Abstract
Numerous reports have highlighted distinct clinical and biological characteristics between right-sided and left-sided colon cancers. Despite this research on commonalities and divergences among colorectal cancers (CRC) at different locations remain sparse. In order to elucidate gene expression disparities across various intestinal regions in CRC, we sourced several data series from the Gene Expression Omnibus (GEO) database. We isolated 344 tumor and 54 normal cases spanning seven locations: sigmoid, ascending, caecum, rectum, transverse, descending, and recto-sigmoid. Cell biological functions were assessed utilizing CCK8, EdU assays, Transwell assays, and wound healing assays. We identified location-related differentially expressed genes (DEGs) and their functional enrichment. TNFSF4 exhibited variances between the transverse and descending regions, allowing for discrimination of tumor tissues across four locations (sigmoid, ascending, caecum, and descending) via immune cells. Moreover, we identified 16 hub genes that collectively indicate commonality among the seven different tissue origins. Furthermore, 6 location-related genes, including DCBLD2, GTF3A, GSS, PDK1, TEAD3 and EGR2 were identified for targeted interventions. In vitro experiments indicated that increased GZMB and decreased IER3 reduced CRC cell proliferation, migration, and invasion. In summary, 16 hub genes and 6 location-related genes enhance our understanding of the potential molecular mechanisms underlying CRC spatial heterogeneity. Furthermore, GZMB and IER3 might be potential targets for CRC therapy.
Similar content being viewed by others
Introduction
Colorectal carcinoma (CRC) is the second most common cause of cancer-related deaths in the United States (U.S.) and a major contributor to cancer mortality, particularly among males aged 20–49 years1,2. A study conducted in the U.S. has shown a sharp increase in CRC incidence3mirroring similar trends in the prevalence of gastrointestinal cancers in East Asia4. Notably, recent research indicates a global trend toward earlier diagnosis of CRC, termed early-onset CRC (diagnosed in patients under 50 years old). By 2030, it is estimated that this subtype will account for more than 10% of colon cancer cases and over 20% of rectal cancer cases5,6,7. In China, CRC mortality rates have gradually increased from 1973 to 20168. Additionally, genetic susceptibility plays a crucial role in the development of CRC9.
Multiple reports have revealed distinctions between left-sided and right-sided colon cancers regarding clinical mortality rates10pathological characteristics11histology and molecular profiles12gut microbiome composition13proteomic, tumor microenvironment features14genomic expressions15prognosis model development16and single-cell omics characteristics17. Furthermore, an anatomical classification dividing CRC into proximal and distal colon subtypes has been identified, representing unique tumor subtypes that require tailored therapeutic approaches18. However, the implementation of personalized antineoplastic treatment demands their integration into the framework of precision oncology.
The current landscape in CRC diagnosis and treatment highlights the urgent need for more accurate and specific biomarkers. Existing single or multi-gene markers often lack the sensitivity and specificity necessary for reliable diagnosis and prediction of tumor progression. To address this limitation, we investigated the association between tumor location subtypes and their underlying molecular mechanisms in CRC. Our study aims to refine CRC classifications based on tumor anatomical location and to identify more precise biomarkers. To this end, we categorized CRC into seven distinct subgroups and conducted comprehensive bioinformatics analyses, including differential transcriptomic profiling, functional enrichment, transcription factor prediction, and immune landscape characterization. These analyses aim to identify both core and location-specific biomarkers, providing valuable insights into their potential roles in precision oncology.
Building upon prior studies on spatial heterogeneity and biomarker discovery in CRC, our work adopts an integrative approach to better understand tumor biology. By aligning tumor location with molecular features, this study contributes to the development of more individualized therapeutic strategies and advances the application of precision medicine in CRC.
