Abstract
The role of adipose-derived stromal stem cells (ADSCs) in BLCA progression is unclear. We investigated the effects of invasion, stemness, Epithelial-mesenchymal transition (EMT), and drug resistance of BLCA cells co-cultured with ADSCs for a long period of time. Cells were divided into six groups: ADSCs group, ADSCs: T24 group (10:1, 3:1 and 1:1 groups), ADSCs-derived conditioned medium group (CM) and T24 cell group (T24), and cells in each group were cultured to 14 days, and puromycin (puro) was added to the co-cultured cell to remove ADSCs cells without puro resistance, and then the function of T24 cells (10:1-COC, 3:1-COC and 1:1-COC) after co-culture was studied; CCK-8 assay, Transwell, Wound healing, Flow cytometry, RNA-sequencing, qRT-PCR and Western Bloting assay were used to detect cell proliferation, invasion, migration, apoptosis and cellular mRNA and protein expression levels, respectively. We unexpectedly found enhanced stemness and drug resistance of BLCA cells after prolonged contact culture with ADSCs. T24 cells after co-culture mediated cell proliferation, invasion, EMT, stemness, drug resistance and immune escape by up-regulating MDM2, mt-P53 and PD-L1, compared to CM and T24 groups. The inhibitor Atezo and CP-31,398 eliminated mt-P53 and PD-L1-mediated T24 cell drug resistance and stemness, respectively. This study demonstrated that after prolonged co-culture of BLCA cells with ADSCs, the stemness, drug resistance, and immune evasion of T24 cells were dramatically enhanced, suggesting that long-term resident ADSCs in the bladder cancer tumor microenvironment play a procarcinogenic role.
Similar content being viewed by others
Introduction
Bladder cancer (BLCA) is a prevalent malignancy associated with high morbidity and mortality rates1,2,3. At the time of diagnosis, approximately 70% of patients present with nonmuscle-invasive BLCA (NMIBLCA), 25% with muscle-invasive BLCA (MIBLCA), and 5% with distant metastases. NMIBLCA is characterized by a high recurrence rate, with 40-70% of cases experiencing recurrence and 10-30% progressing to MIBLCA within the first five years. The five-year overall survival rate for patients with metastatic disease is only 5%4,5. Significant advancements have been made in the treatment of BLCA over the past few years; however, the prognosis for patients with metastatic disease remains dismal.
The standard first-line chemotherapy regimen for BLCA is M-VAC (methotrexate, vincristine, doxorubicin, and cisplatin). However, the efficacy of bladder perfusion chemotherapy remains inadequate, and effective intravesical therapies are not achieved, contributing to poor prognoses6. Although patients may experience effective remission with conventional postoperative adjuvant chemotherapy, the 5-year recurrence rate continues to be alarmingly high, ranging from 50 to 70%7. Tumor recurrence is a significant factor that adversely impacts patient survival8. One of the primary contributors to the high postoperative recurrence rate is the resistance of BLCA cells to chemotherapeutic agents9. Once a tumor develops drug resistance, it severely limits treatment options and negatively affects patient survival outcomes. Current research on drug resistance mechanisms, including abnormal noncoding RNA function, the formation of bladder cancer stem cells, increased drug efflux, and autophagy10,11, offers promising insights for bladder cancer treatment. However, the exact mechanisms of drug resistance remain unclear.
The results of several meta-analyses have demonstrated a clear association between body mass index (BMI) and an elevated risk of ovarian, renal, and BLCAs12,13,14. Moreover, obesity may serve as a risk factor for the recurrence of BLCA15. Research indicates that stromal stem cells (MSCs) can stimulate tumor growth, invasion, metastasis, and drug resistance by interacting with cancer cells or secreting factors that promote epithelial‒mesenchymal transition (EMT)16. Consequently, investigating the associations between adipose-derived MSCs and tumors is crucial.
The interaction between adipose-derived stromal stem cells (ADSCs) and the tumor microenvironment (TME) is still not well understood. It is likely that cancer-associated fibroblasts (CAFs) arise from local fibroblasts or mature adipocytes. Similarly, ADSCs exhibit plasticity toward differentiation into CAFs and may represent the primary population from which CAFs develop, incorporating multiple potential procarcinogenic mechanisms17,18. Notably, ADSCs can transdifferentiate into CAFs and possibly into cancer-associated adipocytes (CAAs), thereby increasing the population of CAFs and CAAs18. Seo et al.19 reported that lipids derived from ADSCs, such as palmitic acid, stearic acid, oleic acid, and linoleic acid, fulfill the demands for synthesizing large quantities of cell membranes and lipid-mediated signaling during the rapid growth and metastasis of tumor cells, increasing their migration and contributing to tumor progression and survival. However, the procarcinogenic roles of ADSCs within the BLCA microenvironment warrant further investigation to better understand the factors driving malignant tumor progression.
The structure of the bladder is characterized by a thick adipose layer. A preliminary analysis conducted by Rizkalla et al.20, which examined 200 consecutive biopsy and transurethral resection of bladder tumor (TURBT) samples (both benign and cancerous), revealed that 37% of these samples contained adipose tissue, predominantly located in the lamina propria or within both the lamina propria and muscularis propria. On the basis of these findings, we hypothesized that when BLCA arises, the tumor region recruits nearby ADSCs to the tumor site, thereby enhancing the malignancy of BLCA cells through interactions between ADSCs and the BLCA. In our study, we demonstrated that ADSCs within the microenvironment regulate MDM2, PD-L1, and p53 through cell‒cell contact, which mediates T24 cell invasion, EMT, stemness, immune evasion, and drug resistance.
Methods
This study was conducted in accordance with the Declaration of Helsinki. All the methods and experimental protocols approved by the Haikou Affiliated Hospital, Xiangya School of Medicine, Central South University Biomedical Ethics Committee, with the decision dated 16.11.2021 and numbered SC20210499. Informed consent was obtained from all participants and/or their legal guardians.
Cell culture and grouping
-
(1)
T24 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, whereas ADSCs were maintained in OriCell® Adult Adipose Stromal Stem Cell Complete Medium (Refer to Table S.1 for details of all reagent sources). All the cell lines were incubated at a consistent temperature and humidity within 5% CO2 at 37 °C.
-
(2)
Cell groups included the following groups: the T24 alone culture group (T24), the ADSC-sourced CM group, and the ADSC: T24 coculture groups at ratios of 10:1, 3:1, and 1:1, in addition to the ADSC-only culture group ( Fig.S1).
-
(3)
CM was collected from ADSCs during the logarithmic growth phase and filtered through a 0.2 μm filter into a new centrifuge tube.
-
(4)
Cell labeling was performed by loading T24 cells with a stably expressed red fluorescent protein (RFP) gene and ADSCs with a green fluorescent protein (GFP) gene; both vectors included a puromycin resistance gene.
