Abstract
In 2019, the severe acute respiratory syndrome coronavirus 2 virus (SARS-CoV-2) started to spread globally and caused the COVID-19 pandemic. SARS-CoV-2, like other members of the Coronaviridae, has a single-stranded, positive sense RNA genome about 30 kb in length, which is translated to generate 16 non-structural proteins (NSPs); a set of sub-genomic mRNAs encode the structural and accessory proteins. The ORF1a precursor includes NSP1-11 and is processed by virus-encoded proteases to produce the mature proteins. We recently identified a short, highly conserved motif (YCPRP) within the structural protein precursor of foot-and-mouth disease virus (FMDV), a member of the Picornaviridae. This motif is conserved among picornaviruses and is found as (W/F/Y)-x-P-R-(P/A). The motif has a major influence on the processing of the FMDV capsid precursor (P1-2A) by the viral protease 3Cpro. We have now identified a similar motif (WVPRA) within the NSP2 of SARS-CoV-2. Interestingly, this motif is required for the efficient processing of the NSP1-NSP2 junction by the SARS-CoV-2 protease PLpro (NSP3) and a single amino acid substitution within the motif can abrogate cleavage of this junction. We hypothesise that this motif acts, within NSP1-NSP2, to enable this precursor to fold correctly and allow efficient processing of the NSP1/NSP2 junction.
Similar content being viewed by others
Introduction
In December 2019, a previously unknown coronavirus caused an outbreak of a respiratory disease among people in the city of Wuhan in China1. Since then, the disease has spread globally and caused the COVID-19 pandemic. The causative virus is called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), it primarily affects the respiratory system causing influenza-like symptoms such as coughing, fever and in more severe cases, breathing problems2.
SARS-CoV-2 belongs to the family Coronaviridae, these viruses have a single stranded, positive-sense, RNA genome (26 to 32 kb in length), which is bound by the nucleocapsid (N) protein and enclosed within a lipid envelope that incorporates the other viral structural proteins, namely the spike (S), envelope (E) and membrane (M) proteins. The SARS-CoV-2 RNA genome is capped at its 5´-terminus, polyadenylated at its 3´-terminus and is infectious by itself following entry into cells3. The virus family is classified into four genera, alpha-, beta-, gamma-, and delta- coronaviruses4and these four genera are further subdivided into four lineage subgroups (A, B, C and D)5. The SARS-CoV-2 (beta-CoV, subgroup B) genome shows a sequence identity of 80.0% to that of SARS-CoV (beta-CoV, subgroup B), 56.9% to the Middle East Respiratory Syndrome (MERS) CoV (beta-CoV, subgroup C), 51.6% to Human coronavirus (HcoV)-HKU1 (beta-CoV, subgroup A) and 50.5% to HcoV-C43 (beta-CoV, subgroup A)6. The human alphacoronaviruses HcoV-NL63 and HcoV-229E have a sequence identity of 48.7% and 47.7%, respectively, to the SARS-CoV-2 genome6.
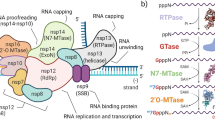
Coronavirus replication takes place within an extensive membranous network of virus-modified vesicles derived from the endoplasmic reticulum within the cytoplasm7. The first 20 kb of the RNA genome, from the 5´-terminus, serves as an mRNA for the synthesis of two large polyproteins from two partially overlapping reading frames termed ORF1a and ORF1b. The latter sequence is only accessed following programmed ribosomal frameshifting7. The two polyproteins, termed pp1a and pp1ab, are cleaved by virus-encoded proteases present within the polyproteins, which results in the production of 16 mature non-structural proteins (NSPs), called NSP1 to NSP16 (Fig. 1). The proteins encoded by ORF1a are largely involved in the inhibition of the host cellular innate immune responses, whereas the proteins encoded by ORF1b are required for genome replication, e.g. the RNA dependent RNA polymerase. The pp1a includes two viral proteases. These are the papain-like protease (PLpro or NSP3) and the 3C-like protease (3CLPro or NSP5). The 3CLPro is responsible for cleavage of the two viral polyproteins at 10 different sites while the PLpro (NSP3) cleaves the pp1a polyprotein at 3 other sites, namely the junctions between NSP1/NSP2, NSP2/NSP3 and NSP3/NSP4 7, see Fig. 1.
Schematic representation of the SARS-CoV-2 genome organization. The RNA genome is translated from two overlapping reading frames, ORF1a and ORF1b, to generate two different polyproteins, pp1a and pp1ab, the latter is generated following a programmed ribosomal frameshift that occurs within the overlap between ORF1a and ORF1b. The two polyproteins are cleaved by the virus-encoded proteases PLpro (NSP3) and 3CLpro (NSP5) into a total of 16 non-structural proteins (NSPs). The sites cleaved by the PLpro (marked with red arrows) and 3CLpro (marked with yellow arrows) are indicated. Downstream of the ORF1b, the RNA genome encodes 10 more proteins, which are translated from sub-genomic mRNAs, including the structural proteins (spike (S), envelope (E), membrane (M) and nucleocapsid (N)) and several accessory proteins. The conserved motif (WVPRA) is found within NSP2 of pp1a and is marked on the lower part of the figure. Created in BioRender. Kristensen, T. (2025) https://BioRender.com/s80a354.
