Abstract
TMBIM6, a transmembrane BAX inhibitor motif containing 6, located in the endoplasmic reticulum, is linked to various cellular processes and cancer progression. Previous investigations showed a substantial association between TMBIM6 and survival in patients diagnosed with various cancer types. However, the lack of extensive studies addressing the correlation between TMBIM6 and gliomas necessitates a comprehensive investigation to explore its potential as a prognosis marker for glioma. This study investigates TMBIM6’s prognostic value by using data from TCGA (The Cancer Genome Atlas), GEO (Gene Expression Omnibus), and CGGA (Chinese Glioma Genome Atlas) databases, along with histopathological analysis of tissue microarray slide. Survival analysis confirmed the prognostic significance of TMBIM6 in glioma, while co-expression analysis identified positively and negatively correlated genes with TMBIM6, and Enrichment analysis suggested TMBIM6’s association with protein processing in the ER and NOD-like receptor signaling pathways. A strong correlation was observed between TMBIM6 expression and immune infiltration, especially with M2 macrophages. Additionally, hsa-miR-128-3p was identified as an upstream regulator of TMBIM6. These findings highlight TMBIM6’s potential as a prognostic biomarker for glioma, offering new insights into its role in glioma progression.
Similar content being viewed by others
Introduction
Gliomas are a broad class of brain tumors that originate from glial cells and can vary in prognosis and aggressiveness in the development of tumors. Glioblastoma (GBM) is the most prevalent and severe form of brain cancer in adults, which accounts for a considerable proportion of primary brain tumors. It has a global prevalence rate of 3 per 100,000 individuals1,2,3. GBM is identified by the presence of necrosis and endothelial proliferation. The World Health Organization’s (WHO) classification system for brain tumors categorizes GBM as a grade IV tumor due to these characteristics4. Furthermore, low-grade gliomas (LGG) from glial cells have slower growth, occur less frequently than GBM, and have a slightly better prognosis5,6. Most patients with LGGs are known to progress to high-grade gliomas, which eventually result in death7.
Transmembrane BAX inhibitor motif containing 6 (TMBIM6) is a gene mapped to human chromosome 12q12-q138 and has been recognized as a regulator of Bax-mediated cell death in yeast9,10. Also, TMBIM6 is strongly correlated with cancer metastasis and progression, making it a potential biomarker for multiple cancer types, including breast cancer, Non-small cell lung cancer (NSCLC), and Laryngeal squamous cell carcinoma (LSCC) further reinforces its significance in cancer research11,12. TMBIM6 has been identified as a downstream target of RBM15-mediated m6A modification in LSCC, suggesting its involvement in the progression of LSCC13. Interestingly, TMBIM6 demonstrated a significant correlation with survival in patients diagnosed with breast invasive carcinoma (BRCA), sarcoma (SARC), cervical squamous cell carcinoma (CESC), lung adenocarcinoma (LUAD), pancreatic adenocarcinoma (PAAD), esophageal carcinoma (ESCA), lower grade glioma (LGG), head and neck squamous cell carcinoma (HNSC), and skin cutaneous melanoma (SKCM), highlighting its potential usefulness as a prognostic biomarker11. Considering the differences in the developmental process of glioma, TMBIM6 could play an important role in prognosis as it is known to regulate apoptosis and calcium homeostasis. Previous reports suggest that long non-coding RNA plasmacytoma variant translocation 1 (PVT1) upregulates TMBIM6 in Glioblastoma by sponging miR1301, which in turn encourages the invasion and proliferation of cancer cells while preventing apoptosis14. However, there is a lack of comprehensive studies on the connection between TMBIM6 and gliomas.
Furthermore, identifying novel biomarkers like TMBIM6 is essential for the early detection, diagnosis, and treatment of glioma. Such markers provide insight into tumor biology and can guide personalized therapies. Given its association with molecular alterations in GBM and LGG, TMBIM6 holds promise as valuable prognostic and therapeutic biomarker15.
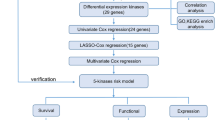
This study thoroughly investigated the correlation between TMBIM6 and glioma using integrated bioinformatics analysis of publicly available datasets, including The Cancer Genome Atlas Program (TCGA), Gene Expression Omnibus (GEO), Genotype-Tissue Expression (GTEx) project, and Chinses Glioma Genome Atlas (CGGA). Differential expression, co-expression, survival, immune infiltration, and pathway enrichment analyses were conducted to assess the prognostic and functional relevance of TMBIM6. Candidate miRNAs regulating TMBIM6 were also predicted. Experimental validation included immunohistochemistry on tissue microarrays, as well as qPCR and MTT assays following TMBIM6 siRNAs and miR-128 mimic transfection in U87-MG cells. Our findings aim to enhance understanding of glioma pathogenesis and support TMBIM6 as a potential prognostic biomarker and therapeutic target.
Results
Differential expression of TMBIM6 in different cancers
The differential expression of TMBIM6 in different cancer types compared to normal conditions helps us understand the crucial molecular changes driving tumorigenesis. Firstly, the expression level of TMBIM6 in different cancer types was assessed using GEPIA2. This result shows that TMBIM6 was significantly higher in Breast cancer (BRCA), Glioblastoma (GBM), Lower grade Glioma (LGG), Stomach adenocarcinoma (STAD), Pancreatic Adenocarcinoma (PAAD), Colon Adenocarcinoma (COAD), Rectum Adenocarcinoma (READ), and Ovarian Serous cyst Adenocarcinoma (OV) (Fig. 1a). Further, the expression level of TMBIM6 in glioma was assessed by BEST webtool (Fig. 1b, c) and R program (Fig. 1d, e). These analyses revealed that TMBIM6 is abundantly expressed in gliomas.
Differential expression analysis of TMBIM6 in glioma. (a) TMBIM6 expression level in different cancers in GEPIA2 tool based on TCGA and GTEx data. (b,c) TMBIM6 expression level checked in GBM and LGG compared to normal by BEST web tool. (d,e) TMBIM6 expression in GBM and LGG compared to normal tissue based on TCGA and GEO gene expression data analyzed in R software.
The prognostic value of TMBIM6 in gliomas
To determine the relationship between TMBIM6 expression levels and prognosis, survival curves of the TMBIM6 gene were plotted using TCGA survival data. The analysis revealed that high expression of TMBIM6 was associated with decreased survival, whereas low expression was associated with increased survival in both the TCGA and CGGA datasets (Fig. 2a, b). The correlation between TMBIM6 and IDH1 mutations in gliomas may affect the prognosis and treatment. IDH1 mutations, common in 70–80% of low-grade gliomas but only 5–15% of glioblastomas, produce the oncometabolite 2-hydroxyglutarate, which alters DNA methylation and promotes tumorigenesis16,17,18. Findings showed TMBIM6 mRNA expression was higher in IDH1 status WT gliomas in TCGA and in non-1p/19q codeletion in the CGGA cohort (Fig. 2c, d). These observations suggest the prognostic significance of TMBIM6 in glioma.