Puromycin screening of postculture tumor cells
Following continuous culture of the coculture groups for 14 days, postculture T24 cells were isolated from the coculture conditions with 3 µg/mL puromycin. The cells were designated the COC group (i.e., 10:1-COC, 3:1-COC, and 1:1-COC). T24-COC cells from the different ratios were then collected.
CCK-8 assay for cell activity
The cells from each group in the logarithmic growth phase were digested with trypsin, counted, and plated in a 96-well plate with an appropriate number of cells (n = 4). The strains were cultured for 1, 3, 5, or 7 days. The medium was subsequently replaced with fresh medium containing 10% CCK-8 reagent, and the cells were cultured for an additional 3 h. The 96-well plate was then placed in an enzyme labeler, and the OD value of each group was measured at 450 nm.
Transwell cell invasion assay
The cells were digested, resuspended in medium containing 1% bovine serum albumin, and counted. They were inoculated into transwell chambers according to their respective groupings. Complete medium at a concentration of 10% was added to the lower square-well plate, and the assembly was placed in an incubator for 24 h. Afterward, the original medium was discarded, and the wells were washed once with PBS. A 4% paraformaldehyde solution was added to each well, and the samples were fixed for 20 min at room temperature. Following fixation, the solution was discarded, and the wells were washed once with tap water. Next, 0.1% (1 mg/mL) crystal violet solution was added for staining for 2 min, and the wells were subsequently washed once with tap water. Nonmigrated cells inside the chamber were gently removed with a cotton swab, and the chambers were left to air dry. The membrane at the bottom of the chamber was carefully removed with tweezers, the bottom side was placed up on a slide, and the sample was sealed with neutral resin glue. Cell observation and counting were performed under a microscope.
Wound healing experiment
Each group of cells was digested with trypsin and adjusted to a density of 2.5 × 105 cells/mL. The cells were inoculated in the center of the dish within the insert area (n = 3) and mixed thoroughly, and 1.5 mL of base medium without fetal bovine serum was added to the outer area to prevent drying. The following day, the cells were observed under a microscope to confirm adherence to the wall. The old medium was aspirated, and the insert was carefully removed with tweezers to create a scratch in the center. The chambers were then washed gently with PBS 1–2 times. Subsequently, 1.5 mL of basic 1640 medium/adult adipose MSC medium without fetal bovine serum was slowly added to the chamber, and the setup was placed in an incubator at 37 °C for continued incubation. Peripheral cell migration to the central scratch area was observed at 0 h, 12 h, and 24 h, with fixed position images taken for recording. The migration rate of the cells in each group was analyzed via ImageJ software.
Immunoblotting experiment
The well-cultured cell culture medium from each group was discarded, and protein lysate, along with phosphatase and protease inhibitors, was added to each dish. Adherent cells were then scraped off via a cell scraper and transferred to EP tubes. The cells were lysed via ultrasonic lysis, followed by centrifugation at 12,000 rpm, yielding the total cell protein in the supernatant. The protein concentration of each sample group was determined via the BCA method. Denatured proteins were reheated at 95 °C for 2–5 min prior to sample loading and allowed to return to normal temperature. Electrophoresis was performed at 160 V for 40 min, and membrane transfer was conducted via a semidry transfer method with the parameters set at 15 V for 1 h. Primary and secondary antibodies were incubated sequentially. Finally, the ECL Luminescent Liquid Kit was used for image development, and the gray values of the lanes were analyzed via ImageJ software to evaluate differences among groups.
qRT‒PCR assay
The cells were collected at each time point, total cellular RNA was extracted, and the concentration was adjusted to 500 ng/µL. Reverse transcription was then performed according to the instructions provided with the reverse transcription kit. Sequences of primers used in the qRT‒PCR experiments is shown in Table S. 2. The reaction conditions were as follows: 50 °C for 15 min; 85 °C for 5 s; and storage at 4 °C indefinitely. Following the synthesis of cDNA, the concentration for each group was adjusted to approximately 200 ng/µL, and the qRT‒PCR mixture was prepared according to the kit guidelines. The qRT‒PCR procedure included the following steps: Step 1 at 95 °C for 20 s (1 cycle); Step 2 at 95 °C for 10 s, 60 °C for 20 s, and 95 °C for 15 s (40 cycles); and Step 3 at 60 °C for 60 s and 95 °C for 16 s (1 cycle). At the conclusion of the procedure, the relative expression levels of each gene were calculated via the 2−ΔΔCT method.
Cell formation assay
The cells were digested, counted, and seeded at a density of 5,000 cells per well in low-adhesion 6-well culture plates. Every three days, 500 µL of fresh culture medium devoid of fetal bovine serum was added. Tumor spheroid formation in each group was observed under a microscope on days 3, 7, and 12, and photographs of five randomly selected fields of view were taken to calculate the number of spheres formed.
Colony formation assay
Cells in the logarithmic growth phase were selected and inoculated into 6-well plates at a density of approximately 1,000 cells per well (n = 3). The next day, the adherent cells were washed twice with PBS, fresh basal medium (2 mL) was added, and the mixture was incubated for 7 days. Once the cell clones were visible, the old medium was discarded, and the cells were washed with PBS. Subsequently, 1 mL of 0.1% crystal violet was added to each well for a staining period of 20 min at room temperature. Afterward, the cells were thoroughly washed with PBS to remove excess dye, and the 6-well plate was allowed to dry in the fume hood. Photographs were taken via a microscope, and the number of cell clones per well was counted.
Reactive oxygen species (ROS) detection
Cells in the logarithmic growth phase were seeded at a density of 5,000 cells per well in a 96-well plate (n = 3) and cultured for 7 days. Following the instructions of the Reactive Oxygen Species Assay Kit, DCFH-DA was diluted to a final concentration of 10 µmol/L. After the cell culture medium was removed, the diluted DCFH-DA was added, and the cells were incubated for 20 min in a cell culture incubator. The cells were then washed three times with serum-free cell culture medium. The fluorescence was measured via a microplate reader, with the excitation wavelength set at 488 nm and the emission wavelength set at 525 nm. A fluorescence intensity image of a random field of view was captured via a fluorescence microscope.
Dosing to calculate the IC50
Piroxicam hydrochloride (THP) was dissolved in DMSO, and a concentration gradient of 0, 25, 50, 100, 200, 400, and 800 ng/ml was prepared. Similarly, erlotinib was dissolved in DMSO, and concentrations of 0, 0.5, 2, 5, 10, 20, and 40 nM/ml were used. Five thousand cells were inoculated into each well of a 96-well plate (n = 6), and the plates were placed in an incubator. After the cells were allowed to adhere for 24 h, the medium was replaced with drug-containing medium, and the mixture was incubated for an additional 24 h. Subsequently, the drug-containing medium was aspirated, and basal medium supplemented with 10% CCK8 reagent was added. The mixture was incubated for 3 h, after which the absorbance was measured at 450 nm using an enzyme marker to calculate the IC50 values for each group of cells.
