Abstract
Hepatic stellate cells (HSCs) are liver-specific mesenchymal cells that play a crucial role in liver formation and regeneration, as well as in different pathological diseases. However, the limited source of primary HSCs (pHSCs) and the suboptimal functionality of induced HSCs (iHSCs) by existing methods restrict their application in biomedical modeling. We developed a de novo differentiation method to generate iHSCs under simulated liver microenvironment in vitro, thereby enhancing the function of the differentiated cells. These iHSCs exhibited key HSC functions, including the expression of α-smooth muscle actin, collagen, and the capability to store Vitamin A. RNA sequencing further revealed that the present iHSCs converged more closely to pHSCs with very similar transcriptional profile compared to the established conventional induction. Additionally, the novel HSC-specific marker genes, FBLN5, NID2, and SVEP1 were identified by RNA sequencing and gene expression assay. In conclusion, our novel differentiation approach enables the generation of iHSCs with phenotypic and functional traits similar to those of pHSCs. The generation of highly functional iHSCs may make it more feasible to accurately simulate the liver-specific multicellular microenvironments, thus providing new perspectives on the modeling of physiological regenerative processes and disease progression in the liver, as well as useful tools for creating of new therapeutic strategies.
Similar content being viewed by others
Introduction
One of the main pathogenic processes of liver fibrosis is thought to be the activation of hepatic stellate cells (HSCs)1. Located in the area between hepatocytes and sinusoidal endothelial cells, HSCs are an essential part of the liver’s non-parenchymal interstitial cells and carry out a variety of tasks in the liver2. Under physiological conditions, HSCs display mature HSC markers such as platelet-derived growth factor receptors (PDGFRA and PDGFRB), activated leukocyte cell adhesion molecule (ALCAM), and neural cell adhesion molecule (NCAM), and they accumulate Vitamin A metabolites3,4,5. However, in response to hepatic injury, activated HSCs unavoidably lead to liver fibrosis6. If untreated, fibrosis could further develop into cirrhosis and hepatocellular carcinoma7.
Regarding the necessity of establishing a reliable experimental platform for modeling the dynamic process of liver fibrosis, as well as screening and evaluation of anti-fibrotic drugs, functional HSCs were eagerly demanded. However, human primary hepatic stellate cells (pHSCs) are difficult to obtain, exhibit limited proliferative capability, and cannot maintain a quiescent state in vitro8. These limitations hinder a comprehensive understanding of their functional characteristics and activation phenotype. As substitute, several immortalized HSC lines, such as LX-2 and JS-1, are commonly used to model fibro genic gene expression, such as in HSCs during viral hepatitis infection9,10 In recent years, induced pluripotent stem cells (iPSCs) have been proven to be an ideal source for differentiating into functional induced HSCs (iHSCs), relying on the combination of growth factors and small chemical molecules. However, all the reported studies failed to generate fully functional cells that closely resemble pHSCs5,11,12,13,14. One major reason may be the lack of the complex and finely regulated liver microenvironment that governs HSC development.
The properties of a cell are often determined by the intrinsic microenvironment in which it resides. For example, macrophages are highly flexible and versatile immune cells that exhibit unique functional states in different tissues and organs. Macrophages are mainly found in the circulation as monocytes. However, they undergo tissue-specific differentiation into Kupffer cells once they reach the liver. In liver, HSCs constitute the main supportive stromal population and are mainly responsible for Vitamin A accumulation and fibrosis15,16. On the other hand, stellate cells in the pancreas have a role in both controlling and regulating the tumor microenvironment and pancreatic fibrosis17. As a comparable population of stellate cells found in the central nervous system, astrocytes sustain the blood–brain barrier, aid in neuronal metabolism, and take part in neuroinflammatory reactions18. These physiological and functional differences illustrate the critical role of distinct tissue microenvironments in determining the specific fate of stellate cells, highlighting the importance of remodeling the liver specific microenvironment during HSC differentiation and maturation.
In this study, a de novo approach is developed that enables the differentiation of iHSCs from iPSCs. By mimicking the embryonic development of the liver and recapitulating key signaling interactions between different hepatocyte cell types, it would be possible to efficiently induce functional HSCs.
Results
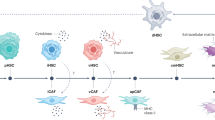
Hepatic micro-environment and iHSC generation
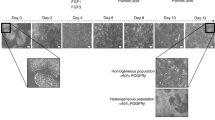
The equilibrium of HSCs is greatly supported by the interactions between various types of liver cells (Fig. 1a). To promote the differentiation of iHSCs, an in vitro differentiation method was designed to reconstruct the hepatic microenvironment (Fig. 1b, Fig S1). This protocol is designed based on the approach proposed by Taniguchi Lab and has demonstrated consistent stability across multiple validations19. First, iPSCs differentiate into definitive endoderm, and then into hepatic endoderm. Pluripotency steadily decreases during differentiation, while mesodermal and endodermal genes such as CXCR4, SOX17, T, and MIXL1 show increased expression (Fig. 1c). At this stage, cells predominantly exhibit a dense, polygonal morphology (Fig. 1d). These cells expressed fetal hepatic parenchymal cell marker genes including albumin (ALB), alpha-1 antitrypsin (AAP) and alpha-fetoprotein (AFP) (Fig. 1e). Interestingly, the specific markers of liver non-parenchymal cell types were also detected, with CD34 and CD31 for endothelial cells, CK19 and EPCAM for cholangiocytes, LYVE1 and STAB2 for liver sinusoidal endothelial cells (LSECs), and VIM for mesenchymal cells (Fig. 1f). which implied that, a hepatic multi-lineage microenvironment was generated synchronously and may facilitate for HSC development. On this basis, we developed a sequential protocol integrating growth factor supplementation, small-molecule compounds, and iterative passaging to gradually deplete parenchymal hepatocytes, and expand niche-resident mesenchymal stromal cells, and eventually differentiation of these microenvironment-primed mesenchymal stromal cells into iHSCs (Fig. 1b).