In an earlier study on foot-and-mouth disease virus (FMDV) capsid precursor processing, we identified a highly conserved motif of 5 amino acids (YCPRP), which is present within the VP1 region of the P1-2A capsid precursor8 of this picornavirus. This motif (with few variations) was also identified in the VP1 of other picornaviruses, including Cardioviruses (which have FCPRP), Hepatoviruses (with YFPRP) and Enteroviruses (with WCPRP). Note, these different forms of this motif each start with an aromatic amino acid (aa) residue (Y, F or W). Single amino acid substitutions within this motif of the FMDV P1-2A resulted in a capsid precursor that was highly resistant to proteolytic cleavage by the FMDV 3C protease (3Cpro) at each of the internal sites8i.e. at the junctions between VP0/VP3 and VP3/VP1, the first of these being more than 400 aa away from the YCPRP motif. This was surprising since, in contrast, we have previously demonstrated that cleavage of these junctions is independent of each other and that blocking cleavage at one site does not prevent cleavage of the other sites9thus indicating the crucial role of this motif in the precursor processing. Furthermore, single substitutions within this conserved motif resulted in the loss of virus infectivity8,10. Thus, it was suggested that this motif is crucial for the correct folding of the capsid precursor to allow subsequent processing into the mature capsid proteins. We hypothesized that this motif may serve as an important binding site for protein chaperones, which may be involved in achieving the correct folding of the precursor8,10 prior to cleavage. Several studies have reported the involvement of such chaperones, e.g. Hsp70 and Hsp90, in picornavirus capsid processing11,12.
Interestingly, we have now identified a similar, but distinct, motif (WVPRA) in the NSP2 protein of SARS-CoV-2. From sequence alignments, we found that this motif is also present (and thus conserved) in other betacoronaviruses, within subgroup A and B, including SARS-CoV, MERS-CoV, human CoV OC43, bovine CoV, equine CoV and several bat coronaviruses (Fig. 2). In this study, we have shown that this motif, within the NSP2 protein of SARS-CoV-2, has high importance for the cleavage of the NSP1-NSP2 junction by PLpro (NSP3) even though it is located far away in the linear sequence (Fig. 3).
Alignment of partial NSP2 amino acid sequences containing the conserved motif from 9 different betacoronaviruses. The alignment was performed using ClustalW in Geneious 9.0.5. The entire ORF1a was included in the alignment but just a short region of the NSP2 sequence is shown here. The amino acids are numbered based on the sequence of the SARS-CoV-2 NSP2. The conserved motif is highlighted in different colors depending on their amino acid properties, i.e. W and F have similar properties (aromatic and non-polar) and thus have a similar color. V, L, I and A have similar properties to each other (aliphatic and non-polar) so also have a similar color. Only the P residue (which has distinct properties as an imino acid) is totally conserved. The R-V/I change is rather non-conservative. A consensus sequence from the alignment is shown in the bottom of the Figure where x represents any amino acid. The sequences used were as follows: SARS-CoV-2 (Beta/subB) (GenBank accession number QWC81719.1), SARS-CoV (Beta/subB) (RefSeq accession number YP_009944365.1), Bat coronavirus 279/2005(Beta/SubB) (GenBank accession number GCA_031121315.1), Bat SARS coronavirus HKU3(Beta/SubB) (GenBank accession number QND76018.1), MERS-CoV (Beta/SubB) (GenBank accession number AVN89452.1), Murine hepatitis virus(Beta/subA) (GenBank accession number AWB14623.1), Human coronavirus OC43(Beta/subA) (GenBank accession number GCA_003972325.1), Equine coronavirus(Beta/subA) (GenBank accession number UGN73921.1) and Bovine coronavirus(Beta/subA) (GenBank accession number UZT75406.1).
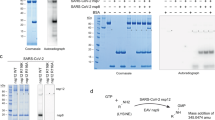
Overview of the coding capacity of plasmids used in the transient expression assays. (a) The coding sequence for the SARS-CoV-2 NSP1-NSP2 plus a C-terminal Flag-tag and a stop codon were inserted into the pGEM-3Z plasmid, containing the T7 promoter upstream of the insert. The lengths of both NSP1 and NSP2 (in terms of the number of amino acid residues) are indicated, and the location of the conserved motif (WVPRA) is marked. This plasmid is referred to as the wt. (b) The NSP3-mCherry construct, encoding a functional PLpro, was a gift from Bruno Antonny (see Materials and Methods)14, a T7 promoter sequence was inserted into this plasmid, upstream of the coding sequence for NSP3, as described in Materials & Methods. Created in BioRender. Kristensen, T. (2025) https://BioRender.com/d9lyyyv.