Hoechst detection of apoptosis
For Hoechst detection of apoptosis, 1,000 cells per well were seeded in a 24-well plate during the logarithmic growth phase. Medium containing 5 nM/ml erlotinib was added, and the mixture was incubated for 24 h. The medium was discarded, and the cells were washed once with PBS. Then, 1× Hoechst reagent was added dropwise to each well to stain the nuclei for 10 min at room temperature in the absence of light. The cells were washed three times with wash solution. A random field of view was subsequently examined under a fluorescence microscope to assess the nuclear morphology.
Detection of apoptosis by double staining with Annexin V/PI
The cells from each group in the logarithmic growth phase were collected, digested and centrifuged with trypsin. The cells were gently washed twice with PBS. The cells were resuspended in binding buffer, Annexin V-FITC was added, and the mixture was incubated for 10 min at room temperature in the dark. The cells were washed once more and then resuspended in binding buffer, after which propidium iodide solution was added. The cells were analyzed via flow cytometry (FACS) to determine the proportions of live, dead, early apoptotic, and late apoptotic cells.
Cellular transcriptome sequencing experiments
RNA extraction and library construction
Total RNA was isolated and purified via TRIzol reagent according to the manufacturer’s protocol. The quantity and purity of each RNA sample were quantified via a NanoDrop ND-1000 spectrophotometer. RNA integrity was evaluated with a Bioanalyzer 2100, which requires a RIN greater than 7.0, and the results were further validated through electrophoresis on a denaturing agarose gel. Poly(A) RNA was purified from 1 µg of total RNA via Dynabeads Oligo(dT)25-61005 via two rounds of purification. The poly(A) RNA was subsequently fragmented into smaller pieces with the Magnesium RNA Fragmentation Module at 94 °C for 5–7 min. The cleaved RNA fragments were then reverse-transcribed into cDNA via SuperScript™ II Reverse Transcriptase. This cDNA was then utilized to synthesize U-labeled second-strand DNA via the use of E. coli DNA polymerase I, RNase H, and dUTP solution. An A-base was added to the blunt ends of each strand to facilitate ligation with indexed adapters, which feature a T-base overhang for binding to the A-tailed fragmented DNA. Single- or dual-index adapters were ligated to the fragments, and size selection was conducted with AMPure XP beads. Following heat-labile UDG enzyme treatment of the U-labeled second-strand DNAs, the ligated products were amplified via PCR under the following conditions: an initial denaturation step at 95 °C for 3 min, followed by 8 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s, with a final extension at 72 °C for 5 min. The average insert size for the final cDNA library was 300 ± 50 bp. Finally, 2 × 150 bp paired-end sequencing (PE150) was conducted on an Illumina NovaSeq™ 6000 according to the manufacturer’s recommended protocol.
Bioinformatics analysis of RNA-seq data
fastp software (https://github.com/OpenGene/fastp) was used to eliminate reads with adapter contamination, low-quality bases, and undetermined bases via default parameters. The sequence quality was subsequently verified via fastp. The reads were aligned to the Homo sapiens GRCh38 reference genome via HISAT2 (https://ccb.jhu.edu/software/hisat2). The mapped reads for each sample were assembled with StringTie (https://ccb.jhu.edu/software/stringtie) using default settings. All the transcriptomes from the samples were merged to create a comprehensive transcriptome via gffcompare (https://github.com/gpertea/gffcompare/). Following the generation of the final transcriptome, StringTie was used to estimate the expression levels of all the transcripts. For the mRNA expression levels, StringTie was used to calculate the FPKM (FPKM = [total_exon_fragments/mapped_reads(millions) × exon_length(kB)]). Differentially expressed mRNAs were identified on the basis of a fold change greater than 2 or less than 0.5, along with a parametric F test comparing nested linear models (p value < 0.05) via the R package edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html).
Statistical analyses
The present study was statistically analyzed via SPSS 22.0 and GraphPad Prism 8.0 software. The data are presented as the means ± standard deviations (‾x ± s). Comparisons between two groups were conducted via a t test, whereas one-way ANOVA was used for comparisons among multiple groups. A p value of less than 0.05 was considered indicative of a significant difference.
Results
ADSCs promote T24 cell proliferation and inhibit apoptosis
To investigate the morphological changes in cells following the coculture of ADSCs and T24 cells, we transfected ADSCs with GFP and T24 cells with RFP. On the third day of coculture, T24 cells exhibited a round or oval morphology, whereas ADSCs exhibited a shuttle or spindle shape. As the culture period increased, T24 cells rapidly proliferated by the seventh day, with ADSCs surrounding the tumor cells. By the fourteenth day of coculture, the tumor cells continued to proliferate rapidly, resulting in significantly increased cell cluster sizes, characterized by irregular tumor cells located at the center of the clusters. Stem cells adhered to the surface of these tumor clusters, forming a network-like connection between them. Over time, the presence of stem cells diminished, potentially indicating their transformation into CAFs, accompanied by a reduction in stem cell infiltration within the tumor mass (Fig. 1A).
Next, we assessed cell activity and demonstrated that the growth rate of cell viability in the mixed-ratio group was greater than that in the CM, T24, and ADSC groups, with the 10:1 mixed group exhibiting the most significant increase. To further investigate the alterations in T24 cell properties following coculture, we utilized T24 (T24puro+) cells, which contain the puromycin resistance gene, for coculture with normal ADSCs. Upon reaching day 14 of culture, we added 3 µg/mL puromycin to the culture medium (at this concentration, all the ADSCs had died, as shown in Figure S2) to eliminate the ADSCs and obtain cocultured T24 cells, referred to as the COC group. Compared with wild-type T24 cells, the T24 cells in the COC group exhibited polygonal or spindle-shaped growth along with increased numbers of pseudopodia (Fig. 1R). The proliferation capacity of these cells was greater than that of the T24 group, both in the mixed culture (Fig. 1B) and in the T24 cells post coculture with ADSCs (Fig. 1C), with the most pronounced difference observed between the 10:1-COC group and the 10:1 coculture group. Consistent results were also obtained in the 5637 and UM-UC-3 cell lines (Fig.S6), while the effect of ADSCs medium on T24 proliferation was excluded (Fig.S7).
