In comparison with the clinical markers, a diagnostic model composed of these key miRNAs provided accurate diagnoses, demonstrating a stronger correlation between the CagA-positive group and expression levels of these key miRNAs, while the expression patterns of clinical markers were inconsistent with that of CagA-positive group in GC (Fig. 6B-D). In conclusion, the identified key miRNAs exhibited significant diagnostic value for detecting GC through analysis of blood samples. These findings also suggested that while bioinformatics predictions may be valuable for diagnosis purposes, results obtained through different analytical approaches may not always align completely with each other.
The expression levels of key miRNAs and clinical markers in CagA-positive patients’ blood specimens by RT-qPCR detection. (A) CagA was remarkably overexpressed in human GC blood specimens compared to the normal controls (P < 0.05). (B) The expression pattern of five key miRNAs in CagA-positive blood samples from GC patients, which showed up-regulation of hsa-miR-124-3p and hsa-miR-29b-3p, as well as down-regulation of hsa-miR-125-3p, hsa-miR-4276, and hsa-miR-575. (C) The clinical markers expression levels including Ki67, E-Cadherin, CK19, and Her-2 in CagA-positive blood samples, while the expression of E-Cadherin and Her-2 showed down-regulated. (D) The clinical markers expression levels including AFP, CEA, CA19-9, CA125 and CA72-4 in CagA-positive blood samples. None of them determined overexpression.
Discussion
In the research, GSE113486, GSE112264 and GSE113740 datasets were initially analyzed using bioinformatics tools to identify key blood miRNAs in gastric carcinoma. RRA analysis were performed to rank the corresponding miRNAs. The top 20 differentially expressed blood miRNAs (10 up-regulated and 10 down-regulated) were selected as the aims for predicting target genes and their relative biological activities. Among these, there were 691 up-regulated differentially expressed genes and 1074 down-regulated genes that might be directly silenced by these 20 blood miRNAs. GO and KEGG enrichment analyses of the differentially expressed genes were conducted using DAVID to explore the biological processes and signalling pathways primarily involved in this system. Based on this analysis, a PPI network of target genes suggested five crucial blood miRNAs: hsa-miR-124-3p, hsa-miR-125a-3p, hsa-miR-29b-3p, hsa-miR-4276, and hsa-miR-575. The subsequent qRT-PCR results obtained from human blood samples validated the expression levels of these pivotal miRNAs in relation to pTNM stages, which exhibited a consistent pattern with that of clinical markers in GC. Survival curve analysis further supported the significant roles of these key blood miRNAs in gastric carcinoma. ROC analysis along with cross-validation using H. pylori-induced miRNAs as well as comparative study with clinical markers all indicated that these key miRNAs could have valuable implications for diagnosing GC.
The currently utilized tumour markers exhibit limited sensitivity and specificity, while the absence of a universally accepted “gold standard” further compounds this issue. Simultaneously, relying on a single tumour marker proves challenging in accurately capturing the intricate nature of tumours. Consequently, identifying effective markers and employing combined detection methods will serve as an efficacious approach to enhance the diagnostic value of tumour markers. MiRNAs have emerged as pivotal regulators in intricate biological processes, encompassing the pathogenesis of cancer27. Human malignancies exhibit distinctive characteristics such as sustained proliferation signals, activation of invasion and metastasis, angiogenesis, and evasion from immune destruction; miRNAs can actively participate in each of these features28,29. During carcinogenesis, miRNAs are involved in various signal pathways, including but not limited to miR-135a, miR-21, miR-218, miR-221/222, miR-375, and miR-451.They function as modulators (e.g. miR-148a, miR-155, miR-181b, miR-218, miR-374b-5p, and miR-499), epigenetic regulators (e.g. miR-34b/c, miR-129, and miR-212), and influencers of drug resistance (e.g. let-7a, miR-106a, miR-148a-3p, miR-495-3p, and miR-508-5p)30. The expression profiles of miRNAs in different cancer cells and tissues can be detected in the circulation, including blood, plasma and other body fluids. Mature miRNAs demonstrate remarkable stability in body fluids and possess high specificity for different cancer states, establishing them as potential non-invasive tumour marker. Consequently, blood miRNAs are increasingly recognized as invaluable biomarkers for the diagnosis and treatment of cancers31,32,33,34. There have been advances in the investigation of blood miRNAs in gastric carcinoma. For instance, the study conducted by Tasuku Matsuoka et al. in 2018 unveiled the up-regulation of several miRNAs, including miR-101, miR-106b, miR-125a, miR-129, miR-130b, miR-148b, miR-181c, miR-199a, miR-21, miR-23a, miR-27a, miR-29a, miR-212, miR-215, miR-218, miR-222-221, miR-331, miR-335, miR-370, miR-375, miR-449, miR-486, and miR-512, in blood samples as potential specific markers for diagnostic/prognostic/therapeutic of GC4. Similarly, in 2020, Soudeh Ghafouri-Fard et al. provided a comprehensively summary of the oncogenic roles of miR-17, miR-130, and miR-181 as well as the tumor suppressor functions of miR-124, miR-128, miR-27b, miR-29 family, miR-218, miR-34, miR-429, and miR-497 in blood samples from patients with gastric carcinoma15. In addition, a 2021 study from Mona Noohi et al. demonstrated an elevation in blood miR-21 level among the gastritis patients infected by H. pylori35. To summarized, despite the crucial blood miRNAs profiles may vary in gastric carcinoma and require identification, all aforementioned studies have consistently demonstrated the significant potential of circulating miRNAs as prognostic and diagnostic biomarkers in GC, thereby positioning them as promising therapeutic targets36,37.