Prognosis of TMBIM6 in glioma. (a,b) The Kaplan-Meier curves of TMBIM6 in glioma patients based on TCGA and CGGA data respectively. (c) The correlation between the TMBIM6 mRNA expression and IDH1 mutation status in the TIMER2.0 database. (d) The correlation between the TMBIM6 mRNA expression and 1p/19q codeletion and non-codeletion by CGGA data.
Identification of co-expressed genes with TMBIM6
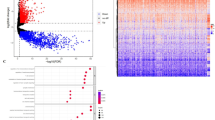
Co-expressed genes reveal complex molecular networks that regulate tumorigenesis in conjunction with TMBIM6. Analyzing these correlations provides valuable knowledge on disease mechanisms and potential therapeutic targets that may contribute to enhanced management strategies. Hence, data was mined from Linkedomics databases to screen for proteins interacting with TMBIM6. The results of the Linkedomics database showed that 4402, 5995 genes were significantly positively correlated with TMBIM6 and 6001, 6059 genes were negatively correlated (FDR < 0.01) in GBM and LGG respectively (Fig. 3a, d, Supplementary File 1, 2). The heat map shows the top 50 genes positively co-expressed with TMBIM6 in GBM (Fig. 3b) and LGG (Fig. 3e). In addition, top 50 negatively co-expressed genes with TMBIM6 in GBM (Fig. 3c) and in LGG (Fig. 3f) were presented through heatmap.
Identification of TMBIM6 co-expressed genes in glioma using LinkedOmics. (a) Volcano plot displaying TMBIM6 co-expressed genes in glioblastoma (GBM). (b,c) Heatmaps presented top 50 positively and negatively correlated genes with TMBIM6 in GBM. (d) Volcano plot displaying TMBIM6 co-expressed genes in lower-grade glioma (LGG). (e,f) Heatmaps presented top 50 positively and negatively correlated genes with TMBIM6 in LGG.
Enrichment analysis of co-expressed genes with TMBIM6
Enrichment analysis of co-expressed genes is crucial for discovering biological pathways and functional correlations, which could offer important insights into biomarker identification. Thus, GO and KEGG analysis were performed on the identified co-expressed genes using the LinkInterpreter module. In GBM, GO functional annotation analysis indicated that TMBIM6 predominantly involved in biological regulation and metabolic process, as shown in biological process categories (Fig. 4a). In terms of cellular components, TMBIM6 highly associated with membrane and nucleus (Fig. 4b). Additionally, molecular function analysis revealed that TMBIM6 participates in nucleic acid binding, ion binding and protein binding activities (Fig. 4c). KEGG pathway analysis presented that TMBIM6 was related to Protein processing in the endoplasmic reticulum, Ribosome, Peroxisome, NOD-like receptor signaling pathway, Alzheimer’s disease, lysosomes, phagosomes, Leishmaniasis pathways (Fig. 4d) in GBM. Similarly, In LGG samples, GO fuction annotation showed that TMBIM6 is highly associated with biological regulation and metabolic process (Fig. 4e), linked to the membrane and nucleus (Fig. 4f), and involved in protein binding, ion binding and nucleic acid binding activities (Fig. 4g). Further, KEGG pathway analysis showed that TMBIM6 was related to protein processing in the endoplasmic reticulum, NOD-like receptor signaling pathway, phagosome, lysosome, Cell adhesion molecules, peroxisome, hematopoietic cell lineage pathways (Fig. 4h) in LGG.
Association between TMBIM6 and immune cells
The analysis of immune cell infiltration in the Tumor microenvironment (TME) reveals the relationship between immune infiltration and TMBIM6 in glioma. A series of tests was conducted to investigate these relationships in GBM and LGG. xCELL algorithm, a ssGSEA-based method, analyzed the abundance of 34 types of immune and stromal cells in GBM and LGG. The analysis revealed that TH2 cells, macrophages, Tgd cells, Macrophage M2, Endothelial cells, CD4 memory T cells, macrophages, and monocytes were significantly correlated with high expression of TMBIM6. However, low TMBIM6 expression is correlated to only CD4 + Tcm, Eosinophil, and Tregs (Fig. 5a, b). Further, the CIBERSORT algorithm demonstrated a significant correlation between higher levels of TMBIM6 and macrophage M2 in LGG (Fig. 5c). Moreover, immune and stromal scores in GBM and LGG distinctly associated with TMBIM6 high expression (Fig. 5d, e).
Association between TMBIM6 and Immune cells. (a,b) The association between high/low expression of TMBIM6 with various immune cells analyzed by the xCELL algorithm in glioblastoma and lower-grade glioma. (c) correlation between M2 macrophages and TMBIM6 in lower grade glioma by CIBERSORT algorithm. (d,e) The correlation between high/low expression of TMBIM6 and Immune and stromal scores respectively in glioblastoma and lower-grade glioma.
Go and KEGG analysis of TMBIM6-related immune-modulatory genes
To investigate the possible immunomodulatory role of TMBIM6 in glioma, a number of studies were carried out using the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) and the TISIDB repository portal. In glioblastoma we identified 12 immunoinhibitory genes (ADORA2A, CD160, CD274, CD96, IDO1, IL10RB, LAG3, PDCD1, PDCD1LG2, PVRL2, VTCN1) and 13 immunostimulatory genes (CD48, CD86, CXCL12, KLRK1, LTA, NT5E, TNFRSF13C, TNFRSF25, TNFSF13, TNFSF13B, TNFSF4, TNFSF9) (Fig. 6a, Supplementary file 3). In case of lower grade glioma, we found 18 immunoinhibitory genes (ADORA2A, CD160, CD96, CSF1R, HAVCR2, IDO1, IL10, KDR, LAG3, LGALS9, PDCD1, PDCD1LG2, PVRL2, TGFB1, TGFBR1, TIGIT, VTCN1) AND 26 Immunostimulatory genes (C1orf54, CD27, CD276, CD28, CD40, CD48, CD86, CXCR4, ENTPD1, IL2RA, IL6R, KLRK1, MICB, NT5E, PVR, TMIGD27, TNFRSF14, TNFRSF25, TNFRSF4, TNFSF13, TNFSF13B, TNFSF4, TNFSF9) were statistically significant (Fig. 6c, Supplementary file 3). TMBIM6-related immunomodulators were then exposed to WebGestalt’s enrichment analysis. The KEGG analyses of these genes indicate that the immunological processes caused by TMBIM6 are associated with the JAK-STAT signaling pathway, pathways in cancer, PI3K-AKT signaling pathway, Th17 cell differentiation, and cytokine receptor interaction (Fig. 6b, d).