In terms of protein expression, the cell cycle-related proteins CDK2, CDK6, and cyclin B1 were upregulated, whereas the cell cycle inhibitors P21 and P27-KIP were significantly downregulated compared with those in the T24 and CM groups (Fig. 1D-I). Additionally, the expression of the apoptotic proteins Bax, caspase-9, and caspase-3 was reduced, whereas the expression of the antiapoptotic protein BCL-2 was increased (Fig. 1J-N). Furthermore, colony formation, as well as cell invasion and migration, was significantly enhanced in the COC group (Fig. 1O-Q, Fig. S3). Annexin V/PI double staining, Hoechst nuclear staining, and ROS assays revealed a decrease in the apoptosis rate, reduced nuclear fragmentation, and decreased ROS expression, respectively (Fig S4). Following coculture, T24 cell morphology was significantly altered, characterized by polygonal or spindle-shaped growth, elongated cells, increased numbers of pseudopodia, and a tendency toward epithelioid transformation (Fig. 1R). Therefore, we hypothesized that ADSCs play a gain-of-function role in T24 BLCA cells. On the basis of these experimental results, we selected the 10:1-COC group for subsequent validation experiments.
A is the cell proliferation morphology diagram, Red is T24 cells, Green is ADSCs, and BF is Bright Field; B is the cell proliferation curves of T24 group, CM group and each mixed culture group (10:1-coculture group, 3:1-coculture group and 1:1-coculture group); C is the cell proliferation curves of T24 group, CM group and T24 group after co-culture (10:1-COC group, 3:1-COC group and 1:1-COC group), respectively. D-I are the expression of cell proliferation-related proteins (CDK2, CDK6 and cyclin B1) and cell cycle inhibitory proteins (P21 and P27-KIP) and their columnar statistical results; J-N are the expression of apoptosis and anti-apoptosis-related proteins (Bax, caspase9, caspase3 and BCL-2) and their columnar statistical results; O-Q are graphs of clone formation, migration and invasion columnar statistical analysis of T24 cells after co-culture, respectively; R is the morphology comparison of T24 cells before and after co-culture, and COC is the morphology of T24 after co-culture, with the cells in elongated shape. * p < 0.05, **p < 0.01, ***p < 0.001,n = 3. The Bar value is 50 μm.
ADSCs promote EMT in T24 cells
To further elucidate the signaling pathways through which ADSCs influence T24 cells, we conducted mRNA transcriptome sequencing. The results revealed that the coculture (10:1-COC) group was significantly enriched in numerous genes associated with the PI3K-AKT signaling pathway, as well as pathways related to the cell cycle, bladder cancer, apoptosis, Hippo signaling, MAPK signaling, TGF-β, Wnt/β-catenin, and P53 (Fig. 2A). Each of these pathways is closely associated with essential processes such as cell proliferation, stemness, EMT, and drug resistance. Specific EMT regulation occurs through several signaling pathways, including the TGF-β, Wnt/β-catenin, Hedgehog, and Notch pathways. These pathways stimulate the activation of transcription factors such as Snail, Slug, and ZEB1/221.
Furthermore, our analysis of the cell cycle, BLCA, apoptosis, and P53 pathways via Venn diagrams revealed that a total of 11 gene sets related to the cell cycle and BLCA were significantly enriched in connection to cell proliferation, with the P53 gene being coenriched among the four gene sets (Fig. 2B-C). Prior research has shown that nearly all MIBLCA cases result in the deletion of cell cycle checkpoints driven by mutations in TP53, RB1, and/or ATM, along with alterations that impact their regulators (e.g., E2F3 and MDM2 expression)8,10. Consequently, we focused on CDKN1A, MDM2, and P53 to investigate whether coculture influences the expression of these genes.
We subsequently examined the EMT pathway. Our findings demonstrated that ADSCs upregulated Snail, Slug, and N-cadherin but downregulated E-cadherin through direct cell‒cell contact, thereby inducing EMT in T24 cells (Fig. 2D-H). These findings suggest that coculture leads to epigenetic modifications in T24 cells, resulting in EMT remodeling and altered cellular morphology (Fig. 1R). The expression of these genes was significantly increased in the COC group, indicating that ADSCs residing in the microenvironment have the potential to promote the tendency of BLCA cells to undergo EMT.
mRNA transcriptome sequencing analysis and EMT-related protein expression. A is the KEGG pathway enrichment bubble plot; B is the VENN plot, i.e., the gene intersection analysis of Cell cycle pathway, Apoptosis pathway, P53 pathway and Bladder cancer pathway; C is the VENN plot of the intersection analysis of Cell cycle pathway and Bladder cancer pathway and their co-varying gene sets (total 11 genes); D-H are the expression of EMT-associated proteins (Snail, Slug, N-cad and E-cad) and the results of their histogram statistical analysis. *p < 0.05, **p < 0.01, ***p < 0.001, n = 3.
ADSCs regulate the migration and invasive capacity of T24 cells through MDM2
MDM2 is recognized as a marker of poor prognosis across various tumors, and its overexpression is associated with the growth and migration of BLCA22. In this study, we assessed the role of MDM2 in the proliferation and metastasis of T24 cells following coculture. Our analysis revealed that the mRNA and protein expression levels of MDM2 in the COC group were significantly greater than those in the T24 and CM groups, as determined by qRT‒PCR and Western blotting (Fig. 3A-C).
To further investigate cell functionality, we inhibited MDM2 via the inhibitor Nutlin-3. 2 µM Nutlin-3 did not change the cell state; whereas the proliferation ability of cells was significantly decreased at 10 µM Nutlin-3. Therefore, we chose 10 µM concentration as the experimental concentration. Treatment with 10 µM Nutlin-3 effectively negated the upregulatory effects of ADSCs on MDM2 protein expression (Fig. 3D-E), as well as on T24 cell proliferation (Fig. 3F), invasion, and migration due to the T24-COC group (Fig. 3G-K). MDM2 is a crucial negative regulator of the tumor suppressor TP53, which functions primarily to inhibit cell cycle arrest and apoptosis via the TP53-dependent pathway. The oncogenic role of MDM2 is attributed primarily to its capacity to obstruct the transcriptional activity of TP53. However, recent findings indicate that MDM2 also regulates multiple signaling pathways that promote tumorigenesis through TP53-independent mechanisms, which are correlated with adverse clinical outcomes23. This study further suggested that ADSCs facilitate T24 cell proliferation, invasion, and migration through the modulation of MDM2.
ADSCs regulate the migration and invasion ability of T24 cells through MDM2. A-C is the analysis of MDM2 gene mRNA and protein expression in each cell group; D-E is the relative expression of MDM2 protein after the addition of the inhibitor Nutlin-3 at a concentration of 10 µM in the 10:1-COC group; and F is the proliferative activity of the cells at 24 h and 48 h with the addition of 2 µM Nutlin-3 and 10 µM Nutlin-3 in the 10:1-COC group, respectively; G-H are comparisons of cell invasion in the 10:1-COC group before and after the addition of 10 µM Nutlin-3 versus the T24 group; I-J are comparisons of cell migration in the 10:1-COC group before and after the addition of 10 µM Nutlin-3 versus the T24 group at 12 h and 24 h. *p < 0.05, **p < 0.01, ***p < 0.001, n = 3.