Materials and methods
Plasmid design
The SARS-CoV-2 sequence (from Wuhan) was obtained from NCBI (RefSeq: NC_045512.2). The cDNA corresponding to the NSP1-NSP2 coding sequence (ca. 2500 nt) was modified by adding the sequence for a C-terminal Flag-tag followed by two stop codons. The entire coding sequence was flanked by restriction sites: BamHI (upstream of the coding sequence) and XhoI, XbaI and ApaI downstream of the Flag-tag. The sequence was inserted into the vector pcDNA3.1 (customer designed and produced by GenScript, USA Inc). Following receipt, the plasmid was transformed into chemically competent Top10 Escherichia coli (E. coli) cells, amplified and purified using a Plasmid Midi Kit (Qiagen).
The pcDNA3.1 plasmid with its NSP1-NSP2 cDNA insert and the empty pGEM-3Z vector (Promega) were digested with BamHI and XbaI and the products were separated by gel electrophoresis and purified using a GeneJet Gel Purification Kit (Thermo Fisher Scientific). The insert and the linearized pGEM-3Z vector were joined using T4 DNA ligase (Thermo Fisher Scientific) and transformed into chemically competent E. coli cells, amplified and plated onto LB agar plates containing carbenicillin. Colonies were screened for the presence of the insert by digestion with BamHI and XbaI. Plasmids with the required structure were amplified and purified as described above.
Variants of the plasmids were prepared using site-directed mutagenesis13. Briefly, fragments were amplified using Phusion® High-Fidelity DNA Polymerase (Thermo Fisher Scientific) and the pGEM-3Z containing the wt Flag-tagged NSP1-NSP2 coding sequence as template. For each modification, one primer with the desired mutation was used together with a wt primer and template to generate a megaprimer (ca. 900 bp), see Supplementary Table 1. The megaprimers were gel purified using the GeneJet Gel Purification Kit (Thermo Fisher Scientific) and used in a mega PCR together with the template. The mega PCR products were digested with DpnI and transformed into chemically competent E. coli cells. Plasmid DNA from individual colonies was screened by Sanger Sequencing (LGC BiosearchTechnologies, Berlin, Germany) for the correct modifications and positive clones were amplified and purified as above.
The plasmid NSP3-mCherry, encoding a functional PLpro, was a gift from Bruno Antonny (Addgene plasmid # 165131 ; http://n2t.net/addgene:165131 ; RRID: Addgene_165131)14. The plasmid was modified to contain the T7 promoter sequence upstream of the NSP3 sequence (see Supplementary Table 1) by site-directed mutagenesis as described above.
Transient expression assays
Baby hamster kidney 21 (BHK-21) cells were seeded into 6-well plates approximately 24 h before starting the transfection assay. The BHK-21 cells were 80–90% confluent when they were infected with the recombinant vaccinia virus, termed vTF7-3, which expresses the T7 RNA polymerase15. All the NSP1-NSP2 constructs were in the pGEM-3Z vector, which contains the T7 promoter upstream of the coding sequence. Starting from the NSP3-mCherry plasmid14we modified it to contain the T7 promoter upstream of the NSP3 coding sequence. After a 1 h incubation with vTF7-3 in 5% CO2 at 37 °C, the medium containing this vaccinia virus was removed and the cells were transfected with the required plasmid DNAs using FuGENE 6 (Promega) as described previously16. For each well, 1000 ng of the various NSP1-NSP2 plasmids was used, either alone or in combination with 50 ng of the T7-NSP3-mCherry construct. The cells were incubated in 5% CO2 at 37 °C overnight. The next day, the medium was removed and the cells were lysed with 500 µl Buffer C (20mM Tris-HCl (pH 8.0), 125 mM NaCl and 0.5% NP-40). After harvesting, the cell lysates were clarified by centrifugation at 13,000 x g for 10 min at 4 °C.
Immunoblot analysis
Prior to immunoblotting, cell lysates were mixed with an equal volume of 2 x Laemmli sample buffer (Bio-Rad) containing 25 mM dithiothreitol (DTT) and boiled for 5 min. The proteins were separated by SDS-PAGE (Any KD Bis-Tris gels (Bio-Rad)) and transferred to a PVDF membrane (Bio-Rad). PBS containing 0.1% Tween (PBST) and 5% bovine serum albumin (BSA) was used as blocking buffer and dilution buffer for both the primary and secondary antibodies. The primary antibody used was anti-Flag rabbit polyclonal antibody (1:1000, Proteintech 20543-1-AP) and the secondary antibody was HRP-conjugated goat anti-rabbit IgG (H + L) (1:500, Thermo Fisher Scientific 32460). Following these incubations, a chemiluminescence kit (Pierce ® ECL Western Blotting Substrate, Thermo Fisher Scientific) was used to detect the target proteins. Images were captured using a Chem-Doc XRS system (Bio-Rad) as described previously8.