Analysis of TMBIM6-related immunomodulatory genes. (a) Heatmaps showing the immunoinhibitory (left), and immunostimulatory (right) genes in Glioblastoma. (b) KEGG analysis of TMBIM6- related immunomodulatory genes via WEbGestalt in GBM. (c) Heatmaps showing the Immunoinhibitory (left), and Immunostimulatory (right) genes in Lower grade glioma. (d) KEGG analysis of TMBIM6- related immunomodulatory genes via WEbGestalt in LGG.
Forecasting and assessment of potential miRNA candidates
The role of non-coding RNAs (ncRNAs) in controlling gene expression has been well recognized. Several target gene prediction algorithms, including PITA, RNA22, miRmap, microT, miRanda, PicTar, and TargetScan, were utilized to determine the upstream binding miRNAs of TMBIM6. Those candidate miRNAs identified in at least two databases were designated as TMBIM6 miRNAs and used for subsequent analysis. Following the identification of 13 miRNAs (Fig. 7a), an expression correlation study revealed that 5 miRNAs had a strong negative connection with TMBIM6 in Gliomas (Fig. 7b, Supplementary Fig. 2). 5 miRNAs were found significantly downregulated (Fig. 7b, d, Supplementray Fig. 2) and then the prognostic value also ascertained of these 5 miRNAs (Supplementary Fig. 3), among 5 miRNAs hsa-miR-128-3p showed its overexpression was positively linked with the prognosis of the patients, as illustrated in (Fig. 7c). Mechanistic suppression of TMBIM6 by miR-128-3p was experimentally validated by transient transfection in U87-MG cell line by checking mRNA level of TMBIM6 (Fig. 7e, Supplementary Table 6) which causes great impact on U87-MG cell’s viability (Fig. 7f, Supplementary Table 7). All these findings suggest that hsa-miR-128-3p might be a potential TMBIM6 regulating miRNA in glioma.
Identification of candidate miRNA of TMBIM6. (a) 13 miRNA candidates of TMBIM6 were identified by 7 programs (b) and 5 predicted negatively correlated miRNAs of TMBIM6 in glioma analyzed by StarBase. (c) The Kaplan Meier curves predicted significantly downregulated miRNA in glioma by StarBase. (d) Quantitative real-time PCR (qPCR) analysis of TMBIM6 mRNA expression in U87-MG cells at 48 h after transfection with has-miR-128 mimic alone or in combination with miR-128-3p inhibitor or miR-128-5p inhibitor. GAPDH was used as an internal control. (e) Cell viability of U87-MG cells were assessed by MTT assay at 48 h of post-transfection with the same conditions as in (d). Data were presented as mean ± SEM of three independent experiments. Statistical significance was determined using one way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
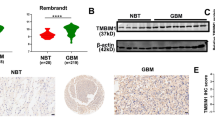
Experimental validation by immunohistochemistry
TMBIM6 expression in gliomas and normal brain tissues was initially evaluated using the HPA database. In normal brain tissue, TMBIM6 expression was not detected in glial cells. In contrast, three out of four LGG (low-grade glioma) samples showed medium levels of TMBIM6 expression, and six out of ten GBM (glioblastoma multiforme) samples exhibited low to medium expression levels (Fig. 8a). To validate these findings, we performed Immunohistochemistry (IHC) on a Tissue Microarray (TMA) of glioma patient samples. Consistent with the database results, IHC analysis confirmed the increased expression of TMBIM6 in glioma tissues compared to normal brain tissues (Fig. 8b). A paraffin-embedded human liver tissue slide was used as a positive control for TMBIM6 IHC staining (Fig. 8b). Furthermore, we investigated the expression of the M2 macrophage marker CD163 by IHC analysis. We observed significantly higher expression of CD163 in both GBM and LGG tissues compared to normal brain tissues (Fig. 8c). A human lung tissue slide served as the positive control for CD163 staining (Fig. 8c). This result is consistent with our immune infiltration analysis, which demonstrated that elevated TMBIM6 expression is significantly associated with increased M2 macrophage infiltration in gliomas. Previous studies have similarly reported that M2 macrophage infiltration promotes tumor progression and enhances cancer cell survival and metastasis19,20. To further strengthen our findings, we conducted additional functional validation experiments. Knockdown of TMBIM6 using siRNA in the U87-MG glioma cell line led to a significant reduction in TMBIM6 mRNA expression (Fig. 8d) and a marked decrease in cell viability as assessed by MTT assay (Fig. 8e). These results support the role of TMBIM6 in promoting glioma cell survival and further validate its oncogenic potential in glioma progression.
Experimental validation by immunohistochemistry and TMBIM6’s siRNAs tranfection. (a) TMBIM6 expression was not detected in normal brain cortex tissue, 3 of 4 lower-grade glioma (LGG) cases showed expression of TMBIM6, and 6 of 10 high-grade glioma (GBM) showed expression of TMBIM6. (b) Immunohistochemical analysis of TMBIM6 protein expression in Glioma compared to normal by glioma patients’ microarray slide. Healthy human Liver tissues were used as positive control. (c) M2 macrophage’s marker CD163 expression in Glioma compared to normal brain by immunohistochemical analysis on glioma patients’ microarray slide. Healthy human lung tissues were considered as positive control for CD163. (d) Quantitative real-time PCR (qPCR) analysis of TMBIM6 mRNA expression in U87-MG cells 48 h after transfection with two different TMBIM6 siRNAs and a negative control siRNA. GAPDH was used as an internal control. (e) Cell viability of U87-MG cells determined by MTT assay 48 h post-transfection under the same conditions as in (d). Data are presented as mean ± SEM of three independent experiments. Statistical significance was determined using Student’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
Discussion
Prognostic biomarkers in patients with glioma are essential in guiding treatment decisions and forecasting disease outcomes. The identification of biomarkers could offer crucial information about the molecular features of cancers, which enables the development of individualized and focused treatment plans. Previous approaches for searching biomarkers for breast and ovarian tumors appreciated us to dig the association of TMBIM6 in glioma21,22. For instance, markers such as IDH mutations and 1p/19q codeletion offer vital information about tumor categorization and patient prognosis. Hence, it is essential to possess biomarkers specific to glioma to obtain early identification and make appropriate treatment decisions. This study orchestrated with different bioinformatics analyses to explore the role of TMBIM6 in tumor progression and prognosis in glioma. The study results suggest that TMBIM6 can serve as a prognostic biomarker in glioma.