ADSCs promote PD-L1 expression in T24 cells to regulate tumor cell stemness
Bladder cancer stem cells (BCSCs) represent a distinct subpopulation of cells characterized by high tumorigenicity, drug resistance, and metastasis, all of which are critical to the development, metastasis, and recurrence of BLCA24. Currently, the genetic basis and origin of human BCSCs remain unknown. Surface molecules identified to be associated with tumour stem cells include CD133 and CD44, stem cell stemness-related genes include NANOG, OCT4 and SOX225,26, with PD-L1 expression noted to enhance tumor stem cell characteristics. In this study, we aimed to investigate the changes in the stemness of T24 cells following coculture with ADSCs. Our assessment of tumor stem cell-related genes and proteins revealed that the mRNA and protein levels of NANOG, OCT4, and CD44 were significantly elevated in the COC group compared with those in the T24 group (Fig. 4A-E). Transcriptome analysis via weighted gene coexpression network analysis (WGCNA) revealed that CD274 (PD-L1) was notably upregulated in the COC group, prompting us to validate these findings in the 10:1-COC group. We discovered that PD-L1 expression in the 10:1-COC group was approximately five times greater than that in the T24 group (Fig. 4F-H). To further investigate the impact of PD-L1 on stemness, we introduced a PD-L1 blocker, Atezo, which is known for its effectiveness in inhibiting PD-L1. Our experiments demonstrated that Atezo significantly inhibited the proliferation of 10:1-COC cells in a dose-dependent manner. Specifically, the number of colonies formed by 10:1-COC cells was closed to halved at an Atezo concentration of 5 µg/mL and nearly eliminated at 20 µg/mL (Fig. S5), with the IC50 was 3.88 µg/mL. Therefore, the 1 µg/mL Atezo-treated co-cultured cell group was selected as the Atezo group for subsequent experiments. Additionally, the mRNA expression levels of stemness-related genes were decreased following the administration of Atezo (Fig. 4I). In the sphere formation assay, the PD-L1 inhibitor group remained composed of scattered single cells by day 3, whereas a significant reduction in both the number and size of the tumor spheres was evident by day 7 (Fig. 4J). On the basis of our findings, we conclude that T24 cells enhance their stemness and EMT characteristics through increased PD-L1 expression following coculture with ADSCs.
ADSCs promote the stemness of T24 cells by expressing PD-L1. A-C are the analysis of relative expression of NANOG, OCT4 and CD44 mRNA in cells of T24, CM and COC groups; D-E are the analysis of relative expression of NANOG, OCT4 and CD44 proteins in cells of T24, CM and COC groups; and F-H are the analysis of relative expression of mRNA and proteins of PD-L1 in T24 and 10:1-COC groups. S1 and S2 are the passaged 1st and passaged 2nd generation of T24 after co-culture, respectively. I is the relative expression of mRNA and protein of stemness factors Nanog, OCT4, and CD44 at 1 µg/mL of Atezo in the 10:1-COC group; N is the results of 3-day and 7-day cell formation experiments at 1 µg/mL of Atezo in the 10:1-COC group; Control is the results of 3-day and 7-day cell formation experiments in the unaddressed group. N is the result of cell formation assay on day 3 and day 7 in the 10:1-COC group at an Atezo concentration of 1 µg/mL. Control is the undosed cells in the 10:1-COC group, * p < 0.05, ** p < 0.01, *** p < 0.001, n = 3.
ADSCs promote drug resistance in T24 cells by regulating p53
As a tumor suppressor gene, p53 plays a crucial role in regulating the cell cycle and apoptosis to eliminate abnormal cells, thereby preventing cancerous transformation. Notably, over 50% of malignant tumors carry mutations in the p53 gene, and all p53 genes in T24 cells derived from BLCA are of the mutant type (mt-p53). Wild-type p53 is instrumental in decelerating or regulating cell division through mechanisms such as cell cycle arrest, promotion of apoptosis, maintenance of genomic stability, and inhibition of tumor angiogenesis. However, upon mutation, the spatial conformation of the p53 gene is altered, resulting in the loss of its regulatory functions related to cell growth, apoptosis, and DNA repair. Consequently, the p53 gene transitions from an antioncogene to an oncogene27. Mutant p53 not only loses its original functional capabilities but also has a dominant negative effect on the activity of wild-type p53. Furthermore, it acquires new oncogenic functions, termed “gain-of-function (GOF) functions”, which are strongly associated with the malignant and drug-resistant characteristics of tumors28.
To investigate the effect of ADSCs on drug resistance in T24 cells within the microenvironment, we selected two commonly used clinical drugs: THP and erdatinib. Following a 24-h period of drug treatment, we assessed the growth of T24 cells across different groups and calculated the maximum half-maximal inhibitory concentration (IC50). The findings indicated that both THP and erdatinib inhibited the proliferation of BLCA cells in a dose-dependent manner; however, the COC and CM diminished the drug sensitivity of T24 cells. The IC50 values for THP in the T24, CM, 10:1-COC, 5:1-COC, and 1:1-COC groups were 149.2 ng/ml, 198.2 ng/ml, 220.8 ng/ml, 202 ng/ml, and 129.6 ng/ml, respectively. The IC50 values of erdatinib were 5.138 nM/ml, 16.46 nM/ml, 22.94 nM/ml, 16.68 nM/ml, and 4.078 nM/ml. Notably, no significant change in drug sensitivity was observed in the 1:1-COC group (Fig. 5A-B).
We subsequently analyzed the expression of mt-p53 across the different groups and found that mt-p53 was significantly upregulated in the COC group compared with the T24 group, with the most notable increase observed in the 10:1-COC group (Fig. 5C-E). We treated various cell groups with CP-31,398, a styryl quinazoline analog that is the first small molecule demonstrated to reactivate mutant p53. This compound is known for its ability to protect wild-type p53 from thermal denaturation and to restore wild-type function in some p53 mutants. After administering CP-31,398 at a concentration of 15 µg/ml for 24 h to the 10:1-COC group, we observed a restoration of the previously downregulated expression levels of P21 and Bax. Additionally, mt-p53 expression returned to levels comparable to those of wild-type T24, with no statistically significant differences between the experimental groups and the T24 group (Fig. 5F-G). Similarly, drug sensitivity increased in each group, with the IC50 values for the T24, CM, and 10:1-COC groups for the two drugs as follows: the IC50 values of THP were 103.5 ng/ml, 138.9 ng/ml, and 141.4 ng/ml, respectively, whereas the IC50 values of erdatinib were 3.37 nM/ml, 5.944 nM/ml, and 6.5 nM/ml, respectively (Fig. 5H-I).