Statistics
Each experiment was independently repeated three times, with separate transfections and western blots. Band intensities were quantified using pixel count in ImageJ (version 1.54 g, National Institute of Health, USA). For background correction, an area of the same size as the band was selected within the negative control lane and subtracted from the pixel count from the band of interest. To normalize for differences in overall expression between samples, each lane was set to 100%, and the relative percentage of each of the two bands was calculated. The mean values of the triplicates were plotted as bar graphs, with error bars representing the standard deviation (SD). The statistical significance was calculated between the amount of NSP2 protein in the NSP1-2 (wt) + PLpro sample and the amount of NSP2 protein in each of the NSP1-2 mutants + PLpro using an unpaired parametric t-test with Welch’s correction in GraphPad Prism (version 10, San Diego, CA, USA ).
Results
Deletion of the WVPRA motif alone prevents processing of the NSP1-NSP2 junction by PLpro
In order to assess the importance of the WVPRA motif within NSP2 for protein processing, a small deletion, removing just the sequence encoding this 5 amino acid motif, was introduced into the NSP1-NSP2 precursor coding sequence, this construct is referred to as NSP1-NSP2 (NSP2 Δ243–247). This construct generated a product very similar in size to the wt protein in the absence of PLpro (Fig. 4, lane 3). A faint band slightly smaller than the full-length NSP1-NSP2 precursor was observed in both the wt and the deletion mutant NSP1-NSP2 in the absence of PLPro. This likely represents a low level of non-specific cleavage (Fig. 4, lane 1 and 3). It is clear that in the presence of PLpro the NSP1-NSP2 (wt) was efficiently processed. However, it was observed that this NSP1-NSP2 (NSP2 Δ243–247) mutant precursor was highly resistant to cleavage (Fig. 4, lane 4) by PLpro although this 5 residue deletion was over 240 residues away from the cleavage site. The experiment was repeated three times with separate transfections and western blots and resulted in very similar results. The additional experiments are shown in Supplementary Figs. S2 and S3. Band intensities for the precursor and NSP2 cleavage product from each experiment were quantified and the relative percentage of each of the two bands was calculated. The mean values of the triplicates were plotted as a bar diagram and are shown in Fig. 4. The amount of processed NSP2 protein from the NSP1-NSP2 (NSP2 Δ243–247) mutant co-expressed with PLpro was much lower than that produced from the NSP1-NSP2 (wt) under the same conditions (P ≤ 0.005).
A short, conserved motif within SARS-CoV-2 NSP2 is required for processing of the NSP1/NSP2 junction by the PLpro (NSP3). Cell lysates from BHK cells infected with vTF7-315 and transfected with plasmids that express the SARS-CoV-2 NSP1-NSP2-Flag (wt or with a small deletion, removing the conserved motif) either alone (odd numbered lanes) or with the plasmid encoding the SARS-CoV-2 PLpro (even numbered lanes) were analyzed by immunoblotting using rabbit anti-Flag antibodies, followed by anti-rabbit HRP-conjugated secondary antibodies and a chemiluminescence detection kit. Molecular mass markers (kDa) are indicated on the left. A negative control (no DNA) was included (lane 5). The uncut membrane is shown in Supplementary Fig. S1. This experiment has been repeated three times in total with separate transfections and western blots and resulted in very similar results. The additional two experiments are shown in Supplementary Figs. S2 and S3. Band intensities for each experiment were quantified using pixel count in ImageJ. Each lane was set to 100%, and the relative percentage of each of the two bands was calculated. The mean values of the triplicates were plotted, and the bar diagram with error bars representing the standard deviation (SD) are shown on the right side of the figure.