Induction of iHSCs in hepatic micro-environment. a, schematic diagram of the liver microenvironment. b, schematic diagram of the differentiation process from hiPSCs to iHSCs. c, changes in the expression of pluripotency, mesoderm, and endoderm markers during iPSC differentiation. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001. d, parenchymal cell morphology. e, Expression of liver parenchymal cell-related genes at MH stage. *p < 0.05, **p < 0.01, **** p < 0.0001 and ns = not significant. f, hepatic non-parenchymal related gene expression at MH stage. Results are presented as the mean ± standard deviation (SD) from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001 and ns = not significant.
To generate iHSCs under special conditions in this experiment until day 18, HSCs were not stimulated to transition. Hepatic parenchymal cells were gradually eliminated, while the population of mesenchymal cells increased. With the progressive expansion of mesenchymal cells, the culture conditions were further adjusted to facilitate HSC differentiation. After five days of induction, the cell morphology gradually shifted to a characteristic spindle-shaped appearance, exhibiting the morphological features of HSCs (Fig. 2a).
Confirmation of iPSCs-derived functional iHSCs. a, image of iHSCs. Scale bar, 100 μm. b, dynamic expression of iHSC-associated genes at different stages. c, dynamic expression of PDGFRB during differentiation. d, expression levels of HSC characteristic markers compared to LX2 cell and NPC. e, expression of α-SMA and collagen. Scale bar: 100 μm. f, Vitamin A storage was verified under UV light and the proportion of Vitamin A+ cells was determined by flow cytometric analysis. Negative control: mesenchymal stem cells (MSCs). Scale bar: 50 μm. Results are presented as the mean ± standard deviation (SD) from three independent experiments. ns indicates no significant difference. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The detection and confirmation of iPSCs-derived functional iHSCs
To assess the iHSCs, the common characteristics were verified by morphology, gene expression, flow cytometry, immunocytochemistry and Vitamin-A expression. Genes such as VIM (vimentin), PDGFRB (platelet-derived growth factor receptor β), ALCAM (activated leukocyte cell adhesion molecule), ACTA2 (α-smooth muscle actin), and COL1A1 (collagen type I α1 chain) are well known highly expressed in HSCs, and their expression were found increased significantly during the differentiation process, indicating that an HSC-like phenotype was successfully induced (Fig. 2b). However, the induction of DES, an important HSC marker, was relatively low in our iHSCs. Additionally, the proportion of PDGFRB+ cells, also showed significant increase, as detected by FACS, further supporting the activated state of these cells (Fig. 2c). In comparison to primary non-parenchymal cells (NPCs) and HSC cell line, LX-2, iHSCs showed superior morphological and molecular identities (Fig. 2d). This method can also be applied to multiple cell lines of iPSCs (Fig. S2).
To give a more precise description and visualization of the cells, immunofluorescence staining was performed. These iHSCs showed both collagen and α-SMA (α-smooth muscle actin) expression, two typical markers of HSCs (Fig. 2e). α-SMA is commonly used to indicate the activation state of HSCs, while collagen is associated with the collagen synthesis and usually massively secreted during liver fibrosis. Thus, typical HSC characteristics were confirmed. To take advantage of this characteristic, we exposed iHSCs to ultraviolet light and compared them to mesenchymal stem cells (MSCs). The findings demonstrated that iHSCs had the common function of Vitamin A storage, while quite different from MSCs (Fig. 2 f, Fig. S3 a-b).
Additionally, our results revealed progressive upregulation of canonical activation markers (DES, ACTA2, and COL1A1) across passages (P1-P4), demonstrating a molecular transition toward an activated state that similar to primary HSC behavior in vitro (Fig. S3c). Despite molecular changes, iHSCs maintained characteristic morphology before P4, with no observable transformation into myofibroblast-like shapes typically associated with advanced activation (Fig. S3d). However, after P4 they entered replicative senescence with markedly diminished expansion potential. These findings collectively demonstrated that while iHSCs undergo molecular activation closely mirroring primary cells, they retained sufficient phenotypic stability through P4, and may serve as a reproducible model system for controlled HSC activation studies.