Biological data is a valuable resource for modern medical research, offering an unparalleled understanding of human health and disease. It helps uncover complex molecular mechanisms responsible for diseases like cancer. Here, data analyses via multiple bioinformatic tools identify significant differences in TMBIM6 expression between glioma patients and normal individuals. (Fig. 1a-d). This differential expression in TMBIM6 indicated that it may play a major role in glioma progression and proliferation. Consequently, several studies were carried out to investigate the potential function of TMBIM6 in GBM, where it could serve as a marker. The analysis of TMBIM6 expression and its correlation with patient survival showed that lower expression levels were associated with better survival rates (Fig. 2a, b). Moreover, TMBIM6 high expression was positively correlated with IDH1 status and 1p/19q codeletion (Fig. 2c, d). These observations create more confidence to propose the prognostic role of TMBIM6 in glioma. Remarkably, IDH1 mutations and 1p/19q codeletion provide critical insights into the prognosis of patients23. As observations demonstrate a strong association between TMBIM6 and IDH mutations and 1p/19q codeletion, TMBIM could serve as a marker.
Therefore, co-expression analysis was conducted to understand the function of TMBIM6 in the context of glioma, and genes that are positively and negatively co-expressed with TMBIM6 were identified. Further, KEGG pathway analysis findings showed TMBIM6 positively associated with protein processing in the endoplasmic reticulum and NOD-like receptor signaling pathway (Fig. 5d, h). The heightened expression of specific genes linked to protein processing in ER demonstrates a notable association with the progression of glioma. Interestingly, analysis of patient survival data from TCGA by OncoLnc tool confirms a substantial decrease in survival probabilities due to elevated expression of genes such as CUL1, HSP90B1, HSPA5, PDIA4, SEC61G, and SIL1 in gliomas (Supplementary Fig. 1). Remarkably, these genes have a substantial impact not just on gliomas but also on a number of cancers, influencing crucial functions in cancer cells such as survival, proliferation, migration, and invasion24,25,26,27,28,29,30,31. Notably, PDIA4 and SIL1, akin to TMBIM6, function within the PI3K/AKT/mTOR pathway, contributing significantly to glioma progression11,31,32. These findings underscore the potential utility of TMBIM6 as a prognostic biomarker in glioma management strategies, reflecting its promise in guiding therapeutic interventions. Research evidenced that the NOD-like receptor signaling pathway is associated with glioma33. Previous reports by Alzahrani et al. suggest that in the case of GBM, upregulation of NOD-like receptors is linked with poor prognosis and accelerates the epithelial-mesenchymal (EMT) signaling34. Consequently, the NOD-like receptor signaling pathway may play a crucial role in initiating and progressing of glioma, suggesting it might be a potential target for further investigation.
Immune cells are an essential part of the glioma microenvironments and constitute up to 50% of the total mass35. In this study, infiltration analysis results showed that the high expression of TMBIM6 significantly correlated with macrophages M2, endothelial cells, Tgd cells, cd4 + memory T cells, Monocytes, and TH2 cells (Fig. 5a, b). Previous studies have indicated the significant involvement of Macrophages M2 in the progression of glioma36,37,38. Consequently, the relationship between TMBIM6 and macrophage M2 in LGG was evaluated using the CIBERSORT algorithm. The analysis showed a positive correlation of TMBIM6 with macrophage M2. Endothelial cells within brain tumors are crucial for tumor metastasis and angiogenesis, shaping the tumor microenvironment, and highlighting the critical role in tumor progression and pathobiology39,40,41. These observations on the data suggest TMBIM6 might have a significant function in the progression of glioma and metastasis.
Furthermore, the immune and stromal scores were assessed in conjunction with the expression levels of TMBIM6 in gliomas. Stromal cells, such as cancer-associated fibroblasts and endothelial cells, are implicated in influencing the rigidity, invasive potential, and metastatic dissemination of tumors42. In this investigation, a striking augmentation in stromal score has been discerned concomitant with a notable increase in TMBIM6 gene expression within metastatic brain tumors, particularly in GBM and LGG (Fig. 5d, e). The significant increase in stromal score indicates a direct correlation between the expression of TMBIM6 and the disturbances in the stromal microenvironment in GBM and LGG. Interestingly, despite the comparatively lower metastatic propensity of LGG in contrast to GBM, the augmented immune score also persists, emphasizing the substantial influence of TMBIM6 expression on the immune composition within the tumor microenvironment (Fig. 5d). Furthermore, the positive correlation observed between TMBIM6 gene expression and tumor angiogenesis factors (Supplementary file1,2), such as vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF), underscores the potential significance of TMBIM6 as a pivotal regulator of angiogenic processes in glioma pathogenesis43. Moreover, by fostering an environment that is conducive to tumor growth and metastasis, M2 macrophages contribute significantly to the survival, proliferation, and spread of cancer cells19. Tumor cells frequently attract and activate these macrophages, which then secrete growth factors, enzymes, and anti-inflammatory cytokines that encourage angiogenesis, tissue remodeling, and immune suppression19. M2 macrophages promote metastasis by allowing tumor cells to infiltrate surrounding tissues and enter the bloodstream through the breakdown of the extracellular matrix20. They also inhibit the function of immune responses such as cytotoxic CD8 + T cells, which would typically target and eliminate tumor cells; as a result, the tumor is able to elude the immune system and spread more quickly44. Our analysis suggested that TMBIM6 expression is positively correlated with M2 macrophage infiltration in Glioma (Rho = 0.055) (Fig. 5c). In tissue microarray slide of Glioma patients and normal cerebrum samples, M2 macrophage marker i.e., CD163 was significantly upregulated in GBM and LGG patients (Fig. 8c) which follows a similar pattern of expression level of TMBIM6 in GBM and LGG (Fig. 8b). Considering the suggested function of TMBIM6 in orchestrating stromal dynamics and M2 macrophage recruitment, TMBIM6 could be an ideal candidate for predicting the prognosis of glioma patients.
Research suggests that ncRNAs, such as miRNAs, cirRNAs, and long non-coding RNAs (lncRNAs), may promote the invasive and metastatic potential of glioma, thereby impacting the progression of cancer45. In this study, 5 miRNAs had a strong negative relationship with TMBIM6 in LGG (Supplementary Fig. 2). Of those 5 miRNAs, hsa-miR-128-3p was identified as the upstream regulator of TMBIM6. Also, Patients with LGG who had higher levels of hsa-miR-128-3p miRNA had a better prognosis. Research has demonstrated that hsa-miR-128-3p inhibits the proliferation, invasion, and differentiation by PI13/AKT signaling pathway46while TMBIM6 elevated the cancer progression via mTORC2 and AKT activation11. In our study, miR-128 mimic transfection along with matured 3p and 5p inhibitor has shown that significant down regulation of TMBIM6 in response with miR-128-3p (Fig. 7d). Consequently, these results provide strong evidence that miR-128-3p has the potential to function as a tumor-suppressive miRNA of TMBIM6 in glioma.
On the contrary, the heterogeneity of gliomas poses a significant challenge in their management, as these tumors exhibit diverse genetic, histopathological, and immunological characteristics47. This variability is not only observed between different patients but also within different regions of the same tumor, making it difficult to develop universal biomarkers or treatments47,48. The dynamic nature of gliomas, with changes in tumor behavior over time and across different areas, further complicates treatment strategies47. As a result, personalized approaches that consider the unique profile of each tumor are essential for effective glioma management49. Even though, a continuous search for an effective biomarker as like as TMBIM6 could open a new path to overcome the hurdles of glioma management.