On the basis of these findings, we suggest that coculture with ADSCs increases mt-p53 expression levels, induces cell cycle arrest, reduces apoptosis, and affects drug sensitivity in T24 cells. while mt-p53 activator (CP-31398) counteracted these effects.
ADSCs promote drug resistance in T24 cells by regulating mt-p53. A-B are the IC50 results of the two drugs in T24, CM, 10:1-COC, 5:1-COC and 1:1-COC groups: the IC50s of THP were 149.2 ng/ml, 198.2 ng/ml, 220.8 ng/ml, 202 ng/ml, and 129.6 ng/ml, respectively; and those of Erdatinib were 5.138nM/ml, 16.46nM/ml, 22.94nM/ml, 16.68nM/ml and 4.078nM/ml; C-E are mRNA and protein expression analyses of mt-p53 in cells of T24, CM, 10:1-COC, 5:1-COC and 1:1-COC groups; F-G are expression analyses of P21, Bax and mt-p53 in T24, CM and 10:1-COC groups after the addition of activation molecule CP-31,398. H-I is the IC50 results of two drugs in T24, CM and 10:1-COC groups after the addition of CP-31,398: the IC50 of THP was 103.5ng/ml, 138.9ng/ml, 141.4 ng/ml. the IC50 of Erdatinib was 3.37nM/ml, 5.944nM/ml, 6.5nM/ml respectively. *p < 0.05, ** p < 0.01, *** p < 0.001, n = 3.
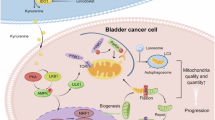
CAFs are increasingly recognized as heterogeneous and ubiquitous stromal cells within the TME29. The primary sources of CAFs include fibroblasts, adipocytes, pericytes, endothelial cells, mesenchymal cells, and epithelial cells30. However, the role of ADSCs in BLCA remains unclear. In this study, we demonstrated robust intercellular interactions between MSCs and tumor cells through various ratios of ADSCs in mixed culture studies using T24 cells. The cocultured T24 cells exhibited a gain-of-function phenomenon characterized by enhanced proliferation, increased invasion and migration capabilities, EMT, decreased apoptosis, and significant upregulation of oncogenes. The underlying mechanisms may involve the induction of stemness in tumor cells, enhanced EMT, and immune evasion through elevated PD-L1 expression. Furthermore, these interactions may regulate cellular invasion and metastatic potential via MDM2, thereby contributing to drug resistance through the gain-of-function of mt-p53 (Fig. 6). Consequently, ADSCs may represent a procancer risk in the BLCA microenvironment.
Discussion
BLCA is one of the most prevalent malignant tumors of the urinary system, and its high rates of metastasis and drug resistance present significant challenges in clinical treatment. Therefore, it is crucial to explore the mechanisms underlying metastasis, invasion, and drug resistance in BLCA to identify potential therapeutic targets. CAFs are the predominant nonmalignant cells present in BLCA, forming abnormal regions within the tumor. These cells are associated with increased tumor invasiveness, chemotherapy resistance, and reduced responsiveness to immune checkpoint inhibitor therapy2,31,32,33. In this study, we found that ADSC coculture with BLCA cells significantly enhanced the proliferation, stemness, invasion, and drug resistance of BLCA cells.
Tumor growth is significantly influenced by the surrounding microenvironment, known as the tumor mesenchyme34. MSCs, as components of this tumor mesenchyme, can drive tumor progression through paracrine signaling or the activation of specific signaling pathways. MSCs, including CAFs, participate in the formation of various cell types within the TME35. Our findings demonstrated that tumor cells exhibited enhanced proliferation, increased expression of stemness markers, heightened invasive capabilities, and abnormal activity of the oncogenes MDM2 and mt-P53 following coculture with ADSCs. These findings suggest that the interaction of ADSCs with BLCA cells promotes their differentiation into CAFs, thereby exacerbating the malignancy of T24 cells.
CSCs maintain their stem cell-like properties through the expression of various transcription factors and surface markers. Key stemness transcription factors include KLF2, OCT4, SOX2, and NANOG, whereas surface markers such as CD133, CD44, CD138, CD34, and nerve growth factor receptor (NGFR) are used36. Recent studies have shown that the overexpression of CSC markers is associated with tumor recurrence, invasion, and treatment resistance across various cancers. For example, elevated levels of NANOG enable tumor cells to adopt stemness characteristics, facilitating tumor expansion, metastasis, and recurrence, which are closely associated with the progression of numerous cancer types37,38. OCT4 serves as a critical regulator of stemness, whereas cells overexpressing CD44 exhibit various CSC characteristics. The expression of CD44 has been shown to induce CSC phenotypes, such as self-renewal, EMT capacity, and metastatic recurrence, which are correlated with clinical prognosis39. Our study revealed that T24 cells underwent epigenetic modifications and differentiated into BCSCs upon coculture, suggesting that this population of cells may facilitate BLCA metastasis and recurrence.
Programmed cell death ligand 1 (PD-L1) is an immunosuppressive molecule that inhibits T-cell activation, contributing to tumor progression40. Research indicates that high levels of PD-L1 expression are not only associated with an increase in tumor stem cell populations but also correlate with the expression of stemness markers, which facilitate the self-renewal of these stem cells19,41,42. Notably, compared with low PD-L1 expression, elevated PD-L1 expression in pancreatic and colorectal tumors is associated with a greater prevalence of CD44+/CD133+ CSCs. This increased expression is also correlated with increased expression of stemness genes, which are crucial for maintaining CSC self-renewal41,42. Epithelial cells exhibiting both PD-L1 and E-cadherin expression on their surfaces swiftly form tumor foci at significant distances from the primary tumor microenvironment in nude mice, demonstrating features of immune evasion and enhanced stemness properties43. Our study further confirmed that T24 cells after co-culture acquired strong PD-L1 expression, which was positively correlated with tumor cell stemness, and that tumor cell stemness were attenuated after inhibition of PD-L1 expression. These findings suggest that ADSCs within the microenvironment may promote epigenetic remodeling of BLCA cells through PD-L1-mediated modulation of their stemness and EMT, thereby contributing to immune escape mechanisms.