The similarities and differences between iHSCs and conventional induction by RNA sequencing
To further evaluate the transcriptional features, Bulk-RNA sequencing was performed. The gene expression data from primary HSCs (pHSCs), aHSCs and qHSCs data from Miyajima’s lab (including aHSC_4M, aHSC_FF, qHSC_4M, and qHSC_FF), and our own iHSC gene expression data was compared14. The principal component analysis (PCA) results show that the in vitro differentiated iHSCs exhibit highest similarity to pHSCs in terms of gene expression profiles (Fig. 3a). Hierarchical clustering based on the global gene expression heatmap showed that, compared to aHSCs and qHSCs induced by conventional methods, iHSCs exhibited the most similar expression with pHSCs (Fig. 3b). Additionally, based on liver-related datasets from the EMBL’s European Bioinformatics Institute (E-MTAB-7407) and genes previously reported to be associated with HSCs, we identified and compiled a list of 72 highly HSC-specific genes. These genes exhibit strong relevance to HSC identity and function, serving as critical markers for evaluating the similarity between iHSCs and pHSCs. The heatmap of these HSC-related gene expression, arranged from high to low, demonstrates that at the gene expression level, iHSCs exhibit a high degree of similarity to pHSCs, such as the high expression of IGFBP5, COL1A1, MMP2, and LAMB1 (Fig. 3c). Correlation analysis further revealed that iHSCs exhibit the highest similarity to pHSCs, highlighting the effectiveness and superiority of this in vitro differentiation system (Fig. 3d). Although some differences between iHSCs and pHSCs could be found in the volcano plot (Fig. 3e), there was no significant difference in most in typical genes of pHSCs, including COL1A1, DES, LRAT et al.. Moreover, qRT-PCR results further confirmed that some typical HSC genes are expressed at relatively higher levels in iHSCs compared to those induced by Miyajima’s method (Fig. 3f), as well as LX2 (Fig. 2d).
The similarities and differences between iHSCs and the HSC induced from conventional methods. a, PCA analysis of iHSC, aHSC, qHSC, and pHSC. b, Heatmap comparison of all genes expresssion. c, Heatmap comparison of HSC-associated genes expression. d, Correlation Analysis. e, Volcano plot comparison between iHSCs and HSCs. f, qRT-PCR comparison of classic genes expression between the HSCs induced by M (Miyajima)_Lab and this work. Results are presented as the mean ± standard deviation (SD) from three independent experiments. ns indicates no significant difference. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001. Note: HSC refers to primary HSC, aHSC: activated HSC, qHSC: quiescent HSC.
The key components of the TGF-β signaling pathway were robustly expressed in iHSCs, including the ligand (TGFB1), receptors (TGFBR1, TGFBR2), downstream transducers (SMAD2, SMAD3, SMAD4, SMAD7), and a set of canonical fibrogenic targets such as COL1A1, COL1A2, ACTA2, TIMP1, CTGF, PAI-1, SKIL and TGIF (Fig. S4). This comprehensive expression profile confirmed the functional integrity of TGF-β signaling in iHSCs. Furthermore, comparison with qHSCs revealed that iHSCs expressed significantly elevated levels of profibrotic genes such as ACTA2, COL1A1, and TIMP1 (Fig. 3C), suggesting a pre-activated state that may be primed for rapid response to fibrogenic stimuli like TGF-β1. Importantly, when compared with pHSCs, iHSCs exhibited highly similar expression patterns of TGF-β–responsive genes, especially COL1A and ACTA2 (Fig. 3C). This functional similarity underscores the fidelity of iHSCs in modeling the transcriptional responses typical of pHSCs, validating their reliability as an in vitro model for investigating hepatic fibrogenesis and TGF-β–mediated fibrogenic signaling.
Overall, our iHSCs in vitro differentiation system is capable of inducing cells with gene expression features similar to those of pHSCs, providing a promising model for simulating the biological behaviors of HSCs in vitro.
Novel critical HSCs biomarkers and validation
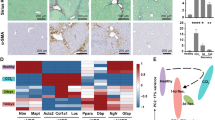
To further validate the similarity of iHSCs and pHSCs, we identified additional HSC-specific marker genes. First, we collected the HSC population datasets. Single-cell RNA sequencing data from the Human Fetal Liver, Skin, and Kidney dataset (E-MTAB-7407) were used to analyze gene expression profiles20. The Uniform Manifold Approximation and Projection (UMAP) was performed to visualize HSCs in comparison with other major liver cell types, including hepatocytes, T cells, and endothelial cells, thereby revealing their relative spatial distribution within the overall cellular lineage (Fig. 4a). Our analysis identified a total of 26 non-specific marker genes, which were not only expressed in HSCs but also in fibroblasts and smooth muscle cells, including ADCY5, DBH, ELN, HGF, MYL9, GGT5, GEM, and NEXN. In addition, we confirmed six HSC-associated marker genes previously reported: ACTA2, COL1A1, DES, PDGFRB, ALCAM, and NGFR. Notably, we identified a series of novel candidate HSC marker genes, including ASPN, BGN, COL6A3, COL5A1, COL25A1, COLEC11, DCN, FBLN5, HHIP, IGFBP5, LAMB1, NID2 and SVEP1 et al. (Fig. 4b). These newly discovered markers may contribute to a more comprehensive understanding of the molecular features of HSCs and provide valuable references for future studies in this field. Among them, BGN, COL6A3, COLEC10, SVEP1, DCN, HHIP, IGFBP3 and LAMB1 had extremely high specificity in HSCs, providing more sensitive biomarkers for HSCs (Fig. 4 c).