In conclusion, a series of multidimensional analyses were performed to investigate the role and prognosis efficiency of TMBIM6 in glioma. The higher expression of TMBIM6 in glioma plays a crucial role in promoting tumor progression and metastasis by activating or inhibiting multiple signaling pathways, lowering overall survival in glioma patients. In addition, TMBIM6 is strongly associated with tumor immune and stromal microenvironment, influencing the regulation of immune infiltration in glioma. Together, the study provides novel insights into the association of TMBIM6 with factors influencing the prognosis of GBM and LGG, suggesting its potential applications as a prognostic biomarker for glioma. Previous research has established that TMBIM6 is a gene implicated in cancer progression11. The influence of TMBIM6 in metastatic tumors such as GBM and LGG demonstrates that inhibiting TMBIM6 could represent a significant advancement in the treatment of these two types of brain tumors. According to the findings of Hyun-Kyoung Kim et al., the contribution of TMBIM6 antagonists in the treatment of various cancers is noteworthy11. Considering this contribution, TMBIM6 antagonists may be utilized as a co-therapy alongside conventional glioma therapies in the future11. However, still there are limitations in our study, the gene expression dataset had a lower number of healthy samples than tumor samples, which meant more research was needed to keep the sample size in balance. This study is based on bioinformatic approaches and immunohistochemical detection, more research is needed to clarify the pathological roles of TMBIM6 and the underlying molecular mechanism by subsequent experiments.
Methods
Data collection and preprocessing
TCGA is an open-access database with data from over 11,000 samples across 39 tumor types50 that were accessed to obtain data on TCGA-GBM and TCGA-LGG, RNA-seq STAR-Counts gene expression data and clinical information were retrieved via TCGAbiolinks using the R program (version 4.3.2), within TCGAbiolinks, GDCquery_clinic and summarizedExperiment functions were used to search, download, and analyze clinical data51. The collected data were processed and normalized by R software for downstream analysis. In addition, GEO, an international public data repository for microarray and NGS functional genomic data52 was accessed using the GEOquery function and retrieved the gene expression and clinical data of GSE4290. Then the dplyr function preprocessed the collected data and performed further analysis. Similarly, CGGA, a data portal featuring 2000 primary and recurrent glioma samples from a Chinese cohort53was used to collect FPKM gene expression and clinical data on glioma (dataset ID: mRNAseq_693). The GEO and TCGA data were accessed and downloaded on 10 September, 2023 and CGGA data was collected on 15 January, 2024.
Expression analysis
First, we observed the expression of TMBIM6 in different cancers compared to normal tissues using Gene Expression Profiling Interactive Analysis 2 (GEPIA2), an online tool for analyzing transcriptional profiles by using the TCGA database and the GTEx projects (http://gepia.cancer-pku.cn/)54. Secondly, the web tool Biomarker Exploration of Solid Tumors (BEST) was used to investigate the expression level of TMBIM6 in glioma and normal tissues (https://rookieutopia.hiplot.com.cn/app_direct/BEST/). Additionally, expression levels in glioma and normal tissues were compared using edgeR and limma functions within the R program. Besides, the immunohistochemical images from the Human Protein Atlas (HPA) database were used to analyze the differential expression level of TMBIM6 in GMB compared to normal tissues55.
Survival prognosis analysis
TMBIM6 gene expression and overall survival were examined in patients with TCGA-GBM and TCGA-LGG. The survival data of GBM and LGG were acquired from Oncolnc, and subsequently performed analysis of overall survival by using GraphPad Prism. Similarly, overall survival analysis was performed using CGGA survival data. The samples with partial information were removed from the analysis. The FPKM CGGA data were again used to analyze the TMBIM6 expression in relation to the IDH mutation status and 1p/19q codeletion.
Co-expression analysis
To determine the genes that are co-expressed with TMBIM6 in glioma, we performed an analysis using the LinkedOmics database (http://www.linkedomics.org/login.php). This database includes multiomics information from 11,158 participants from the TCGA program, comprising 32 types of cancer56. The TMBIM6 co-expressed genes were identified using a Spearman correlation test. The LinkFinder module was used to analyze the association within and across data types. The collected data was analyzed to identify both positively and negatively co-expressed genes.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analysis
The LinkInterpreter module was used to perform Gene Set Enrichment Analysis (GSEA), which includes the GO database from the Cellular Component (CC), Biological Process (BP), Molecular Function (MF), and KEGG pathways.
Immune cell association analysis
The collected CGGA data was preprocessed and separated into two categories based on the level of TMBIM6 expression (High/low). Next, grouped data was analyzed to confirm the association between the immune and stromal cells and the level of TMBIM6 expression by using the xCELL algorithm, a bioinformatics tool that analyzes cell type enrichment from gene expression data57. Here, the investigation was focused on analyzing 34 specific types of immune and stromal cells in glioma. In addition, the immunological and stromal scores were calculated. The association between M2 macrophages and TMBIM6 expression in glioma TMBIM6 was analyzed by TIMER2, (https://cistrome.shinyapps.io/timer/) is useful for exploring the molecular characteristics of tumor-immune interactions and the levels of immune-infiltration tumors from 32 different forms of cancer58. Consecutively, to determine the associations between the immunomodulators and TMBIM6 expression in glioma we used the immunomodulator module of TISIDB (http://cis.hku.hk/TISIDB/), which is a user-friendly web portal that combines several forms of cancer-immunology research data59. Besides, enrichment analysis of relevant immunomodulators was performed using the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt).
miRNA prediction analysis
Multiple target gene prediction algorithms, including PITA, RNA22, miRmap, microT, miRanda, PicTar, and TargetScan were used to determine the upstream binding miRNAs of the TMBIM6. Keeping the study objective in focus, only the projected miRNAs frequently appearing in more than two programs listed previously were considered for further analysis. The projected miRNAs were thought to be TMBIM6 candidate miRNAs. Thus, StarBase was used for correlation analysis for miRNA-TMBIM6.