Identification of critical residues within the WVPRA motif for NSP1-NSP2 processing
To identify individual residues of high importance within the WVPRA motif, single amino acid substitutions were introduced into it. The wt and the mutant precursors were expressed within cells, either in the absence or presence of the PLpro. Transfections of these constructs followed by western blotting were repeated three times and resulted in very similar results. The results of one experiment are shown in Fig. 5, and the additional experiments are shown in Supplementary Figs. S5, S6, S8 and S9. Band intensities for the precursors and products in each experiment were quantified, and each lane was set to 100%, and the relative percentage of each of the two bands was calculated. The mean values were plotted and are shown in Fig. 5. The NSP1-NSP2 (wt) and each of the mutants all generated the expected, similarly sized, products in the absence of the PLpro (Fig. 5, odd numbered lanes). A faint band slightly smaller than the full-length NSP1-NSP2 precursor was observed in both the wt and the mutants in the absence of PLPro. This likely represents a low level of non-specific cleavage (Fig. 5, odd numbered lanes). In the presence of PLpro, the NSP1-NSP2 (wt) was cleaved to generate the Flag-tagged NSP2, as above (Fig. 5a and b, lane 2). However, there was no processing observed for the NSP1-NSP2 (NSP2 W243A) mutant due to the presence of PLpro (P ≤ 0.0005) (Fig. 5a, lane 4). In contrast, some processing was observed for both the NSP1-NSP2 (NSP2 W243Y) (Fig. 5a, lane 6) and NSP1-NSP2 (NSP2 W243F) (Fig. 5a, lane 8) precursors in which rather conservative amino acid substitutions had been made (W, Y and F are all aromatic residues whereas A, in contrast, is a small non-polar residue). However, although some processing was observed, the amount of processed NSP2 produced from the NSP1-NSP2 (NSP2 W243Y) was significantly lower than the amount of the NSP2 observed from the wt precursor (P ≤ 0.005). Processing, at a similar level as in the wt, was observed for the NSP1-NSP2 (R246A) mutant (no significant difference) (Fig. 5b, lane 4) (with a change in a different residue within the motif) in the presence of PLPro. Cleavage was also observed with the NSP1-NSP2 (P245A) mutant in the presence of PLPro, however it is noteworthy that for this mutant more unprocessed NSP1-NSP2 precursor remained compared to the wt, and the amount of processed NSP2 was significantly lower in the NSP1-NSP2 (P245A) mutant compared to the wt in the presence of PLPro (P ≤ 0.005). The NSP1-NSP2 (P245G) mutant yielded less cleaved NSP2 compared to both the wt and the NSP1-NSP2 (P245A) mutant, indicating less efficient cleavage in the presence of PLPro (P ≤ 0.005, compared to the wt).
Identification of amino acid residues within the conserved WVPRA motif within the SARS-CoV-2 NSP2 that are required for efficient processing of the NSP1/NSP2 junction by PLpro (NSP3). Cell lysates from BHK cells infected with vTF7-315 and transfected with plasmids that express the SARS-CoV-2 NSP1-NSP2-Flag (wt or with single amino acid substitutions within the conserved WVPRA motif) either alone (odd numbered lanes) or with the plasmid encoding the SARS-CoV-2 PLpro (even numbered lanes) were analyzed by immunoblotting as in Fig. 4. Molecular mass markers (kDa) are indicated on the left. A negative control was included (lane 9). The NSP1-NSP2-Flag and the NSP2-Flag products are indicated on the right side of the western blot picture and were detected using anti-Flag-antibodies. Panel (a) shows the wt and various substitutions of the NSP2 W243 and a negative control. Panel (b) shows the wt, the NSP2 R246A, the NSP2 P247A, NSP2 P247G and a negative control. The uncut membranes are shown in Supplementary Figs. S4 and S7. This experiment has been repeated three times in total with separate transfections and western blots and resulted in very similar results. The additional experiments are shown in Supplementary Figs. S5, S6, S8 and S9. Band intensities for each experiment were quantified using pixel count in ImageJ. Each lane was set to 100%, and the relative percentage of each of the two bands was calculated. The mean values of the triplicates were plotted, and the bar diagram with error bars representing the standard deviation (SD) are shown on the right side of panels a and b.
Discussion
The function of the NSP2 in coronaviruses is not well defined. It has been proposed that the SARS-CoV NSP2 is involved in disrupting intracellular host signaling through interaction with two host proteins termed prohibitin 1 and 217. The NSP2 coding sequence has been completely deleted from the RNA genome of Murine Hepatitis Virus (MHV) and SARS-CoV, and these mutant viruses could still be rescued, proving that NSP2 is not essential for virus growth in cell culture18. However, these viruses showed attenuated viral growth and RNA synthesis compared to the wt18. One study has suggested that the SARS-CoV-2 NSP2 stimulates translation. In HEK293T-cells, expression of the NSP2 protein increased the protein synthesis rate from mRNAs using both cap- and IRES-dependent translation initiation mechanisms19. Another study showed that SARS-CoV-2 NSP2 impedes translation of Ifnb1 transcripts, which encode type I interferon β (IFN-β), by interacting with the GIGYF2 protein20. This interaction increases the binding of the GIGYF2 protein to the mRNA cap-binding protein 4EHP, thereby repressing translation of the Ifnb mRNA. Furthermore, analysis of the NSP2 structure, obtained using cryo-electron microscopy and structure prediction, revealed a highly conserved zinc ion-binding site, as present in a number of RNA binding proteins such as RNA polymerases and ribosomes, suggesting a possible role for NSP2 in such interactions21.
We recently identified a short motif (YCPRP) within the FMDV P1-2A precursor (VP1 in the mature capsid protein) and found it to be conserved among all FMDVs. Furthermore, this motif was also highly conserved across many members of the Picornavirus family, including within cardioviruses, hepatoviruses and enteroviruses. When the motif was modified in the context of FMDV, processing of the FMDV P1-2A precursor by the 3Cpro was blocked8. This was consistent with earlier studies by Ryan et al.,22 indicating that truncation of the P1-2A capsid precursor blocked processing in vitro. Furthermore, another earlier study had observed a similar effect with the poliovirus capsid precursor (P1) processing by 3CDpro, when the C-terminus of P1 (including the closely related WCPRP motif) was removed then processing of the residual P1 was blocked23.