Results
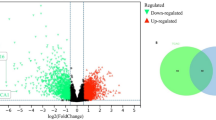
Up-regulated and down-regulated differentially expressed genes (DEGs)
First, overall workflow of this study is depicted in Fig. 1. Next, according to this workflow, an expression matrix consisting of 133 genes was obtained after removing batch effects from the five data series. Among the 344 samples, seven groups were categorized based on their anatomical collection locations. Differential gene expression analysis revealed distinct sets of DEGs for each location. 29 DEGs were identified in sigmoid location (Fig. 2A), comprising 16 upregulated and 13 downregulated genes. From tumors in the ascending region, 27 DEGs were identified (Fig. 2B), with 15 genes upregulated and 12 downregulated. In the caecum area (Fig. 2C), 16 genes exhibited increased expression, while 15 genes displayed decreased expression, resulting in a total of 31 DEGs. In rectum cancer (Fig. 2D), 14 genes were overexpressed and 14 genes were under-expressed. In the transverse region, 32 DEGs were found (Fig. 2E), with 17 genes showing increased expression and 15 showing decreased expression. In the descending region (Fig. 2F), 15 genes had increased expression and 13 genes had decreased expression. The recto-sigmoid neoplasms yielded 30 DEGs (Fig. 2G), including 13 upregulated and 17 downregulated genes. Detailed records of all these DEGs are provided in Table 1.
Gene ontology (GO) enrichment analysis of DEGs
Hierarchical clustering heatmaps of DEGs presented the top 10 genes with the most significant variations in expression, both upregulated and downregulated across rectum, transverse colon, descending colon, and recto-sigmoid, caecum, sigmoid colon, and ascending colon (Fig. 3 A–G). It is noteworthy that CSPG4, DOCK10, GSN, NR3C2, NR5A2, PDK4, SELENBP1, and STAB1 exhibited consistently low expression across different regions. In contrast, CCT5, CENPE, COL2A1, DNMT1, GTF3A, GZMB, IER3, and IFITM1 were overexpressed in all seven colorectal regions. Furthermore, TP53 showed high expression (P < 0.05) in six regions, namely sigmoid, ascending, caecum, transverse, rectum, and recto-sigmoid, but was not identified as differentially expressed in the descending colon (Fig. 3 A–G). Collectively, these findings reveal distinct region-specific expression patterns among the seven tumor subgroups, underscoring the spatial heterogeneity of gene expression in CRC compared to the normal tissue. Based on the GO enrichment analysis in the sigmoid region (Fig. 3H, I), the most enriched functions were related to the extracellular region and extracellular space. Genes enriched in the extracellular region exhibited positive fold changes, while those in the extracellular space showed negative trend. In the ascending intestines (Fig. 3J, K), genes were mainly involved in protein binding and extracellular exosome functions, with differential trends across these categories. The most significant upregulated pathway was rRNA transcription, while the most prominent downregulation was observed in the reactive oxygen species (ROS) metabolic process. In the caecum region, six genes (HADHB, PDK4, GZMB, ABAT, TP53, and IER3) were enriched in mitochondrion function-related pathways (Fig. 4A, B), displaying varying directions of fold change. DEGs such as CCT5, TP53, GNAQ, and GSN in the rectum were associated with the regulation of protein stability (Fig. 4C, Supplementary Figure S1A). The most prominent functions in the transverse region were related to the regulation of protein stability (GNAQ, CCT5, GSN and TP53 enriched) and rhythmic process (GNAQ, TP53, EGR2, ID1 enriched) (Fig. 4D, Supplementary Figure S1B), with four DEGs clustering in each category. Only three DEGs in the descending tumor underwent GO analysis (Fig. 4E, Supplementary Figure S1C), with shared functionalities linked to the regulation of the viral life cycle (NR5A2, GSN and IFITM1 enriched). The most important function in the DEGs of recto-sigmoid was involvement in the glucose metabolic process, and the genes enriched in this function were PDK4, TP53, PDK1, and FABP5 (Fig. 4F, Supplementary Figure S1D).
Gene differential expression and Gene Ontology (GO) analysis of DEGs. (A–G) Heatmaps of the top 10 DEGs for (A) Rectum, (B) Transverse, (C) Descending, (D) Recto-sigmoid, (E) Caecum, (F) Sigmoid, (G) Ascending. (H) Lollipop chart for GO analysis of DEGs in the sigmoid. (I) Heatmap of GO analysis for DEGs in the sigmoid. (J) Lollipop chart for GO analysis of DEGs in the ascending colon. (K) Heatmap of GO analysis for DEGs in the ascending colon.