Immunohistochemistry
Two tissue microarray (TMA) slides of brain tumor sections (#GL2082a) were obtained from TissueArray.com. This TMA panel included 48 astrocytoma (lower-grade glioma; LGG), 24 glioblastoma (GBM), and 8 normal brain tissue samples. Paraffin blocks of human liver and lung tissues were used as positive controls for TMBIM6 and CD163, respectively. Positive control samples were procured from the National Biobank of Korea under informed consent and IRB (Institutional Review Board)-approved protocols (IRB approval number: 2025-04-049). Immunohistochemistry of the TMA was carried out as described in the previous report60. The sample underwent deparaffinization and rehydration processes, followed by heat-induced epitope retrieval utilizing Dako Target Retrieval Solution with a pH of 9 for TMBIM6 and pH of 6 for CD163 (Agilent Technologies, CA, USA). Subsequently, the section was treated with Dako REAL peroxidase blocking solution (#S2023, Agilent Technologies, CA, USA) at room temperature. Then, the section was rinsed with TBS-T buffer and blocked using serum-free protein block solution (#X0909, Agilent Technologies, CA, USA) for 15 min. The TMAs were then incubated overnight at 4°C with a 1:100 diluted primary antibody against TMBIM6 (#MA1-41108, Invitrogen, USA) and CD163 (#NB110-59935, Novus Biologicals, USA). The primary antibody was prepared using Dako Antibody Diluent (#S3022, Agilent Technologies, CA, USA). The following day, the section was washed with TBS-T and incubated with Envision + System-HRP labeled Polymer anti-mouse secondary antibody (#K4001, Agilent Technologies, CA, USA) for 1 h. Subsequently, AEC chromogen (#ACD015/ACE500, ScyTek Laboratories, Utah, USA) was used to develop color and stained with hematoxylin. All the observations were captured using an EVOS M5000 fluorescence microscope (ThermoFisher Scientific, USA). All the images were magnified to a scale of 40X and further analyzed using Image J. (NIH, USA).
Cell culture
U87-MG human glioblastoma cells were generously provided by Professor Sung-Ae Hyun from the Department of Advanced Toxicology Research, Korea Institute of Toxicology (KRICT), Daejeon, Republic of Korea. Cell were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin-streptomycin (Gibco, USA), and maintained at 37°C in a humidified atmosphere with 5% CO2.
Transfection with siRNA and miRNA reagents
For transfection, cells were seeded in 6 cm dishes, 96 well plates and 12- well plates and grown to approximately 50–70% confluency. siRNAs targeting TMBIM6 (two distinct sequences) and siRNA negative control, along with miRNA reagents including miRNA negative control, miR-128 mimic, miR-128-3p inhibitor, and miR-128-5p inhibitor, were obtained from Bioneer Corporation, Daejeon, Republic of Korea (Order no.: Q-250403-0040). Transfection were carried out using Lipofectamine® RNAiMAX Reagent (#13778-150, ThermoFisher Scientific, USA) according to manufacturer’s protocol. In brief, the siRNAs or miRNA reagents were diluted in Opti-MEM® Reduced Serum Medium (Gibco, USA) and mixed with Lipofectamine RNAiMAX. After a 10–20 min incubation at room temperature, the complexes were added to the cells. The final concentrations were optimized as per the supplier’s recommendations. Cells were incubated for 48 h post-transfection and subjected to downstream analyses such as qPCR and MTT assay.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from U87-MG cells 48 h after transfection using TRIzol™ Reagent (#15596-026, ThermoFisher Scientific, USA), following the manufacturer’s protocol. RNA concentration and purity were measured by using DeNovix™ Spectrophotometer (#DS-11+, DeNovix Inc., USA). For cDNA synthesis, 1 µg of total RNA was reverse transcribed by using the PrimeScript™ RT Reagent Kit (#RR037A, Takara, Japan), according to manufacturer’s instructions. Quantitative PCR was preformed by using the AMPIGENE® qPCR Green Mix (#ENZ-NUC126-100) on a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, USA). The expression level of TMBIM6 was normalized to GAPDH as the internal control. Relative gene expression was analyzed using the 2(−ΔΔCT) method. All reactions were carried out in triplicate. Primers for human TMBIM6 and GAPDH were purchased from the AccuTarget™ Real-Time PCR Primer Library (#P145301, P267613; Bioneer Corporation, Daejeon, Korea).
MTT assay
Cell viability after the transfection was assessed by using the MTT assay. At 48 h post-transfection, 10 µL of MTT solution (5 mg/mL in PBS) (#475989, Sigma-Aldrich, USA) was added to each well of the 96-well plate containing U87-MG cells. The cells were incubated for an additional 4 h at 37 o C. After incubation, the media containing MTT was carefully removed, and 150 µL of Dimethyl sulfoxide (#D2650, Sigma-Aldrich, USA) was added to each well to dissolve the formazan crystals. Absorbance was measured at 570 nm using a microplate reader (Multiskan SkyHigh, ThermoScietific, USA). Cell viability was calculated as a percentage relative to the negative control group. All experiments were performed in triplicate.
Statistical analysis
All the statistical analyses were performed using GraphPad Prism and R software. Specifically, the Pearson correlation was used during co-expression analysis, while the log-rank test was conducted to create Kaplan-Meier curves during survival analysis. The unpaired t-test and two-way ANOVA were used to compare two groups and multiple groups, respectively. p < 0.05 was considered statistically significant.
Data availability
TCGA-GBM and TCGA-LGG Dataset are obtained by R software from GDC data portal (https://portal.gdc.cancer.gov/). GEO data GSE4290 was obtained by R software from (https://www.ncbi.nlm.nih.gov/geo/). CGGA data were downloaded from the website (http://www.cgga.org.cn/). Multidimensional analysis was performed by different web tool: GEPIA2 (http://gepia.cancer-pku.cn/), for differential expression analysis of TMBIM6 in different cancers.BEST (https://rookieutopia.hiplot.com.cn/app_direct/BEST/), for differential expression analysis of TMBIM6 in GBM and LGG.LinkedOmics (http://www.linkedomics.org/login.php), for co-expression analysis, GO and KEGG analysis.TIMER2 (https://cistrome.shinyapps.io/timer/), for correlation analysis of TMBIM6 with immune cell.TICIDB (http://cis.hku.hk/TISIDB/), to determine the associations between the immunomodulators and TMBIM6 expression.WebGestalt (http://www.webgestalt.org/), for enrinhment analysis.StarBase (http://starbase.sysu.edu.cn/), to predict miRNA prediction and analysis.
References
Yu, H., Wang, M., Wang, X. & Jiang, X. Immune-related matrisomes are potential biomarkers to predict the prognosis and immune microenvironment of glioma patients. FEBS Open. Bio. 13, 307–322. https://doi.org/10.1002/2211-5463.13541 (2023).
Kovic, B. & Xie, F. Economic evaluation of bevacizumab for the First-Line treatment of newly diagnosed glioblastoma multiforme. J. Clin. Oncol. 33, 2296–2302. https://doi.org/10.1200/JCO.2014.59.7245 (2015).
Kfoury, N. et al. Brd4-bound enhancers drive cell-intrinsic sex differences in glioblastoma. Proc. Natl. Acad. Sci. U. S. A. 118 https://doi.org/10.1073/pnas.2017148118 (2021).
Wirsching, H. G., Galanis, E., Weller, M. & Glioblastoma Handb. Clin. Neurol. 134, 381–397. https://doi.org/10.1016/B978-0-12-802997-8.00023-2 (2016).