In this study, utilizing bioinformatics tools, five crucial blood miRNAs (hsa-miR-124-3p, hsa-miR-125a-3p, hsa-miR-29b-3p, hsa-miR-4276, and hsa-miR-575) were identified as potential targets for diagnosis and treatment of gastric carcinoma. These key miRNAs and their respective target genes were involved in the biological processes closely correlant to tumorigenesis. Notably, platelet-derived growth factor receptor signalling pathway and transcription RNA polymerase II have been implicated in gastric carcinoma and development38,39.Additionally, focal adhesion and the PI3K-Akt signalling pathway are intricately linked to carcinogenesis16,40. Furthermore, through the identification of upstream miRNAs regulating target genes, the results obtained from Cytoscape analysis revealed that hsa-miR-124-3p exhibited the highest frequency among key miRNAs in the PPI network. Both examination of human blood specimens and survival curve analysis provided evidence supporting a correlation between hsa-miR-124-3p and gastric carcinogenesis. Additionally, ROC analysis further emphasized the significance of hsa-miR-124-3p in diagnosing gastric cancer. Up-regulation of hsa-miR-124-3p was observed in blood samples from CagA-positive GC patients. Moreover, several genes among the predicted targets of hsa-miR-124-3p exhibit a robust association with GC prognosis. For instance, ANXA5, a member of the Annexin family implicated in tumorigenesis and development across various cancers including GC41,42,43,44, and CAV1 to significantly regulate E-cadherin expression as well as alterations in cell morphology and migration ability of GC cells45,46. The impact of hsa-miR-125a-3p on immunity and carcinogenesis has been investigated through its regulation of tumour-associated signal pathways, including the Hippo pathway47. In this study, aberrant expression of hsa-miR-125a-3p was detected in blood specimens from patients with GC. Importantly, CagA also induced down-regulation of hsa-miR-125a-3p, consistent with its inhibition observed in blood samples from GC patients. Our findings also revealed a potential downstream target of hsa-miR-125a-3p as PRDM1, which exhibited a correlation between its expression and survival probability among GC patients. The involvement of PRDM1 in the regulation of B cell and T cell differentiation, as well as its crucial role in immunosuppression, has been demonstrated to be associated with various types of cancers48,49. Hsa-miR-29b-3p plays a pivotal role in tumorigenesis and metastasis50,51,52,53. Research has demonstrated that hsa-miR-29b-3p exhibits potential as a crucial circulating miRNA with clinical significance in endometrial cancer54. Additionally, activation of PER1, one of the predicted key target genes of hsa-miR-29b-3p in our findings, can effectively impede the progression of pancreatic cancer55. The correlation between expression levels of circulating hsa-miR-29b-3p and gastric cancer prognosis and diagnosis was revealed through GC blood samples analysis in this study. The Up-regulation of hsa-miR-29b-3p was also observed in qRT-PCR results of CagA-positive blood samples from GC patients. Hsa-miR-4276 has been implicated in conferring resistance to influenza A infection in lung epithelial cells56. However, limited research exists regarding the involvement of hsa-miR-4276 in tumorigenesis, as well as its target genes RNF217 and IP6K1. Conversely, among the key nodes identified through Cytoscape analysis, there was a significant enrichment of downstream genes regulated by hsa-miR-4276, suggesting a strong correlation between hsa-miR-4276 and its target genes with GC. This possibility is further supported by patient blood specimens’ check and predictions based on H. pylori-induced miRNAs, as well as the observed down-expression level in CagA-positive blood samples. The protein MSRB3, which has been identified as a crucial regulator of proliferation and migration in GC cells, making it a potential marker for predicting peritoneal metastasis and poor prognosis57,58, was predicted as the target of hsa-miR-575 in this study. Hsa-miR-575 has also been identified to regulate development of gastric cancer by targeting PTEN59. Differential expression of hsa-miR-575 was observed in GC through blood checks and its down-regulation by CagA was also confirmed. The patterns reflecting the correlation with pTNM stages and H. pylori infection in GC patients of these blood-based key miRNAs were also found to be consistent with their expression levels, which seemed to be better than those of clinical markers. Considering the feasibility of combining miRNA markers with clinical blood markers, we compared the diagnostic accuracy for gastric cancer detection using clinical blood markers alone, miRNA markers alone, and the combination of both approaches. The results demonstrated that the highest positive detection rate was achieved when using miRNA markers alone (Figure S9). Therefore the present findings highlight the robustness and significance of the identified blood miRNAs in gastric carcinogenesis and diagnosis, emphasizing their potential as promising biomarkers.