Constructing regulatory networks of transcription factors (TFs) and DEGs
TFs regulate chromatin structure and transcription by recognizing specific DNA sequences, forming a complex system that guides genomic activity19. The regulatory network in the sigmoid region (Fig. 5A) consisted of 9 DEGs regulated by 11 TFs. The ascending subgroup comprised 10 DEGs regulated by 13 TFs (Fig. 5B). The caecum network consisted of 11 DEGs and 14 TFs (Fig. 5C). In the rectum, 11 TFs regulated 9 DEGs (Fig. 5D). Both transverse and recto-sigmoid subgroups had 12 DEGs regulated by 10 TFs (Fig. 5E, F). In comparison to other subgroups, the descending subgroup had the smallest network, comprising 7 TFs that affected 9 DEGs (Fig. 5G). A notable feature of the regulatory network was the role of TP53, often called the “guardian of the genome,” which plays a pivotal role in key biological processes20. To illustrate the disparities in TP53 expression between controls and treated groups, violin plots (Fig. 5H–N) were employed. Analysis of p-values revealed significant variance across tissues, ranked as follows: Transverse (6.5e-13) < Ascending (2.7e-12) < Sigmoid (4.8e-11) < Recto-sigmoid (7.8e-09) < Rectum (1.8e-06) < Caecum (1.5e-05) < Descending (0.0053).
Transcription factors (TFs) analysis of DEGs in the colon. (A–G) Network formed by DEGs and their corresponding TFs: (A) Sigmoid, (B) Ascending, (C) Caecum, (D) Rectum, (E) Transverse, (F) Recto-sigmoid, and (G) Descending. Triangles represent TFs, and ellipses represent genes. (H–N) Differential expression analysis of the transcription factor TP53 across locations: (H) Sigmoid, (I) Ascending colon, (J) Caecum, (K) Rectum, (L) Transverse, (M) Recto-sigmoid, and (N) Descending colon.
Characteristics of immune cell abundance, heterogeneity, and correlation
The abundance of immune cells across the seven groups was analyzed and is depicted (Fig. 6A-G). A prevailing trend across all groups was the low abundance of B cells and comparatively high levels of mast cells and monocytes. Nevertheless, distinct variations in other immune cell types were evident. Heatmaps of immune cell expression (Fig. 7A-G) demonstrated that the identical immune cell types exhibited distinct expression patterns across different tissues. For instance, monocytes exhibited high expression in caecum cancer but low expression in other tumors. Correlation analysis (Supplementary Figure S2A-D) highlighted interactions and regulatory relationships among immune cells.
Immune cell expression and principal components analysis (PCA) in the colon. (A–E) Immune cell expression analysis in cancer samples for (A) Caecum, (B) Rectum, (C) Transverse, (D) Sigmoid, (E) Descending, (F) Recto-sigmoid, and (G) Ascending colon. (H) Expression of immune checkpoint TNFSF4 across various locations. (I–L) PCA of immune cells in tumors for (I) Sigmoid, (J) Ascending, (K) Caecum, (L) Descending colon.
Expression levels of the immune checkpoint inhibitor target TNFSF4 (Fig. 7H) revealed a remarkable disparity between the transverse and descending groups. To assess whether immune cells can distinguish various locations, principal component analysis (PCA) was conducted (Fig. 7I-L). The results demonstrated that four positions can be distinguished based on immune cell composition, while the remaining three locations did not exhibit clear separation (Supplementary Figure S3A-B).
Characteristics of immune cell types
Among the immune cell types, sigmoid tumors (Fig. 8A) could be segmented by the presence of ten cell types, whereas ascending tumors displayed six immune cell types (Fig. 8B). Caecum cancer was associated with ten immune cell types (Fig. 8C), and descending tumors exhibited the highest diversity, with thirteen immune cell type (Fig. 8D). The other three groups did not show any remarkable disparity in immune cell composition (Supplementary Figure S3C-E).
Characteristics of 16 hub DEGs
By identifying the intersection of DEGs across seven cancer tissue locations, a set of 16 hub DEGs was obtained (Fig. 9A). Chromosomal mapping (Fig. 9B) revealed these genes were primarily resided on chromosomes 1–7, 9, 11–15, and 19, with chromosomes 1 and 4 each harboring two DEGs. GO enrichment analysis (Fig. 9C, Supplementary Figure S4B-C) indicated that these genes were mainly associated with several functions such as regulation of viral life cycle (GO: 1903900), regulation of viral process (GO: 0050792), and regulation of chromosome organization (GO: 0033044). A protein-protein interaction (PPI) network, constructed with a low confidence threshold (0.15), identified DNMT1 as the central hub gene (Fig. 9D). In contrast, within the PPI network for all DEGs, TP53 occupied a prominent position (Supplementary Figure S4A). The regulatory network diagram (Fig. 9E) highlighted four DEGs (IER3, DNMT1, COL2A1, and PDK4) regulated by five transcription factors (RELA, TP53, SP1, E2F1, and NFKB1).