Novel critical biomarkers of HSCs and the validation. a, UMAP plot showing the relative positions of HSCs compared to other hepatic cells. b, HSC-related marker genes were selected by integration of single cell RNA sequencing data from different astrocytes. c, UMAP expression plot of 8 novel HSC-specific marker genes. d, qRT-PCR analysis of novel HSC-specific marker genes. Results are presented as the mean ± standard deviation (SD) from three independent experiments. ns indicates no significant difference. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001.
Next, we performed qRT-PCR to evaluate the expression of these selected genes in different cell populations, including iPSC, iHSC, NPC, LX2 and a&qHSCs induced by Miyajima Lab14(Fig. 4 d). The results showed that the expression levels of FBLN5, NID2, LAMB1, IGFBP5 and SVEP1 were significantly higher in iHSCs, compared to Miyajima Lab’s, suggesting the enhanced efficiency of our induction protocol. In particular, FBLN5, NID2, and IGFBP5 were recently reported to be significantly expressed during HSC differentiation and development, which aligned with our current findings21. However, the other two genes, LAMB1 and SVEP1, although exhibiting high expression levels in iHSCs, have not yet been reported up to date, serving as brand-new biomarkers for HSCs in future studies and warrant further investigation.
Discussion
In this study, a de novo approach was developed to generate functional HSC-like cells from iPSCs in vitro. In the simulating liver microenvironment, HSC-like cells developed and expressed typical characteristics, including ACTA2 and PDGFRB expression, as well as Vitamin A storage. Transcriptomic analyses further demonstrated a higher degree of similarity between iHSCs and pHSCs than the previous reported, with HSC-specific genes such as LGALS1 and IGFBP5 even elevated to a level comparable to pHSCs. These results suggest that our approach was successful in recapitulating fundamental identity of HSCs, could address the limited supply of pHSCs and improved the functionality of iHSCs compared to previous protocols. Providing a reliable source of HSC-like cells in vitro, this work lay a foundation for more accurate liver disease modelling and regenerative medicine applications.
In our initial attempts, we followed differentiation methods developed by Miyajima Lab14. In order to increase the effectiveness of differentiation and the functionality of HSCs, we sought to improve these techniques by mixing growth factors with chemical compounds. After repeating the reported method, we found that the expression of HSC characteristics was suboptimal, and several improvements also failed. Thus, we developed a de novo method to generate iHSCs with higher functionality. In 2013, the Taniguchi Lab had developed an in vitro system composed of hepatic lineage and co-cultured with HUVECs and mesenchymal cellss and succeed in the generation of vascularized and functional human liver organoids from iPSCs19. This study implied that a multicellular co-culture microenvironment is necessary to maximize the functions of each cell type. Furthermore, during embryonic development, the intercellular crosstalk was reported to play a significant role in cell differentiation22,23. Therefore, we hypothesized that by simulating the liver developing microenvironment in vitro, we could leverage the interactions between cells and the liver microenvironment to induce the differentiation of HSCs in a more specific manner.
The core innovation of our approach lies in the systematic incorporation of a biomimetic liver microenvironment into the differentiation processing from iPSCs into HSCs for the first time. Previous studies have shown that single-lineage differentiation in isolation often yields cells with incomplete functionality, whereas a multicellular context can significantly improve maturation process19. In liver, parenchymal and nonparenchymal cells were engaged in extensive crosstalk, for example, liver sinusoidal endothelial cells secrete factors like PDGF-BB, which binds PDGFRβ on stellate cells, maintaining proliferation and survival of HSCs during development24. Paracrine signaling between hepatocytes and endothelial cells, such as HGF and Wnt2, coordinates the temporal sequence and functional maturation of HSC differentiation via activation of the Notch pathway25,26. Thus, co-culturing iPSC-derived hepatic endoderm with endothelial and mesenchymal cells, iLSEC for example, can generate vascularized liver organoids with improved function. In addition to matrix cues, cell–cell interactions and soluble signals in the microenvironment play a pivotal role in HSC development and were integrated into our differentiation strategy27. we also sought to recapitulate the key paracrine signals by supplementing the factors known to be involved in liver development and HSC activation. For example, the employ of local signaling molecules such as TGF-β1 and PDGF-BB, further simulated the microenvironmental responses of HSCs during liver development and fibrosis. In contrast to previously reported iHSC induction protocols that predominantly employed single-lineage system11,14, our approach introduced a more comprehensive reconstruction of the liver developmental microenvironment by incorporating critical parenchymal–nonparenchymal cell interactions and essential signaling factors. This strategy not only significantly enhanced the induction efficiency, but also generated iHSCs with gene expression profiles, activation status, and functional characteristics that more faithfully recapitulate those of primary HSCs. Notably, our method facilitated the induction of newly identified HSC-specific marker genes, including FBLN5, NID2 and SVEP1, which have not been consistently detected in cells induced by conventional protocols.