However, further in-depth exploration is warranted. In addition to miRNAs, it has also been reported that eleven cytokines exhibit significant increases in blood samples from patients with GC. To precisely determine the expression levels of key miRNAs and their target genes at different stages of GC, a larger number of blood samples from GC patients are required. The identified blood miRNAs such as miR-21 not included in this model also can serve as controls to assess the significance of the five key miRNAs. Furthermore, it is imperative to investigate the correlation between five key miRNAs and pTNM stages as well as H. pylori infection in gastric carcinoma, as well as elucidate the underlying mechanisms involved.
Data availability
The datasets GSE113486 for this study can be found in the GEO Accession viewer (nih.gov), The datasets GSE112264 for this study can be found in the GEO Accession viewer (nih.gov), The datasets GSE113740 for this study can be found in the GEO Accession viewer (nih.gov), The datasets GSE108307 for this study can be found in the GEO Accession viewer (nih.gov).
References
Wang, Q. Q., Liu, G. L. & Hu, C. H. Molecular classification of gastric adenocarcinoma [J]. Gastroenterol. Res. 12 (6), 275–282 (2019).
Xia, C. F. et al. Cancer statistics in China and united states, 2022: profiles, trends, and determinants [J]. Chin. Med. J-Peking. 135 (5), 584–590 (2022).
Wagner, J. A. & Atkinson, A. J. Measuring biomarker progress [J]. Clin. Pharmacol. Ther. 98 (1), 2–5 (2015).
Matsuoka, T. & Yashiro, M. Biomarkers of gastric cancer: current topics and future perspective [J]. World J. Gastroentero. 24 (26), 2818–2832 (2018).
Li, Y. et al. Evolutionary expression of HER2 conferred by chromosome aneuploidy on Circulating gastric Cancer cells contributes to developing targeted and chemotherapeutic resistance [J]. Clin. Cancer Res. 24 (21), 5261–5271 (2018).
Zhang, J. et al. Exosomal miR-4745-5p/3911 from N2-polarized tumor-associated neutrophils promotes gastric cancer metastasis by regulating SLIT2 [J]. Mol. Cancer, 23(1), (2024).
Zhang, M. et al. Liquid biopsy: Circulating tumor DNA monitors neoadjuvant chemotherapy response and prognosis in stage II/III gastric cancer [J]. Mol. Oncol. 17 (9), 1930–1942 (2023).
Yamamoto, H. et al. Non-Invasive early molecular detection of gastric cancers [J]. Cancers 12 (10), 2880 (2020).
Bhandari, M. P. et al. Volatile markers for Cancer in exhaled Breath-Could they be the signature of the gut microbiota?? [J]. Molecules 28 (8), 3488 (2023).
Li, S. S. et al. Magnetic organic porous polymer as a solid-phase extraction adsorbent for enrichment and quantitation of gastric cancer biomarkers (P-cresol and 4-hydroxybenzoic acid) in urine samples by UPLC [J]. Microchim. Acta. 187 (7), 388 (2020).
Guo, X. H. et al. Risk assessment of gastric cancer in the presence of Helicobacter pylori CagA and hopqii genes [J]. Cell. Mol. Biol. 67 (4), 299–305 (2021).
Hrovatin, K. & Kunej, T. Classification of miRNA-related sequence variations [J]. Epigenomics-Uk 10 (4), 463–481 (2018).
Fabian, M. R. & Sonenberg, N. The mechanics of miRNA-mediated gene silencing: a look under the Hood of MiRISC [J]. Nat. Struct. Mol. Biol. 19 (6), 586–593 (2012).