Characteristics of hub DEGs. (A) Venn diagram of intersecting hub DEGs from seven colon locations. (B) Chromosomal positions of the 16 hub DEGs. (C) GO enrichment analysis of the 16 hub DEGs. (D) Protein-protein interaction (PPI) network for the 16 hub DEGs. (E) Network of the 16 hub DEGs and their TFs.
Differential expression and survival analysis of ten hub genes
Expression analysis of the 16 hub genes across different tissues (Fig. 10A–I) revealed obvious variation in nine genes (COL2A1, GTF3A, GZMB, IFITM1, NR3C2, NR5A2, SELENBP1, DOCK10, and STAB1), with IFITM1 showing the greatest inter-group variability. However, seven genes (GSN, IER3, PDK4, DNMT1, CSPG4, CENPE, and CCT5) showed no discernible diversity in expression (Supplementary Figure S5 A-G). Further validation in the The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression Project (GTEx) databases revealed that 10 genes (GSN, GTF3A, GZMB, IER3, IFITM1, PDK4, CSPG4, STAB1, CENPE, and CCT5) exhibited differential expression between normal and tumor groups (Fig. 10J–S), with six genes did not display statistical significance (Supplementary Figure S5 H-M). Survival analysis (Fig. 10T, U) indicated two genes, NR3C2 and SELENBP1, as being significantly associated with patient prognosis, while the remaining genes did not exhibit survival correlations (Supplementary Figure S5N-W, S6A-C).
Differential expression and survival analysis of ten hub genes. (A-I) Expression of individual genes at different sites: (A) COL2A1, (B) GTF3A, (C) GZMB, (D) IFITM1, (E) NR3C2, (F) NR5A2, (G) SELENBP1, (F) DOCK10, (I) STAB1. *p < 0.05, **p < 0.01, ***p < 0.001. (J–S) Genes expression in normal versus tumor groups: (J) GSN, (K) GTF3A, (L) GZMB, (M) IER3, (N) IFITM1, (O) PDK4, (P) CSPG4, (Q) STAB1, (R) CENPE, (S) CCT5. *p < 0.05. (T, U) Survival analysis of (T) NR3C2 and (U) SELENBP1.
Six exclusive genetic characteristics
Six genes emerged as location-related genes from the Venn diagram analysis of intersected DEGs. These were GSS in the sigmoid position, DCBLD2, FABP5 and PDK1 in recto-sigmoid tissue, and EGR2 and TEAD3 in the transverse region. Expression analysis across the seven tumor sampling sites (Fig. 11A-D) revealed significant inter-group differences for four genes (GSS, PDK1, TEAD3 and EGR2), while DCBLD2 and FABP5 showed no noticeable differences (Supplementary Figure S6D-E). Both GSS and TEAD3 exhibited substantial discrimination between 275 COAD tumor samples and 349 normal samples (Fig. 11E-F), whereas the remaining genes failed to differentiate tumor tissues (Supplementary Figure S6F-I). None of these genes revealed a marked association with overall patient survival (Supplementary Figure S6J-O). In drug sensitivity analysis, the PDK1 gene demonstrated considerable sensitivity to KIN001-244 and BX-912 in MLL2_mut cell lines of COAD (Fig. 11G-H). Drugs BX795 and OSU-03012 did not show sensitivity (Supplementary Figure S6P-Q).
Differential expression and drug sensitivity analysis of six location-related genes. (A-D) Expression of location-related genes: (A) GSS, (B) PDK1, (C) TEAD3, (D) EGR2. *p < 0.05, **p < 0.01, ***p < 0.001. (E, F) Genes expression in normal and tumor groups: (E) GSS and (F) TEAD3. (G, H) Drug sensitivity analysis of PDK1 for (G) KIN001-244 and (H) BX-912.