Studies have demonstrated that the activity of CAFs negatively correlates with tumor prognosis, likely because of their role in regulating tumor growth and metastatic potential across various levels44,45. Murine double minute 2 (MDM2) is recognized as an oncogene, and its expression is upregulated in multiple tumor types. MDM2 serves as a key negative regulator of the tumor suppressor mt-P53, inhibiting cell cycle arrest and apoptosis mainly through mt-P53-dependent pathways23. In high-grade invasive BLCA (≥ T2), MDM2 is closely associated with cancer invasion2,46. Our study demonstrated that MDM2 expression in T24 cells following coculture with ADSCs was significantly upregulated and positively correlated with tumor cell invasion and migration. The inhibition of MDM2 expression resulted in a reduction in these phenomena, suggesting that ADSCs in the microenvironment promote MDM2 expression in T24 cells, thereby influencing the processes of BLCA invasion and metastasis.
Mechanisms involved in the development of drug resistance in BLCA include the abnormal function of noncoding RNAs, the generation of BLCA stem cells, increased drug efflux, and autophagy10,11,47. In addition to losing the oncogenic function of wild-type P53, mutated mt-P53 also has acquired independent oncogenic activity that promotes cancer progression. This gain-of-function activity includes enhancing tumor cell proliferation and metastasis, facilitating the development of drug resistance, and contributing to genomic instability and metabolic reprogramming48. A significant mechanism underlying these processes is the ability of mt-P53 to regulate various EMT-related proteins and transcription factors, thereby promoting the transformation of tumor cells48. MSCs can stimulate tumor growth, invasion, metastasis, and drug resistance by interacting with cancer cells or secreting relevant factors that promote EMT16,49. In our study, we found that mt-P53 expression was significantly elevated and that drug sensitivity was diminished in T24 cells following coculture with ADSCs. When we activated the mt-P53 structure to revert it to the wild-type structure, the T24 cells returned to their initial wild-type state with respect to apoptosis and drug resistance. These findings strong evidence that ADSCs in the microenvironment promote mt-p53 expression in BLCA cells modulates drug resistance in BLCA.
The role of ADSCs in the microenvironment of BLCA remains inadequately understood. Although some studies have indicated that the immunomodulatory potential of TNF-α-stimulated ADSCs is increased during tumorigenesis50, it has also been shown that ADSC-derived exosomes can penetrate tumor tissues. Furthermore, successful in vivo delivery of miR-138-5p has been demonstrated to inhibit the growth of xenograft tumors51, potentially laying the groundwork for MSC-based BC therapies. However, we propose that ADSCs, which reside in the BLCA microenvironment for extended periods, play a complementary role in tumor progression and may serve as a significant source of CAFs within the TME. Our study revealed that cell cocultures produced stronger effects than ADSC-derived conditioned media did.
Overall, our study reveals that BLCA cells have greater stemness, drug resistance and immune escape after prolonged co-culture with ADSCs, suggesting that long-term resident ADSCs in the bladder cancer tumor microenvironment may play a pro-carcinogenic role, and should be highly valued especially in obese patients. Although we did not perform the validation of multiple bladder cancer cells in this study, nevertheless, these results can provide a better reference value for the pro-carcinogenic role of ADSCs in the BLCA microenvironment, and the signaling pathways involved in this process should be a potential target for the treatment of metastasis and drug resistance.
Data availability
The datasets generated during the current study are available in the [GEO: GSE290399] repository. The datasets are scheduled to be publicly available on: Feb 22, 2026. Until then to access the dataset please use this link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE290399.
References
Siegel, R. L. et al. Cancer statistics, 2023. CA Cancer J. Clin. 73 (1), 17–48 (2023).
Dyrskjøt, L. et al. Bladder cancer. Nat. Rev. Dis. Primers. 9 (1), 58 (2023).
Caramelo, B. et al. Cancer-associated fibroblasts in bladder cancer: origin, biology, and therapeutic opportunities. Eur. Urol. Oncol. 6 (4), 366–375 (2023).
Zang, Y. et al. An overview of patients with urothelial bladder cancer over the past two decades: a surveillance, epidemiology, and end results (SEER) study. Ann. Transl Med. 8 (23), 1587 (2020).
Lenis, A. T. et al. Bladder Cancer: Rev. JAMA 324(19):1980–1991. (2020).
Valenza, C. et al. Emerging treatment landscape of non-muscle invasive bladder cancer. Expert Opin. Biol. Ther. 22 (6), 717–734 (2022).
Kamat, A. M. et al. Bladder cancer. Lancet 388 (10061), 2796–2810 (2016).
Motzer, R. J. et al. NCCN guidelines insights: kidney cancer, version 1.2021. J. Natl. Compr. Canc Netw. 18 (9), 1160–1170 (2020).
Witjes, J. A. et al. European association of urology guidelines on Muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur. Urol. 79 (1), 82–104 (2021).
Mari, A. et al. Genetic determinants for chemo- and radiotherapy resistance in bladder cancer. Transl Androl. Urol. 6 (6), 1081–1089 (2017).
Singh, A. & Settleman, J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29 (34), 4741–4751 (2010).
Larsson and Burgess. Causal role of high body mass index in multiple chronic diseases: a systematic review and meta-analysis of Mendelian randomization studies. BMC Med. 19, 320 (2021).
de Andrade, C. T., Rocha, G. Z., Zamuner, M., Dos Reis, R. B. & Reis, L. O. Obesity influence on bladder inflammation and cancer: a cystitis model. Int. J. Clin. Exp. Pathol. 15 (9), 373–379 (2022).
Trevellin, E. et al. Obesity, the adipose organ and Cancer in humans: association or causation?? Biomedicines. 11(5):1319. (2023).
Lin, Y. et al. Association between obesity and bladder cancer recurrence: A meta-analysis. Clin. Chim. Acta. 480, 41–46 (2018).
Kabashima-Niibe, A. et al. Mesenchymal stem cells regulate epithelial-mesenchymal transition and tumor progression of pancreatic cancer cells. Cancer Sci. 104 (2), 157–164 (2013).
Zhang, Y. et al. Stromal progenitor cells from endogenous adipose tissue contribute to pericytes and adipocytes that populate the tumor microenvironment. Cancer Res. 72 (20), 5198–5208 (2012).
Guillaume, V. G. J. et al. The crosstalk between Adipose-Derived stem or stromal cells (ASC) and Cancer cells and ASC-Mediated effects on Cancer formation and Progression-ASCs: safety hazard or harmless source of tropism?? Stem Cells Transl Med. 11 (4), 394–406 (2022).
Seo, J. et al. Metastasis-on-a-chip reveals adipocyte-derived lipids trigger cancer cell migration via HIF-1α activation in cancer cells. Biomaterials 269, 120622 (2021).
Rizkalla, C. N., Srinivas, S. & Sangoi, A. R. Incidence and pitfalls of adipose tissue encountered in urinary bladder biopsy/transurethral resection specimens. Int. J. Surg. Pathol. 19, 10668969241271957 (2024).
Lei, Y. et al. MicroRNAs target the Wnt/βcatenin signaling pathway to regulate epithelialmesenchymal transition in cancer (Review). Oncol. Rep. 44 (4), 1299–1313 (2020).