Sarmiento, J. M., Venteicher, A. S. & Patil, C. G. Early versus delayed postoperative radiotherapy for treatment of low-grade gliomas. Cochrane Database Syst. Rev. 6, CD009229. https://doi.org/10.1002/14651858.CD009229.pub2 (2015).
Penman, C. L., Faulkner, C., Lowis, S. P. & Kurian, K. M. Current Understanding of BRAF alterations in diagnosis, prognosis, and therapeutic targeting in pediatric Low-Grade gliomas. Front. Oncol. 5, 54. https://doi.org/10.3389/fonc.2015.00054 (2015).
Chammas, M., Saadeh, F., Maaliki, M. & Assi, H. Therapeutic interventions in adult Low-Grade gliomas. J. Clin. Neurol. 15, 1–8. https://doi.org/10.3988/jcn.2019.15.1.1 (2019).
Walter, L., Marynen, P., Szpirer, J., Levan, G. & Gunther, E. Identification of a novel conserved human gene. TEGT Genom. 28, 301–304. https://doi.org/10.1006/geno.1995.1145 (1995).
Guarnaccia, L. et al. Angiogenesis in human brain tumors: screening of drug response through a patient-specific cell platform for personalized therapy. Sci. Rep. 8, 8748. https://doi.org/10.1038/s41598-018-27116-7 (2018).
Xu, Q. & Reed, J. C. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell. 1, 337–346. https://doi.org/10.1016/s1097-2765(00)80034-9 (1998).
Kim, H. K. et al. TMBIM6/BI-1 contributes to cancer progression through assembly with mTORC2 and AKT activation. Nat. Commun. 11, 4012. https://doi.org/10.1038/s41467-020-17802-4 (2020).
Shin, Y. et al. TMBIM6-mediated miR-181a expression regulates breast cancer cell migration and invasion via the MAPK/ERK signaling pathway. J. Cancer. 14, 554–572. https://doi.org/10.7150/jca.81600 (2023).
Wang, X. et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J. Exp. Clin. Cancer Res. 40, 80. https://doi.org/10.1186/s13046-021-01871-4 (2021).
Jin, Z. et al. Long non-coding RNA plasmacytoma variant translocation 1 (PVT1) promotes glioblastoma multiforme progression via regulating miR-1301-3p/TMBIM6 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 11658–11665. https://doi.org/10.26355/eurrev_202011_23810 (2020).
Kaluzinska-Kolat, Z., Kolat, D., Kosla, K., Pluciennik, E. & Bednarek, A. K. Molecular landscapes of glioblastoma cell lines revealed a group of patients that do not benefit from WWOX tumor suppressor expression. Front. Neurosci. 17, 1260409. https://doi.org/10.3389/fnins.2023.1260409 (2023).
Li, Y. et al. IDH1 mutation is associated with a higher preoperative seizure incidence in low-grade glioma: A systematic review and meta-analysis. Seizure 55, 76–82. https://doi.org/10.1016/j.seizure.2018.01.011 (2018).
Mu, L. et al. The IDH1 Mutation-Induced oncometabolite, 2-Hydroxyglutarate, May affect DNA methylation and expression of PD-L1 in gliomas. Front. Mol. Neurosci. 11, 82. https://doi.org/10.3389/fnmol.2018.00082 (2018).
Ohka, F. et al. The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS One. 6, e23332. https://doi.org/10.1371/journal.pone.0023332 (2011).
Wang, S. et al. Targeting M2-like tumor-associated macrophages is a potential therapeutic approach to overcome antitumor drug resistance. NPJ Precis. Oncol. 8, 31. https://doi.org/10.1038/s41698-024-00522-z (2024).
Jayasingam, S. D. et al. Evaluating the polarization of Tumor-Associated macrophages into M1 and M2 phenotypes in human Cancer tissue: technicalities and challenges in routine clinical practice. Front. Oncol. 9, 1512. https://doi.org/10.3389/fonc.2019.01512 (2019).
Pullikuth, A. K. et al. Bulk and Single-Cell profiling of breast tumors identifies TREM-1 as a dominant immune suppressive marker associated with poor outcomes. Front. Oncol. 11, 734959. https://doi.org/10.3389/fonc.2021.734959 (2021).
Jiang, Y. et al. ROS-mediated SRMS activation confers platinum resistance in ovarian cancer. Oncogene 42, 1672–1684. https://doi.org/10.1038/s41388-023-02679-6 (2023).
Anand, N., Husain, N., Varshney, R., Malhotra, K. P. & Kaif, M. Molecular classification and stratification of adult diffuse gliomas: A tertiary care center study. J. Carcinog. 20, 20. https://doi.org/10.4103/jcar.jcar_17_21 (2021).
Fan, Y. C. et al. Cullin1 regulates proliferation, migration and invasion of glioma cells. Med. Oncol. 31 https://doi.org/10.1007/s12032-014-0227-x (2014).
Meenakshi Sundaram, D. N., Kc, R. B. & Uludag, H. Linoleic-acid-substituted polyethylenimine to silence heat shock protein 90B1 (HSP90B1) to inhibit migration of breast cancer cells. J. Gene Med. 24, e3419. https://doi.org/10.1002/jgm.3419 (2022).
Li, Y. et al. HSP90B1-mediated plasma membrane localization of GLUT1 promotes radioresistance of glioblastomas. J. Biomed. Res. 37, 326–339. https://doi.org/10.7555/JBR.37.20220234 (2023).
Zhang, C. et al. HSPA5 Could Be a Prognostic Biomarker Correlated with Immune Infiltration in Breast Cancer. Dis. Mark. 7177192. https://doi.org/10.1155/2022/7177192 (2022).
Wang, Z., Zhang, H. & Cheng, Q. PDIA4: the basic characteristics, functions and its potential connection with cancer. Biomed. Pharmacother. 122, 109688. https://doi.org/10.1016/j.biopha.2019.109688 (2020).
Zeng, K. et al. SEC61G assists EGFR-amplified glioblastoma to evade immune elimination. Proc. Natl. Acad. Sci. U. S. A. 120, e2303400120. https://doi.org/10.1073/pnas.2303400120 (2023).
Liu, B. et al. Identification of SEC61G as a novel prognostic marker for predicting survival and response to therapies in patients with glioblastoma. Med. Sci. Monit. 25, 3624–3635. https://doi.org/10.12659/MSM.916648 (2019).
Xu, H., Xu, S., Zhang, R., Xin, T. & Pang, Q. SIL1 functions as an oncogene in glioma by akt/mtor signaling pathway. Onco Targets Ther. 11, 3775–3783. https://doi.org/10.2147/OTT.S167552 (2018).
Wang, M. et al. PDIA4 promotes glioblastoma progression via the PI3K/AKT/m-TOR pathway. Biochem. Biophys. Res. Commun. 597, 83–90. https://doi.org/10.1016/j.bbrc.2022.01.115 (2022).