PDGFRB, one of the reliable membrane markers for detecting HSCs, was expressed in over 80% iHSCs. Additionally, the iHSCs exhibited Vitamin A storage in around 87.1% of the population, and meanwhile expressed both α-SMA and collagen, which are common features of HSCs. Notably, they exhibit high similarity to pHSCs in gene expression profiles, such as LGALS1 and IGFBP5. Interestingly, several novel markers that are commonly expressed in fetal HSCs also raised in iHSCs, including NID2, FBLN5, LAMB1, IGFBP5, and SVEP1, providing potential markers for tracing the early stage of HSC development, and pave the way to further investigating into the biological functions of HSCs. However, further experimental validation and functional studies are still needed.
Despite the overall similarities with pHSCs, the functional characterization of iHSCs has not been verified in vivo. It remains to be determined whether the complex factors involved in vivo, such as hemodynamics and immune cell interactions, may have significant effects on the function of the transplanted iHSCs. Additionally, the role of HSCs in liver homeostasis and pathogenesis remains unclear. It is reported that HSCs not only participate in the fibrosis process but also regulate hepatocyte regeneration and metabolism through the secretion of cytokines including HGF, IL-6, and TGF-β5,27,28. These secreted factors create a complex microenvironment that influences hepatocyte proliferation, apoptosis, and metabolic functions, thereby contributing to liver homeostasis and repair mechanisms. Using 3D and multilineage organoid culture system, the precise functions of iHSCs in liver development, regeneration and illness may be uncovered. Furthermore, the activation status of iHSCs should be emphasized. In a healthy liver, HSCs mostly stay quiescent, with minimal activation markers expression such as ACTA2 and COL1A1; and elevated only when transdifferentiated into activated myofibroblasts during fibrogenesis. The observed moderate expression of ACTA2 and COL1A1 in iHSCs indicated the acquisition of a partially activated phenotype, reflecting the fibrogenic microenvironment established in our culture system. This finding was consistent with well-documented evidence that conventional monolayer culture conditions inherently induce partial HSC activation through multiple mechanisms, including mechanical stress, replicative senescence, and autocrine profibrogenic signaling29. These findings highlight the need for systematic investigation of iHSC activation mechanisms, with particular focus on: 1) culture-induced microenvironmental stresses (matrix stiffness, oxidative stress), and 2) exogenous/autocrine profibrogenic signaling pathways (e.g. TGF-β/Smad, PDGF/PI3K-AKT). Finally, the establishment of iHSCs provides a critical functional module for constructing hepatic parenchymal-stromal interaction models with high physiological similarity. Along with our team’s prior success in deriving liver sinusoidal endothelial cells (LSECs) from hiPSCs30, these achievement may addresses a longstanding technological gap in hepatic organoid research—the lack of integration of isogenic multicellular components, and provided a methodological foundation for developing immunecompatible autologous organoids from a single iPSC donor.
In this study, we established an efficient and reproducible induction strategy for generating iPSC-derived HSCs. This de novo approach holds potential for the stable and scalable production of functional HSCs, opening new avenues for the study of liver disease progression and related cellular microenvironments.
Materials and methods
iPSC culture and maintenance
iPSC cell line WTC-11, a wild-type human male iPSC line (Coriell Institute: # GM25256) under Materials Transfer Agreements, and hiPSC gifted from Jiangsu University, were cultured and maintained in StemFit Basic04 medium (AJINOMOTO, Basic04CT) on Matrix511-silk (Nippi 38,710,131) coated tissue culture plates. Cells were passaged using Accutase (STEMCELL technology, 07,922).
Mesenchymal stem cells (MSC)
Umbilical cord collection and MSC separation are based on the stem cell research ethics approval No. 2023–02 issued by the Affiliated Hospital of Jiangsu University. The complete medium for MSCs consisted of α-MEM (Thermo Fisher, 12,571,063) + 10% FBS (GIBCO, 10,091–148) + 10 ng/ml EGF (R&D Systems, 236-EG) + 10 ng/ml FGF2 (R&D Systems, 233-FB) + 1% PS (GIBCO, 15,140,122). Cells were directly cultured in 10 cm culture dishes and passaged every 2–3 days. For passaging, cells were digested using 0.05% Trypsin–EDTA (GIBCO, 25,300,062), and digestion was terminated with medium containing 10% FBS (GIBCO, 10,091–148) in a 3 × volume. Cells were centrifuged at 1200 rpm for 3 min and passaged at a density of 5 × 103 cells/cm2. Cells between passage 3–4 were used.
Immortalized human HSCs (LX2)
Human hepatic stellate cells (LX2) and the maintenance methods were provided by Duan Lab, Institutes for Life Sciences and School of Medicine, South China University of Technology31.
The complete medium for immortalized human HSCs (LX2) consists of DMEM (Thermo Fisher, 11,885,084) + 10% FBS (GIBCO, 10,091–148) + 1% PS (GIBCO, 15,140,122). Cells were directly cultured in tissue culture dishes and passaged every 2–3 days. For passaging, cells were digested with 0.05% Trypsin–EDTA (GIBCO, 25,300,062), and digestion was terminated with medium containing 10% FBS (GIBCO, 10,091–148) in a 3 × volume. Cells were centrifuged at 1200 rpm for 3 min and passaged at a 1:2 to 1:4 ratio. Cells between passages 3–4 were used for experiments.