Zheng, J. et al. Identification of MDM2 as a prognostic and immunotherapeutic biomarker in a comprehensive pan-cancer analysis: A promising target for breast cancer, bladder cancer and ovarian cancer immunotherapy. Life Sci. 327, 121832 (2023).
Oliner, J. D., Saiki, A. Y. & Caenepeel, S. The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb Perspect. Med. 6 (6), a026336 (2016).
Huang, P. et al. Cancer stem cell-like characteristics of a CD133+ subpopulation in the J82 human bladder cancer cell line. Mol. Clin. Oncol. 1 (1), 180–184 (2013).
Ooki, A. et al. CD24 regulates cancer stem cell (CSC)-like traits and a panel of CSC-related molecules serves as a non-invasive urinary biomarker for the detection of bladder cancer. Br. J. Cancer. 119 (8), 961–970 (2018).
Siddiqui, Z. et al. Synergic effects of cancer stem cells markers, CD44 and embryonic stem cell transcription factor nanog, on bladder cancer prognosis. Br. J. Biomed. Sci. 77 (2), 69–75 (2020).
Aylon, Y. & Oren, M. Living with p53, dying of p53. Cell 130 (4), 597–600 (2007).
Zhou, X., Hao, Q. & Lu, H. Mutant p53 in cancer therapy-the barrier or the path. J. Mol. Cell. Biol. 11 (4), 293–305 (2019).
Cords, L., de Souza, N. & Bodenmiller, B. Classifying cancer-associated fibroblasts-The good, the bad, and the target. Cancer Cell. 42 (9), 1480–1485 (2024).
Biffi, G. & Tuveson, D. A. Diversity and biology of Cancer-Associated fibroblasts. Physiol. Rev. 101 (1), 147–176 (2021).
Gouin, K. H. 3rd et al. An N-Cadherin 2 expressing epithelial cell subpopulation predicts response to surgery, chemotherapy and immunotherapy in bladder cancer. Nat. Commun. 12 (1), 4906 (2021).
Su, S. et al. CD10 + GPR77 + Cancer-Associated fibroblasts promote Cancer formation and chemoresistance by sustaining Cancer stemness. Cell 172 (4), 841–856e16 (2018).
Lee, Y. C. et al. The dynamic roles of the bladder tumour microenvironment. Nat. Rev. Urol. 19 (9), 515–533 (2022).
Tlsty, T. D. & Coussens, L. M. Tumor stroma and regulation of cancer development. Annu. Rev. Pathol. 1, 119–150 (2006).
D’souza, N. et al. MSC and tumors: homing, Differentia-tion, and secretion influence therapeutic potential. Adv. Biochem. Eng. Biotechnol. 130, 209–266 (2013).
Zhao, W., Li, Y. & Zhang, X. Stemness-Related markers in Cancer. Cancer Transl Med. 3 (3), 87–95 (2017).
Lin, T., Ding, Y. Q. & Li, J. M. Overexpression of Nanog protein is associated with poor prognosis in gastric adenocarcinoma. Med. Oncol. 29 (2), 878–885 (2012).
Teng, J. et al. Long non-coding RNA ARAP1-AS1 promotes the progression of bladder cancer by regulating miR-4735-3p/NOTCH2 axis. Cancer Biol. Ther. 20 (4), 552–561 (2019).
Wu, T. et al. Effect of endostatin on Wnt pathway of stem-like cells in bladder cancer in tumor microenvironment. Mol. Biol. Rep. 47 (5), 3937–3948 (2020).
Wang, X. et al. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 9, 5023–5039 (2016).
Mao, X. et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 20 (1), 131 (2021).
Wei, F. et al. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 450, 1–13 (2019).
Jinesh, G. G. et al. Surface PD-L1, E-cadherin, CD24, and VEGFR2 as markers of epithelial cancer stem cells associated with rapid tumorigenesis. Sci. Rep. 7 (1), 9602 (2017).
Kwa, M. Q., Herum, K. M. & Brakebusch, C. Cancer-associated fibroblasts: how do they contribute to metastasis? Clin. Exp. Metastasis. 36 (2), 71–86 (2019).
Wang, Z. et al. Metastasis-associated fibroblasts: an emerging target for metastatic cancer. Biomark. Res. 9, 47 (2021).
Brunelli, M. et al. MDM2 gene amplification as selection tool for innovative targeted approaches in PD-L1 positive or negative muscle-invasive urothelial bladder carcinoma. J. Clin. Pathol. 75 (1), 39–44 (2022).
Zahreddine, H. & Borden, K. L. Mechanisms and insights into drug resistance in cancer. Front. Pharmacol. 4, 28 (2013).
Mantovani, F., Collavin, L. & Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell. Death Differ. 26 (2), 199–212 (2019).
Iser, I. C. et al. Conditioned medium from Adipose-Derived stem cells (ADSCs) promotes Epithelial-to-Mesenchymal-Like transition (EMT-Like) in glioma cells in vitro. Mol. Neurobiol. 53 (10), 7184–7199 (2016).
Ting, H. K. et al. Inflammatory regulation by TNF-α-Activated Adipose-Derived stem cells in the human bladder Cancer microenvironment. Int. J. Mol. Sci. 22 (8), 3987 (2021).
Liu, T. et al. Evaluating adipose-derived stem cell exosomes as MiRNA drug delivery systems for the treatment of bladder cancer. Cancer Med. 11 (19), 3687–3699 (2022).
Acknowledgements
The authors thank the natural science foundation of Hainan Province (822MS202, 822RC861), the National Science Foundation of China (82160531 and 82360500) and Finance science and technology project of Hainan province (ZDYF2025SHFZ016, and ZDYF2022SHFZ296).
Author information
Authors and Affiliations
Contributions
DGH: Writing review & editing, Writing original draft, Supervision, Funding acquisition, Conceptualization, Investigation. NG, YLP: Writing original draft, Investigation, Data curation. ZYN, YHG, MC, XHW, YS: Investigation, Data curation. HC, SLW: Validation, Methodology, Data curation. ZJW, LLZ, HWH: Methodology, Investigation, Formal analysis. YC, SFZ: Writing review & editing, Supervision, Conceptualization.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All methods and experimental protocols were performed in accordance with the regulations of the Biomedical Ethics Committee of Haikou Affiliated Hospital, Xiangya School of Medicine, Central South University (No.SC20210499).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, D., Guo, N., Peng, Y. et al. Long-term resident adipose-derived stromal stem cells in the microenvironment remodeling BLCA cell stemness and EMT promotes bladder cancer progression. Sci Rep 15, 23049 (2025). https://doi.org/10.1038/s41598-025-07387-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-07387-7