Yin, X. F. et al. NLRP3 in human glioma is correlated with increased WHO grade, and regulates cellular proliferation, apoptosis and metastasis via epithelial-mesenchymal transition and the PTEN/AKT signaling pathway. Int. J. Oncol. 53, 973–986. https://doi.org/10.3892/ijo.2018.4480 (2018).
Alzahrani, F. A., Khan, M. F. & Ahmad, V. Recognition of differentially expressed molecular signatures and pathways associated with COVID-19 poor prognosis in glioblastoma patients. Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms24043562 (2023).
Gargini, R., Segura-Collar, B. & Sanchez-Gomez, P. Cellular plasticity and tumor microenvironment in gliomas: the struggle to hit a moving target. Cancers (Basel). 12 https://doi.org/10.3390/cancers12061622 (2020).
Li, M. et al. Tumor-derived exosomes deliver the tumor suppressor miR-3591-3p to induce M2 macrophage polarization and promote glioma progression. Oncogene 41, 4618–4632. https://doi.org/10.1038/s41388-022-02457-w (2022).
Komohara, Y., Ohnishi, K., Kuratsu, J. & Takeya, M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 216, 15–24. https://doi.org/10.1002/path.2370 (2008).
Liu, L. et al. RNA-binding protein DHX9 promotes glioma growth and tumor-associated macrophages infiltration via TCF12. CNS Neurosci. Ther. 29, 988–999. https://doi.org/10.1111/cns.14031 (2023).
Bejarano, L. et al. Interrogation of endothelial and mural cells in brain metastasis reveals key immune-regulatory mechanisms. Cancer Cell. 42, 378–395. https://doi.org/10.1016/j.ccell.2023.12.018 (2024).
He, Q. et al. circ-SHKBP1 regulates the angiogenesis of U87 Glioma-Exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol. Ther. Nucleic Acids. 10, 331–348. https://doi.org/10.1016/j.omtn.2017.12.014 (2018).
Yang, D., Guo, P., He, T. & Powell, C. A. Role of endothelial cells in tumor microenvironment. Clin. Transl. Med. 11, e450. https://doi.org/10.1002/ctm2.450 (2021).
Cai, X. et al. Glioma-Associated stromal cells stimulate glioma malignancy by regulating the tumor immune microenvironment. Front. Oncol. 11, 672928. https://doi.org/10.3389/fonc.2021.672928 (2021).
Jensen, R. L. Growth factor-mediated angiogenesis in the malignant progression of glial tumors: a review. Surg. Neurol. 49, 189–195. https://doi.org/10.1016/s0090-3019(97)00218-8 (1998).
Shen, L. et al. Crosstalk between macrophages, T cells, and Iron metabolism in tumor microenvironment. Oxid. Med. Cell. Longev. 2021 (8865791). https://doi.org/10.1155/2021/8865791 (2021).
Goenka, A. et al. The role of Non-Coding RNAs in glioma. Biomedicines 10 https://doi.org/10.3390/biomedicines10082031 (2022).
Huo, L. et al. miR-128-3p inhibits glioma cell proliferation and differentiation by targeting NPTX1 through IRS-1/PI3K/AKT signaling pathway. Exp. Ther. Med. 17, 2921–2930. https://doi.org/10.3892/etm.2019.7284 (2019).
Tirosh, I. & Suva, M. L. Tackling the many facets of glioblastoma heterogeneity. Cell. Stem Cell. 26, 303–304. https://doi.org/10.1016/j.stem.2020.02.005 (2020).
Sledzinska, P., Bebyn, M. G., Furtak, J., Kowalewski, J. & Lewandowska, M. A. Prognostic and predictive biomarkers in gliomas. Int. J. Mol. Sci. 22 https://doi.org/10.3390/ijms221910373 (2021).
Roncevic, A. et al. Personalized treatment of glioblastoma: current state and future perspective. Biomedicines 11 https://doi.org/10.3390/biomedicines11061579 (2023).
Tomczak, K., Czerwinska, P. & Wiznerowicz, M. The Cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn). 19, A68–77. https://doi.org/10.5114/wo.2014.47136 (2015).
Mounir, M. et al. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 15, e1006701. https://doi.org/10.1371/journal.pcbi.1006701 (2019).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–995. https://doi.org/10.1093/nar/gks1193 (2013).
Zhao, Z. et al. Chinese glioma genome atlas (CGGA): A comprehensive resource with functional genomic data from Chinese glioma patients. Genom. Proteom. Bioinf. 19, 1–12. https://doi.org/10.1016/j.gpb.2020.10.005 (2021).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560. https://doi.org/10.1093/nar/gkz430 (2019).
Uhlen, M. et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 4, 1920–1932. https://doi.org/10.1074/mcp.M500279-MCP200 (2005).
Vasaikar, S. V., Straub, P., Wang, J. & Zhang, B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963. https://doi.org/10.1093/nar/gkx1090 (2018).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220. https://doi.org/10.1186/s13059-017-1349-1 (2017).
Li, T. et al. A web server for comprehensive analysis of Tumor-Infiltrating immune cells. Cancer Res. 77, e108–e110. https://doi.org/10.1158/0008-5472.CAN-17-0307 (2017).
Ru, B. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202. https://doi.org/10.1093/bioinformatics/btz210 (2019).
Hoang, T. H. et al. IBF-R, a botanical extract of Rhus verniciflua controls obesity in which AMPK-SIRT1 axis and ROS regulatory mechanism are involved in mice. J. Funct. Foods. 87 https://doi.org/10.1016/j.jff.2021.104804 (2021).
Acknowledgements
This work was supported by National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (Grant numbers: 2023R1A2C2003446, RS-2023-00225239). The biospecimens (human liver and lung tissue blocks) for this study were provided by the Jeonbuk National University Hospital Biobank, Jeonju-si, Republic of Korea. The Biobank is a member of the Korea Biobank Network, supported by the Ministry of Health, Welfare and Family Affairs, Republic of Korea. All samples were derived from the Natioinal Biobank of Korea were obtained with informed consent under Institutional Review Board- approved protocols (2025-04-049). We gratefully acknowledge that Professor Sung-Ae Hyun, Department of Advanced Toxicology Research, Korea Institute of Toxicology (KRICT), Daejeon, Republic of Korea, for generously providing the U87-MG cell line.
Author information
Authors and Affiliations
Contributions
M.S.K: Conceptualization, Methodology, Investigation, Data curation, Software, Validation, Writing- original draft. M.M.U.R: Conceptualization, Methodology, Investigation, Validation, Writing-review & editing. A.U: Investigation, H.R.K: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing—review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Khatun, M.S., Rashid, M.M.U., Ullah, A. et al. TMBIM6 promotes glioma progression according to integrated bioinformatics and experimental evidence. Sci Rep 15, 21422 (2025). https://doi.org/10.1038/s41598-025-06799-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06799-9