On the basis of the results from both SARS-CoV-2 (as described here) and FMDV8,10, it seems likely that the overall structures of the precursor proteins are adversely affected upon changing or deleting amino acids within these distinct, but closely related, motifs. This would explain why the proteases are not able to recognize the unchanged cleavage sites even when these are located far from the motif in the linear sequence. The short conserved motif (YCPRP) within the C-terminus of FMDV VP1 is extremely important for the cleavage of the capsid precursor P1-2A by 3C[pro8. Deleting this motif within the FMDV capsid precursor completely abrogated its processing even though the cleavage sites at the VP0/VP3 and VP3/VP1 junctions were located more than 400 aa and around 190 aa away from the deleted motif, respectively. Here, we have now shown that the related motif (WVPRA) within NSP2 of SARS-CoV-2 is necessary for efficient processing of the NSP1/NSP2 junction by the PLpro. Deletion of the WVPRA motif resulted in the NSP1-NSP2 precursor being highly resistant to cleavage by PLpro (P ≤ 0.005) (see Fig. 5, lane 4). We have also analyzed more subtle changes in these motifs. The substitution of the first aromatic amino acids in these motifs, Y in the FMDV P1-2A precursor and the W in the SARS-CoV-2 NSP1-NSP2 precursor, to an A had similarly adverse effects on cleavage of the two precursors (P ≤ 0.0005 for the NSP1-NSP2 (NSP2 W243A)). The substitution of the second amino acid in the motif, C to an A, in the FMDV P1-2A precursor, did not have any obvious effect on cleavage, it is also noteworthy that this amino acid is rather non-conserved among different picornaviruses. However, surprisingly, substitution of the third residue P to an A in the FMDV P1-2A precursor, also did not have any apparent effect on the cleavage assays although this amino acid is fully conserved amongst all the picornaviruses that we checked including various cardioviruses, hepatoviruses and enteroviruses8. Furthermore, a mutant FMDV RNA transcript encoding the VP1 P187A substitution did not produce infectious virus following introduction into cells, clearly indicating the importance of this residue. Due to these observations with FMDV and the fact that the residue is conserved in beta-coronaviruses, we also tested the effect on cleavage of changing proline to an alanine in the WVPRA motif. Cleavage was observed for the SARS-CoV-2 NSP1-NSP2 (NSP2 P245A) mutant, however the cleavage efficiency was lower than the wt, as there was still residual precursor detected (Fig. 5b, lane 6, and bar graph) (P ≤ 0.005). To investigate the effect of this residue further, the proline in the WVPRA motif was also changed to a glycine. Less cleavage of the NSP1-NSP2 junction was observed for this mutant (Fig. 5b, lane 8) compared to the alanine substitution (Fig. 5b, lane 6 and bar graph), suggesting that the alanine residue is tolerated better (P ≤ 0.005, compared to the wt). Glycine and proline residues within proteins have unusual properties; proline (as an imino acid) introduces a “kink” into the polypeptide chain and glycine residues allow a much greater degree of chain conformations than other residues. Hence, the proline to glycine substitution had been expected to be less detrimental to the protein structure than the alanine substitution but it appeared that the effect of the P to G substitution on cleavage was greater than the P to A change. The FMDV VP1 R188 residue had a large effect on cleavage of the capsid precursor by the 3C[pro8. Furthermore, the FMDV (VP1 R188A) substitution also adversely affected the efficiency of virus rescue from RNA and a secondary compensatory substitution, W129R in VP2, was observed after a few passages of the recovered virus in cells10. Interestingly, we found that introducing this secondary VP2 (W129R) substitution into the FMDV P1-2A (VP1 R188A) mutant could reverse the block on cleavage, thus showing the importance of the interaction between amino acids within this conserved motif to other sites in the precursor10. The SARS-CoV-2 NSP2 R246 is not as conserved (Fig. 2) as the FMDV VP1 R188. However, due to the marked effect observed in FMDV, the importance of this residue in the NSP2 was also tested by substitution to an alanine. The NSP2 R246A substitution did not have a huge effect on the cleavage of the NSP1-NSP2 precursor (no significant difference) (Fig. 5b, lane 4, and bar graph). However, it seemed that the processing had slowed to some degree, since more precursor remained for this mutant, compared to the wt (Fig. 5b, cf. lane 4 and lane 2).