Non-parenchymal cells (NPCs)
cDNA of hepatic non-parenchymal cell, was gifted from Hui Lab, the Institute of Biochemistry, Chinese Academy of Sciences, Shanghai, China.
Differentiation of hiPSCs into hepatic parenchymal-like cells
Induction to definitive endoderm
On day 0, cells were cultured in RPMI1640 (GIBCO, 11,875,093) medium supplemented with 1% B27 (GIBCO, 17,504,044), 100 ng/mL Activin A (Nacalai Tesque, 18,585–81), and 3 μM CHIR99021. From day 1 to day 2, the medium was replaced with RPMI1640 (GIBCO, 11,875,093) containing 1% B27 (GIBCO, 17,504,044) and 100 ng/mL Activin A (Nacalai Tesque, 18,585–81).
Induction to hepatic progenitor cells
From day 3 to day 8, iPSCs were cultured in KO-DMEM (GIBCO, 10,829,018) as the base medium, supplemented with 20% KSR (GIBCO, 10,828,028), 1% non-essential amino acids (NEAA, GIBCO, 11,140,050), 1% glutamine (ThermoFisher, 35,050,061), 1% DMSO (Solarbio, D8371-50 ml), 1% PS (GIBCO, 15,140,122), and 50 nM β-mercaptoethanol. After digestion of endodermal cells, they were re-plated in this induction medium.
Induction to Mature Hepatocyte Cells
From day 9 to day 18, cells were cultured in SFD medium as the base culture medium, which contained 2.5 mM dexamethasone, 10 ng/ml OSM (Okine, Qk049), 10 ng/ml FGF2 (R&D Systems), 0.407 g/ml nicotinamide, and 20 ng/ml HGF. The SFD medium was composed of 75% IMDM (GIBCO, 12,440,053), 25% Ham’s F-12 K (GIBCO, 21,127,022), 1% N2 (GIBCO, 17,502,048), 1% B27 (GIBCO, 17,504,044), and 450 μM 1-thioglycerol (Sigma, M6145).
Mesenchymal cell isolation from Co-cultured hepatic parenchymal-like cells
Starting from the second stage, mesenchymal cells were purified from impure hepatic parenchymal cells and expanded in large numbers. The specific method is as follows: the medium at this stage was based on SFD medium, supplemented with 2.5 mM dexamethasone, 10 ng/ml IL6 (R&D Systems, 206-IL), 10 ng/ml FGF2 (R&D Systems, 233-FB), 20 ng/ml EGF, 10 ng/ml Wnt3a(R&D Systems, 5036-GMP), 2 μM A83-01, 20 ng/ml HGF(PEPROTECH, 100-39H), and 2% FBS (GIBCO, 10,091–148). The SFD medium was composed of 75% IMDM (GIBCO, 12,440,053), 25% Ham’s F-12 K (GIBCO, 21,127,022), 1% N2 (GIBCO, 17,502,048), 1% B27 (GIBCO, 17,504,044), 450 mM 1-thioglycerol (Sigma, M6145-100ML), 1% GlutaMAX (GIBCO, 35,050,061), 1% PS (GIBCO, 15,140,122), 0.05% BSA (Sigma-Aldrich, V900933-100G) and 0.5 mM ascorbic acid-2-phosphate (AA2P, Sigma). Approximately 3 × 104 (3 × 103/cm2) liver parenchymal cells were isolated and cultured on Matrix511-coated 6-well plates. The medium was replaced every 2 days during the culture. During this process, hepatic parenchymal-like cells were gradually eliminated, and mesenchymal cells were enriched. Serial passages were performed to achieve Mesenchymal cells expansion.
Differentiation of stromal cells into HSC-like cells
Starting from the third stage, mesenchymal cells were further differentiated into HSC-like cells in an HSC induction medium. The composition of the HSC induction medium included StemPro™−34 SFM (ThermoFisher, 10,639,011), 5.4 μM SB-431542 (Selleck, S1067-10 mg), FGF2 (20 ng/ml, R&D Systems), 20 ng/ml VEGF (R&D Systems, 58,097,824), 0.5 μM Dorsomorphin 2HCl (Selleck, S7306-10 mg), and 2% FBS (GIBCO, 10,091–148).
After the hepatic parenchymal-like cells were gradually eliminated and mesenchymal cells were extensively expanded, approximately 3 × 103/cm2 cells were seeded on Matrix511-coated 6-well plates (Nippi, 38,710,131) and cultured with the HSC induction medium. The medium was not changed on the second day post-seeding, and subsequently, the medium was replaced every 2 days for a total of 5 days, after which the cells were harvested for further analysis.
Passaging and cryopreservation of HSC-like cells
HSC-like cells were passaged using 0.05% Trypsin–EDTA (GIBCO, 25,300,062) for digestion. After digestion, the reaction was terminated with medium containing 10% FBS (GIBCO, 10,091–148). The cryopreservation medium, Stem Cell Banker (ZENOAQ) was used for cell freezing.
FACS
FACS was performed using the following primary antibody: anti-PDGFRB (antibody details in supplementary materials). Flow cytometry was conducted using the CytoFLEX (Beckman Coulter). Data were analyzed using FlowJo software (version 10.6.2).