Saliminejad, K. et al. An overview of micrornas: biology, functions, therapeutics, and analysis methods [J]. J. Cell. Physiol. 234 (5), 5451–5465 (2019).
Ghafouri-Fard, S. et al. MicroRNAs in gastric cancer: biomarkers and therapeutic targets [J]. Gene 757, 144937 (2020).
Hu, M. L. et al. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer [J]. Oncol. Rep. 41 (3), 1439–1454 (2019).
Taheri, M. et al. LncRNAs and MiRNAs participate in determination of sensitivity of cancer cells to cisplatin [J]. Exp. Mol. Pathol. 123, 104602 (2021).
Emami, S. S. et al. Evaluation of Circulating miR-21 and miR-222 as diagnostic biomarkers for gastric cancer [J]. J. Cancer Res. Ther. 15 (1), 115–119 (2019).
Su, F. et al. Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications [J]. Genes-Basel, 13(11): 1931. (2022).
Masterson, A. N. et al. A novel liquid biopsy-based approach for highly specific cancer diagnostics: mitigating false responses in assaying patient plasma-derived Circulating MicroRNAs through combined SERS and plasmon-enhanced fluorescence analyses [J]. Analyst 145 (12), 4173–4180 (2020).
Berger, M. F. & Mardis, E. R. The emerging clinical relevance of genomics in cancer medicine [J]. Nat. Rev. Clin. Oncol. 15 (6), 353–365 (2018).
Scionti, F. et al. Integration of DNA microarray with clinical and genomic data [J]. Methods Mol. Biol., : 239–248. (2022).
Kourou, K. et al. Machine learning applications in cancer prognosis and prediction [J]. Comput. Struct. Biotec. 13, 8–17 (2015).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms [J]. Protein Sci. 28 (11), 1947–1951 (2019).
Kanehisa, M. et al. KEGG: biological systems database as a model of the real world [J]. Nucleic Acids Res. 53 (D1), D672–D677 (2025).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes [J]. Nucleic Acids Res. 28, 27–30 (2000).
Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases [J]. Nat. Rev. Drug Discov. 16 (3), 203–221 (2017).
Hanahan, D. Hallmarks of cancer: new dimensions [J]. Cancer Discov. 12 (1), 31–46 (2022).
Van Roosbroeck, K. & Calin, G. A. Cancer hallmarks and micrornas: the therapeutic connection [J]. Adv. Cancer Res. 135, 119–149 (2017).
Favier, A. et al. MicroRNA as epigenetic modifiers in endometrial cancer: A systematic review [J]. Cancers 13 (5), 1137 (2021).
Giannopoulou, N. & Constantinou, C. Recent developments in diagnostic and prognostic biomarkers for colorectal cancer: a narrative review [J]. Oncology-Basel 101 (10), 675–683 (2023).
Schwarzenbach, H. et al. Clinical relevance of Circulating cell-free MicroRNAs in cancer [J]. Nat. Rev. Clin. Oncol. 11 (3), 145–156 (2014).
Turchinovich, A. et al. Characterization of extracellular Circulating MicroRNA [J]. Nucleic Acids Res. 39 (16), 7223–7233 (2011).
Van Den Broek, D. & Groen, H. J. M. Screening approaches for lung cancer by blood-based biomarkers: challenges and opportunities [J]. Tumor Biology, 1–16. (2023).
Nooh, M. et al. Prediction of blood miRNA-mRNA regulatory network in gastric Cancer [J]. Rep. Biochem. Mol. Biol. 10 (2), 243–256 (2021).
Aalami, A. H., Aalami, F. & Sahebkar, A. Gastric Cancer and Circulating micrornas: an updated systematic review and diagnostic Meta-Analysis [J]. Curr. Med. Chem. 30 (33), 3798–3814 (2023).
Jelski, W. & Mroczko, B. Molecular and Circulating biomarkers of gastric Cancer [J]. Int. J. Mol. Sci. 23 (14), 7588 (2022).
Chen, F. X., Smith, E. R. & Shilatifard, A. Born to run: control of transcription elongation by RNA polymerase II [J]. Nat. Rev. Mol. Cell. Bio. 19 (7), 464–478 (2018).
Huang, F. et al. Gastric cancer-derived MSC-secreted PDGF-DD promotes gastric cancer progression [J]. J. Cancer Res. Clin. 140 (11), 1835–1848 (2014).
Zhang, H. S. et al. Gain-of-Function RHOA mutations promote focal adhesion kinase activation and dependency in diffuse gastric Cancer [J]. Cancer Discov. 10 (2), 288–305 (2020).