A recent study has investigated the processing by the SARS-CoV-2 PLpro of various small synthetic peptides corresponding to the junction sequences (P8, P7, P6, P5, P4, P3, P2, P1- P1´, P2´, P3´, P4´, P5´, P6´, P7´, P8´) of NSP1-NSP2 (LMRELNGG-AYTRYVDN), NSP2-NSP3 (NTFTLKGG-APTKVTFG) and NSP3-NSP4 (TKIALKGG-KIVNNWLK)24. Surprisingly, the study reported that the PLPro was not able to process the NSP1-NSP2 junction peptide at all, despite being able to process peptides corresponding to the two other junctions24. The NSP3-NSP4 junction was not very efficiently processed either, whereas the NSP2-NSP3 junction was very effectively processed, and the authors hypothesized that the proline at residue P2´ in the NSP2-NSP3 sequence might influence the effectiveness of the processing. Thus, the P2´ residue at the NSP1-NSP2 junction in NSP2 (Y2P ) was changed to a proline in the peptide to investigate the effect on processing24. Interestingly the small peptide corresponding to the NSP1-NSP2 junction containing the NSP2 (Y2P) substitution was processed very effectively, clearly indicating the importance of this residue. Furthermore, the P2´ residue at the NSP3-NSP4 junction was also changed to a proline in NSP4 (I2P) to investigate the effect on this processing. Interestingly this also greatly enhanced processing24.
It is very likely that there is some kind of interaction, either direct or indirect, between the NSP1-NSP2 junction and the conserved motif which is necessary for correct processing of the junction. The fact that the PLPro was not able to process the wt NSP1-NSP2 junction peptide24 further suggests that there are some other regions in the polyprotein that are necessary for correct processing of this specific junction, and it is very easy to propose that the conserved motif is involved in this. The introduction of the P residue may well influence the structure of the short peptide substrate and hence facilitate its recognition, the conserved motif may function in an analogous manner for the whole protein.
It is interesting to note that similar motifs exist in various different virus families and within different proteins, i.e. in the structural protein (P1-2A or P1) precursor of different picornaviruses and in the NSP2 of multiple betacoronaviruses. Furthermore, the motif seems to have the same important effect on processing in each case. We have also identified similar motifs (of the form Y/F/W-x-P-R-P/A) in other proteins within other viruses, including: human astrovirus, duck astrovirus, canine astrovirus, bovine astrovirus, porcine astrovirus, macacine betaherpes virus 9, Testudinid alphaherpesvirus 3, human papillomavirus 18, human circovirus, Canis familiaris papillomavirus 13, atypical porcine pestivirus 1 and classical swine fever virus. For some of these viruses, the motif is also located within a precursor protein, which is subsequently processed to generate the mature proteins, as with the SARS-CoV-2 NSP2 and FMDV VP1. However, this motif might also affect folding of proteins that are not part of precursor proteins and this could in turn affect interactions of such proteins with other proteins. Moreover, we also found that a similar motif (WGVRP) is present within the Enterobacteria phage K1F. Interestingly, in Enterobacteria phage K1F, this motif is located within a protein previously defined as an intramolecular chaperone. The intramolecular chaperone is located at the C-terminus of the trimeric tail spike protein and is also referred to as the C-terminal domain (CTD)25. This intramolecular chaperone, is part of a larger precursor polyprotein and has been shown to be necessary for correct folding of the precursor polyprotein. After the folding of the precursor, the CTD is cleaved off after a conserved serine residue and this cleavage is necessary to stabilize the native protein complex25.
Originally, an intramolecular chaperone was defined as a pro-region or a pro-sequence, which is covalently linked to the newly synthesized polypeptide26. Here it guides and catalyzes the folding of the protein, but subsequently it is cleaved off and thus does not remain as part of the mature protein. However, it has been suggested by Ma et al.,26 that there is also another type of intermolecular chaperone that is not subsequently removed. These chaperones have been referred to as intramolecular chaperone-like building blocks as they are incorporated into the mature protein26. Both groups of intramolecular chaperones play important roles in protein folding and hence in protein function.
It is very attractive for viruses to produce proteins that have multiple functions, as the viral coding capacity is quite limited. Thus, it is feasible that a protein that functions as an intramolecular chaperone may also be used within the mature virus as is the case for the FMDV VP1 (where the motif is located). However, the SARS-CoV-2 NSP2 is different from the FMDV VP1, as it is not included in the mature virus particle, and furthermore the functions of this protein are not fully defined. It is noteworthy that both FMDV (plus many other picornaviruses) and SARS-CoV-2 (plus many other betacoronaviruses) contain these similar motifs, and that for FMDV and now SARS-COV-2 it has been shown that modification of these motifs in the polyproteins can have a dramatic effect on cleavage of the polyprotein even though the cleavage sites/site are located far away (in the linear sequence) from the site of the conserved motif.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and the supplementary materials. The data generated, used and analyzed in this study are available upon request from the corresponding author.
References
Wu, D., Wu, T., Liu, Q. & Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 94, 44–48 (2020).
Naqvi, A. A. T. et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. BBA Mol. Basis Dis. 1866, 1–17 (2020).
Beyerstedt, S., Casaro, E. B. & Rangel, É. B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 40, 905–919 (2021).
Wu, A. et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328 (2020).