Additionally, since HSCs have the capability to store Vitamin A and emit blue-purple fluorescence, they are detected under 355 nm UV laser or 405 nm violet laser.
qRT-PCR
RNA was isolated using TRIzol™ reagent (Invitrogen, 15596018CN), followed by reverse transcription of up to 1 μg of RNA into cDNA using the Revert-Aid First Strand cDNA Synthesis Kit (ThermoFisher, K1622). Quantitative reverse transcription polymerase chain reaction Polymerase Chain Reaction (qRT-PCR) was performed using TB Green Premix Ex Taq (Takara, RR420A) along with gene-specific forward and reverse primers on ABI 7500 FAST (ThermoFisher). The qPCR primer sequences are listed in Supplementary Table 1. The expression levels of target genes were normalized to the expression levels of the housekeeping gene GAPDH.
Immunofluorescence
Cells were fixed with a 1:1 mixture of methanol and acetone at 4 °C for 30 min, followed by blocking with 10% (v/v) normal donkey serum in PBS for 60 min. The primary antibodies, anti-α-SMA (1:2000) and anti-Collagen (1:2000), were incubated with the cells at 4 °C overnight. Subsequently, secondary antibodies, including CyTM5-conjugated donkey anti-goat (1:500, Jackson, 70,575,147), CyTM3-conjugated donkey anti-mouse (1:500, Jackson, 715,165,150), and Alexa Fluor-488-conjugated donkey anti-rabbit (1:500, Invitrogen, A-21206), were applied to the cells for 60 min. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at room temperature for 1–2 min, followed by washing the cells three times. Fluorescence images were acquired using the Revolve (ECHO) imaging system. Image processing was performed using Image-Pro Plus (v6.0) software.
Single cell RNA datasets
The Seurat package was used to pre-process the dataset. This dataset was obtained from the European Bioinformatics Institute under the accession number E-MTAB-7407 for fetal liver data, and the Harmony package was used to eliminate batch effects. Plots were generated using the R package ggplot2.
Bulk RNA sequencing
RNA sequencing data for HSCs was obtained from the GEO database (GSE232640), as well as RNA sequencing data for cells induced using conventional methods of Miyajima Lab. For the obtained normalized data, differential analysis between multiple samples was performed using the limma package, with a significance threshold of 0.05 and a log-fold change threshold of 1 for filtering. PCA was conducted on the preprocessed gene expression data using the vegan package, and visualization was carried out with the ggplot2 package to intuitively display the distribution features of different samples at the gene expression level. A heatmap was generated using the heatmap package to show the expression patterns of specific genes across different sample groups, clearly presenting the differences and similarities in gene expression. The correlation analysis was performed using the Spearman method via the `cor` function on normalized data to compute the pairwise correlation matrix among samples. The results were then grouped using hierarchical clustering and visualized through a heatmap to illustrate the relationships between different cell populations. Based on the results of the differential expression analysis, a volcano plot was created using the ggplot2 package to visually represent the gene expression differences between sample groups, highlighting the distribution of upregulated and downregulated genes.
Statistical analysis
Except for bulk and scRNA-seq, all data are presented as mean ± standard deviation (SD), and one-way or two-way analysis of variance (ANOVA) with Bonferroni post-hoc analysis was performed using Prism 9 (GraphPad). Significance was determined based on the degree of difference using either the Student’s two-tailed t-test or Welch’s two-tailed t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 were considered significant.
Data availability
The RNA sequencing data generated in this study are available in the NCBI Short Read Archive under the accession number: PRJNA1230113. The data is available at: https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA1230113.
References
Tsuchida, T. & Friedman, S. L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 14, 397–411. https://doi.org/10.1038/nrgastro.2017.38 (2017).
Giampieri, M. P., Jezequel, A. M. & Orlandi, F. The lipocytes in normal human liver. A Quantitative Study. Digestion 22, 165–169. https://doi.org/10.1159/000198640 (1981).
Kamm, D. R. & McCommis, K. S. Hepatic stellate cells in physiology and pathology. J. Physiol. 600, 1825–1837. https://doi.org/10.1113/jp281061 (2022).
Wake, K. “Sternzellen” in the liver: Perisinusoidal cells with special reference to storage of vitamin A. Am. J. Anat. 132, 429–462. https://doi.org/10.1002/aja.1001320404 (1971).
Lai, X. et al. Generation of functionally competent hepatic stellate cells from human stem cells to model liver fibrosis in vitro. Stem Cell Rep. 17, 2531–2547. https://doi.org/10.1016/j.stemcr.2022.09.010 (2022).
Mederacke, I. et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 4, 2823. https://doi.org/10.1038/ncomms3823 (2013).
Vilar-Gomez, E. et al. Fibrosis severity as a determinant of cause-specific mortality in patients with advanced nonalcoholic fatty liver disease: a multi-national cohort study. Gastroenterology 155, 443–457 (2018).
Chang, W. et al. Isolation and culture of hepatic stellate cells from mouse liver. Acta. Biochim. Biophys. Sin. (Shanghai) 46, 291–298. https://doi.org/10.1093/abbs/gmt143 (2014).
Xu, L. et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut 54, 142–151. https://doi.org/10.1136/gut.2004.042127 (2005).