Li, Q. et al. The potential role of miR-124-3p in tumorigenesis and other related diseases [J]. Mol. Biol. Rep. 48 (4), 3579–3591 (2021).
Peng, B. Y. et al. Annexin A5 as a potential marker in tumors [J]. Clin. Chim. Acta. 427, 42–48 (2014).
Romero-López, M. J. et al. miR-23b-3p, miR-124-3p and miR-218-5p synergistic or additive effects on cellular processes that modulate cervical Cancer progression?? A molecular balance that needs attention [J]. Int. J. Mol. Sci. 23 (21), 13551 (2022).
Wang, X. J. et al. AnnexinA5 might suppress the phenotype of human gastric Cancer cells via ERK pathway [J]. Front. Oncol. 11, 665105 (2021).
Luo, Z. et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression [J]. Mol. Cancer. 19 (1), 86 (2020).
Zhang, K. D. et al. Decreased expression of Caveolin-1 and E-Cadherin correlates with the clinicopathologic features of gastric Cancer and the EMT process [J]. Recent. Pat. Anti-Canc. 11 (2), 236–244 (2016).
Wang, J. K., Wang, Z. & Li, G. D. MicroRNA-125 in immunity and cancer [J]. Cancer Lett. 454, 134–145 (2019).
Boi, M. et al. /BLIMP1: a tumor suppressor gene in B and T cell lymphomas [J]. Leuk. Lymphoma. 56 (5), 1223–1228 (2015).
Shen, L. J. et al. Role of PRDM1 in tumor immunity and drug response: A Pan-Cancer analysis [J]. Front. Pharmacol. 11, 593195 (2020).
Amirian, M. et al. Overview of the miR-29 family members’ function in breast cancer [J]. Int. J. Biol. Macromol. 230, 123280 (2023).
Shin, J. et al. Restoration of miR-29b exerts anti-cancer effects on glioblastoma [J]. Cancer Cell Int., 17 (1). (2017).
Ulivi, P. et al. Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal Cancer [J]. Int. J. Mol. Sci. 19 (1), 307 (2018).
Zhao, X. H. et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma [J]. J. Cell. Mol. Med. 22 (1), 655–667 (2018).
Bloomfield, J. et al. Clinical value and molecular function of Circulating MicroRNAs in endometrial Cancer regulation: A systematic review [J]. Cells-Basel 11 (11), 1836 (2022).
Guo, X. Y. et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner [J]. Mol. Cancer. 19 (1), 91 (2020).
Othumpangat, S., Noti, J. D. & Beezhold, D. H. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by MiRNA 4276 [J]. Virology 468, 256–264 (2014).
Ma, X. M. et al. Increased expression of methionine sulfoxide reductases B3 is associated with poor prognosis in gastric cancer [J]. Oncol. Lett. 18 (1), 465–471 (2019).
Zhang, S. M. et al. Identification of key gene and pathways for the prediction of peritoneal metastasis of gastric Cancer by Co-expression analysis [J]. J. Cancer. 11 (10), 3041–3051 (2020).
Wang, Y. N. et al. MicroRNA-575 regulates development of gastric cancer by targeting PTEN [J]. Biomed. Pharmacother. 113, 108716 (2019).
Funding
This research was funded by National Natural Science Foundation of China (No. 81871627, No. 82372599) and Shandong University Student Innovation and Entrepreneurship Training Program (No. 2019325, S202110422208).
Author information
Authors and Affiliations
Contributions
L.X.,W.Q.,X.Z.,Y.X.,Z.R.and W.H.conducted the data curation; Z.J. was responsible for funding acquisition, project administration supervision, original draft writing and review and editing writing; L.X. was in charge of resource acquisition; L.X. and W.Q. was responsible for original draft writing and review and editing writing; W.H. was responsible for review and editing writing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Institutional review board statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Shandong University School of Basic Medical Sciences (No.ECSBMSSDU2021-1-097). All human participants have provided their informed consent forms.
Informed consent
Statement: Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, X., Wang, Q., Xu, Z. et al. Screening and evaluation of specific blood MiRNAs as potential biomarkers in diagnostics of gastric Cancer. Sci Rep 15, 22974 (2025). https://doi.org/10.1038/s41598-025-06773-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06773-5
Keywords
This article is cited by
-
Serum Aberrant Expression of miR-431-5p and Their Diagnostic Value in Parkinson’s Disease
Neurochemical Research (2026)
-
Extracellular vesicle-enclosed microRNAs serve as non-invasive biomarkers in the major upper and lower gastrointestinal tract tumors: a bibliometric analysis (2010–2025)
Discover Oncology (2025)