Murugan, C. et al. COVID-19: A review of newly formed viral clades, pathophysiology, therapeutic strategies and current vaccination tasks. Int. J. Biol. Macromol. 193, 1165–1200 (2021).
Cicaloni, V. et al. A bioinformatics approach to investigate structural and non-structural proteins in human coronaviruses. Front. Genet. 13, 1–12 (2022).
Knoops, K. et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6, 1957–1974 (2008).
Kristensen, T. & Belsham, G. J. Identification of a short, highly conserved, motif required for picornavirus capsid precursor processing at distal sites. PLoS Pathog. 15(1) (2019).
Kristensen, T., Newman, J., Guan, S. H., Tuthill, T. J. & Belsham, G. J. Cleavages at the three junctions within the foot-and-mouth disease virus capsid precursor (P1–2A) by the 3 C protease are mutually independent. Virology 522, 260–270 (2018).
Kristensen, T. & Belsham, G. J. Identification of plasticity and interactions of a highly conserved motif within a picornavirus capsid precursor required for virus infectivity. Sci. Rep. 9, 1–10 (2019).
Macejak, D. G. & Sarnow, P. Association of heat shock protein 70 with enterovirus capsid precursor P1 in infected human cells. J. Virol. 66, 1520–1527 (1992).
Newman, J. et al. The cellular chaperone heat shock protein 90 is required for foot-and-mouth disease virus capsid precursor processing and assembly of capsid pentamers. J. Virol. 92, 1–14 (2018).
Chen, G. J., Qiu, N., Karrer, C., Caspers, P. & Page, M. G. P. Restriction site-free insertion of PCR products directionally into vectors. Biotechniques 28, 498–505 (2000).
Miserey-Lenkei, S. et al. A comprehensive library of fluorescent constructs of SARS-CoV-2 proteins and their initial characterisation in different cell types. Biol. Cell. 113, 311–328 (2021).
Fuerst, T. R., Niles, E. G., Studier, F. W. & Moss, B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83, 8122–8126 (1986).
Belsham, G. J., Nielsen, I., Normann, P., Royall, E. & Roberts, L. O. Monocistronic mRNAs containing defective hepatitis C virus-like picornavirus internal ribosome entry site elements in their 5′ untranslated regions are efficiently translated in cells by a cap-dependent mechanism. RNA 14, 1671–1680 (2008).
Cornillez-Ty, C. T., Liao, L., Yates, J. R., Kuhn, P. & Buchmeier, M. J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 83, 10314–10318 (2009).
Graham, R. L., Sims, A. C., Baric, R. S. & Denison, M. R. The nsp2 proteins of mouse hepatitis virus and SARS coronavirus are dispensable for viral replication. Adv. Exp. Med. Biol. 581, 67–72 (2006).
Korneeva, N. et al. SARS–CoV–2 viral protein Nsp2 stimulates translation under normal and hypoxic conditions. Virol. J. https://doi.org/10.1186/s12985-023-02021-2 (2023).
Xu, Z. et al. SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation. Proc. Natl. Acad. Sci. U. S. A. 119, 1–9 (2022).
Gupta, M. et al. CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes. bioRxiv. https://doi.org/10.1101/2021.05.10.443524 (2021).
Ryan, M. D., Belsham, G. J. & King, A. M. Q. Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology 173, 35–45 (1989).
Ypma-Wong, M. F. & Semler, B. L. Processing determinants required for in vitro cleavage of the poliovirus P1 precursor to capsid proteins. J. Virol. 61, 3181–3189 (1987).
Chan, H. T. H. et al. Studies on the selectivity of the SARS-CoV-2 papain-like protease reveal the importance of the P2′ proline of the viral polyprotein. RSC Chem. Biol. 5, 117–130 (2023).
Schwarzer, D., Stummeyer, K., Gerardy-Schahn, R. & Mühlenhoff, M. Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail Spike and fiber proteins. J. Biol. Chem. 282, 2821–2831 (2007).
Ma, B., Tsai, C. J. & Nussinov, R. Binding and folding: In search of intramolecular chaperone-like Building block fragments. Protein Eng. 13, 617–627 (2000).
Funding
This research was funded by the Danish Veterinary and Food Administration (FVST) as part of the agreement for commissioned work between the Danish Ministry of Food and Agriculture and Fisheries and the University of Copenhagen.
Author information
Authors and Affiliations
Contributions
T.K. and G.J.B. were responsible for conceiving the idea, planning and designing this study. T.K. and P.N. were responsible for the lab work carried out in this study. T.K. and G.J.B. analyzed the results. T.K. wrote the initial draft of the manuscript and G.J.B. read and modified it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kristensen, T., Normann, P. & Belsham, G.J. A conserved motif within the NSP2 of SARS-CoV-2 is required for processing of the distal NSP1/NSP2 junction by NSP3. Sci Rep 15, 25797 (2025). https://doi.org/10.1038/s41598-025-10244-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10244-2