GZMB and IER3 regulate cell proliferation
To ascertain the expression levels of target genes in CRC cells, CCD-841coN cells were use the control group. Relative expression levels of 16 hub genes, including GZMB, IER3, and COL2A1, were assessed in HCT116 and SW480 cells. RT-qPCR results revealed significant upregulation of GZMB and IER3 in CRC cells (Supplementary Figure S7). The elevated expression is closely associated with functional abnormalities observed in these cancer cells. To explore the function role of GZMB and IER3, siRNAs targeting these genes (si-GZMB and si-IER3) were transfected into SW480 cells, and interference efficiency was assessed via Western blotting analysis (Fig. 12A, B). CCK8 and EdU assays demonstrated that GZMB interference led to a significantly lower cell proliferation, suggesting its role in promoting proliferation in CRC (Fig. 12C–E). Conversely, interference with IER3 had the opposite effect, enhancing proliferation (Fig. 12F–H).
Western blot (WB), Cell Counting Kit-8 (CCK8) and 5-ethynyl-2’-deoxyuridine (EdU) detection assays. (A, B) Western blot analysis of gene interference efficiency in SW480 cells for (A) GZMB and (B) IER3. (C) Effects of GZMB knockdown on the proliferation of SW480 cells by CCK-8 assay. (D) Proliferation of GZMB siRNA-transfected cells were detected by EdU. (E) EdU-positive cells component variance analysis. (F) Effects of IER3 knockdown on the proliferation of SW480 cells by CCK-8 assay. (G) Proliferation of IER3 siRNA-transfected cells detected by EdU. (H) EdU-positive cells component variance analysis. Results are presented as means ± SEM.s, *p < 0.05, **p < 0.01.
GZMB and IER3 regulate cell migration and invasion
To explore whether GZMB and IER3 could affect cell migration and invasion, we performed scratch and Transwell invasion assays in SW480 cells following knockdown of GZMB and IER3 genes. The scratch assay demonstrated that GZMB silencing inhibited cell migration (Fig. 13A, B), whereas IER3 knockdown promoted migratory capacity of SW480 cells (Fig. 13C, D). Consistent with these findings, Transwell assays showed that GZMB knockdown suppressed cell invasion (Fig. 13G, H), while IER3 knockdown enhanced invasive behavior in SW480 cells (Fig. 13E, F). Collectively, these results underscore the distinct roles of GZMB and IER2 in regulation of cell migration and invasion in CRC.
Discussion
CRC is characterized by spatial tumor heterogeneity, influencing tumor growth, progression, and treatment strategies21,22. Our research revealed variations in the number of DEGs among tumor sampling locations. Through transcription factor regulatory network analysis, TP53 emerged as a pivotal hub gene, regulated by other TFs and encoding the TP53 protein that, in turn, regulated other genes. Immune cells at four locations (sigmoid, ascending, caecum, and descending) displayed distinguishable features, offering potential insights for developing novel immune-based cancer therapies23.
The six exclusive genes identified (DCBLD2, GTF3A, GSS, PDK1, TEAD3, and EGR2) hold promise as precise molecular markers for targeted interventions. Studies indicates that high expression levels of GZMB and CD68 are independent prognostic factors for CRC24. GZMB has been proposed as a potential therapeutic target for African American CRC patients25Co-expression analysis supports its association with CRC progression26though cellular experiments validating these findings for GZMB has been lacking until now. Our study addresses this gap by elucidating the functional roles of GZMB in CRC progression through cellular experiments. In contrast, research on IER3 in CRC remains limited. Existing studies highlight its role in stress response modulation in colonic epithelial cells and its regulation of NF-κB and Nrf2 pathways27. In tongue cancer, downregulation of IER3 inhibited proliferation, colony formation, migration, and invasion of Tca-8113 cells28. In lung carcinoma, IER3 is linked to T-cell exhaustion and immune regulation29. Similarly, in cervical cancer, IER3 acts as a novel target gene for Tap73β30. In gastric cancer, IER3 + myeloid-derived suppressor cells are implicated in immune suppression and resistance to immune checkpoint inhibitors within the tumor microenvironment31. These findings provide valuable directions for future in-depth exploration, particularly regarding the roles of GZMB and IER3 in CRC progression, treatment resistance, and immune regulation.
In summary, this study represents the first attempt to refine CRC analysis based on sampling locations. We identified 16 hub genes and 6 location-related genes and analyzed their characteristics. Our findings indicate that GZMB exhibits inhibitory effects, while IER3 appears to promote CRC progression. Of course, more experimental evidence is needed to prove their mechanisms of action and therapeutic effects. However, this study has certain limitations that must be acknowledged. First, publicly available databases lack sufficient clinical information regarding sampling locations, resulting in a limited sample size for analysis. Second, the roles of both common and location-related DEGs in tumorigenesis and progression requires further validation through cellular experiments, animal studies, and clinical tissue specimens. The identification of precise biomarkers for CRC and the specific manifestations of PDK1 in drug resistance remain areas for deeper investigation. Future research should incorporate comprehensive clinical data to assess the practical unitality of these genes as biomarkers and therapeutic targets, facilitating the development of more effective, location-based strategies for CRC treatment.