Zhang, Q. et al. Exosomes derived from hepatitis B virus-infected hepatocytes promote liver fibrosis via miR-222/TFRC axis. Cell Biol. Toxicol. 39, 467–481. https://doi.org/10.1007/s10565-021-09684-z (2023).
Coll, M. et al. Generation of hepatic stellate cells from human pluripotent stem cells enables in vitro modeling of liver fibrosis. Cell Stem Cell 23, 101–113 (2018).
Koui, Y. et al. An in vitro human liver model by iPSC-derived parenchymal and non-parenchymal cells. Stem cell Rep. 9, 490–498 (2017).
Miyoshi, M. et al. LIM homeobox 2 promotes interaction between human iPS-derived hepatic progenitors and iPS-derived hepatic stellate-like cells. Sci. Rep. 9, 2072 (2019).
Koui, Y. et al. Development of human iPSC-derived quiescent hepatic stellate cell-like cells for drug discovery and in vitro disease modeling. Stem Cell Rep. 16, 3050–3063 (2021).
Kordes, C., Bock, H. H., Reichert, D., May, P. & Häussinger, D. Hepatic stellate cells: Current state and open questions. Biol. Chem. 402, 1021–1032 (2021).
Chen, G., Weiskirchen, S. & Weiskirchen, R. Vitamin A: too good to be bad?. Front. Pharmacol. 14, 1186336 (2023).
Hrabak, P., Kalousova, M., Krechler, T. & Tomáš, Z. Pancreatic stellate cells-rising stars in pancreatic pathologies. Physiol. Res. 70, S597 (2021).
Lübke, J. H. & Rollenhagen, A. Synapses: Multitasking global players in the Brain. Neuroforum 26, 11–24 (2020).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. https://doi.org/10.1038/nature12271 (2013).
Popescu, D. M. et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371. https://doi.org/10.1038/s41586-019-1652-y (2019).
Martínez García de la Torre, R. A. et al. Trajectory analysis of hepatic stellate cell differentiation reveals metabolic regulation of cell commitment and fibrosis. Nat Commun 16, 1489, https://doi.org/10.1038/s41467-025-56024-4 (2025).
Sun, Y., Chen, C. S. & Fu, J. Forcing stem cells to behave: a biophysical perspective of the cellular microenvironment. Annu. Rev. Biophys. 41, 519–542 (2012).
Hazeltine, L. B., Selekman, J. A. & Palecek, S. P. Engineering the human pluripotent stem cell microenvironment to direct cell fate. Biotechnol. Adv. 31, 1002–1019 (2013).
Yin, C., Evason, K. J., Asahina, K. & Stainier, D. Y. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin. Invest. 123, 1902–1910. https://doi.org/10.1172/jci66369 (2013).
Ding, B. S. et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 468, 310–315. https://doi.org/10.1038/nature09493 (2010).
Geerts, A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin. Liver Dis. 21, 311–335. https://doi.org/10.1055/s-2001-17550 (2001).
Friedman, S. L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 88, 125–172. https://doi.org/10.1152/physrev.00013.2007 (2008).
Kisseleva, T. & Brenner, D. A. Hepatic stellate cells and the reversal of fibrosis. J. Gastroenterol. Hepatol. 21(Suppl 3), S84-87. https://doi.org/10.1111/j.1440-1746.2006.04584.x (2006).
Mederacke, I., Dapito, D. H., Affò, S., Uchinami, H. & Schwabe, R. F. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 (2015).
Tian, S. P. et al. A novel efficient strategy to generate liver sinusoidal endothelial cells from human pluripotent stem cells. Sci. Rep. 14, 13831. https://doi.org/10.1038/s41598-024-64195-1 (2024).
Wang, N. et al. 3D hESC exosomes enriched with miR-6766-3p ameliorates liver fibrosis by attenuating activated stellate cells through targeting the TGFβRII-SMADS pathway. J Nanobiotechnology 19, 437. https://doi.org/10.1186/s12951-021-01138-2 (2021).
Acknowledgements
We would like to thank all the members of the Regenerative Medicine Team at Wuyi University, especially Miss Ji-Yue Yan for their research management and technical support.
Funding
National Natural Science Foundation of China, 82270697, China Foundation For Youth Entrepreneurship and Employment -Incaier Public Welfare Fund, HH25KYHX0003, Haihe Laboratory of Cell Ecosystem Innovation Fund, HH24KYZX0008, the Guangdong Basic and Applied Basic Research Foundation, 2023A1515012574, the Science and Technology Planning Project of Guangdong Province of China, 2021B1212040016, the Jiangsu Provincial Medical Key Discipline Cultivation Unit, JSDW202229
Author information
Authors and Affiliations
Contributions
Conceptualization, YWZ and JYG; writing—original draft preparation, DY and MD; writing—review and discussing, DY, MD and YWZ; supervision and resources supply, YWZ and JYG; funding acquisition, YWZ, JYG and MD; Cellular and molecular experiments DY, MD, and HXM; Single-Cell RNA-seq and bulk RNA sequencing analysis, YMS and WHC (assistant); All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ye, D., Ding, M., Song, YM. et al. Liver developmental microenvironment promotes iHSC generation from human iPSCs. Sci Rep 15, 22338 (2025). https://doi.org/10.1038/s41598-025-09026-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09026-7