Although our study provides valuable insights into molecular variations across CRC subtypes, several key limitations should be acknowledged. A major constraint is the inherent heterogeneity of tumor samples, which may limit the generalizability of our findings and fail to fully capture the complexity of CRC biology. To overcome this, advanced techniques such as single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics could prove invaluable. These approaches offer perspectives on tumor architecture and gene expression dynamics. In addition, scRNA-seq enables the dissection of intratumoral heterogeneity at the single-cell level, allowing for the identification of rare cell populations and key regulatory genes that may be obscured in bulk analyses. In parallel, spatial transcriptomics can elucidate region-specific gene expression patterns within the tumor microenvironment, thereby offering critical insights into the spatial organization of tumor progression and cellular interactions. Future investigations should leverage these advanced technologies to achieve a more granular understanding of CRC pathogenesis. Moreover, further validation of key candidate genes, such as GZMB and IER3, is essential to elucidate their roles in tumor progression, immune modulation, and therapeutic resistance. Integrative, multi-modal analyses that combine scRNA-seq with spatial transcriptomics hold great potential to unravel the complex molecular landscape of CRC and advance the development of personalized therapeutic strategies.
Materials and methods
Data collection and preprocessing
All transcriptomic and clinical data for colorectal cancer were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/)32. Five data series (GSE25071, GSE28814, GSE41258, GSE110223, and GSE110224) were included in the analysis. The focus of our study was to investigate the homogeneity and heterogeneity of tumors across different anatomical locations in CRC. Therefore, we selected datasets that included comprehensive clinical information, particularly with clearly annotated tumor sampling sites. As a result, seven distinct regions within the colon were identified: sigmoid (n = 89), ascending (n = 65), caecum (n = 58), rectum (n = 48), transverse (n = 31), descending (n = 28), and recto-sigmoid (n = 25). The control group consisted of 54 cases. Detailed distributions and source platforms are provided in Table 2. Batch correction of the five data series from four different platforms was performed33 using the “combat” function from the R package “sva”34 and the “limma” package35. Data normalization was performed after batch correction to ensure comparability across datasets.
Identification of DEGs
Differential expression analysis was carried out using the “limma” package, with filtering criteria aet as follows: absolute log2 fold change (logFC) > 1 and adjusted p-value < 0.0536. DEGs were visualized through a volcano plot generated using the “ggplot2” package37clearly distinguishing positive and negative regulator genes with appropriate annotations. A heatmap of the top 10 DEGs was constructed using the “pheatmap” package, enabling clear visualization of gene expression differences.
GO enrichment analyses
GO enrichment analysis classified genes into three main functional categories: Cellular Component (CC), Molecular Function (MF), and Biological Process (BP)38. The “org.Hs.eg.db” package was used to annotate gene IDs39. The “enrichplot” package was utilized to exclude genes with empty IDs39. Enrichment analysis was performed using a significance threshold of p < 0.05 and adjusted p-value < 1. To visually represent the enriched gene functions, a lollipop chart was generated with the “ggpubr” package40highlighting the number of enriched genes in each category. Additionally, a heatmap was generated using the “GOplot” package to display the significance of enriched functional categories41.
Construction of regulatory networks for TFs and DEGs
To construct regulatory networks of TFs and target genes, the Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST, version2, https://www.grnpedia.org/trrust/) database42 was utilized. DEGs from each location were input into TRRUST to retrieve associated transcription factor files. A Perl script was used to process the files into required formats, including node, network, gene list, and TFs list files, for network construction. The processed files were imported into Cytoscape (version 3.9.1)43where the network graph’s layout, color scheme, and node shapes were refined for better visualization and interpretability.
Immune-related analysis of DEGs
The CIBERSORT algorithm44 was employed to analyze immune-related characteristics and immune cell infiltration across all examples, with a threshold of P < 0.05 for sample inclusion. A gene list of immune checkpoint markers was used to predict the responsiveness of various CRC tumor tissues to immune therapy. PCA45 was conducted to determine whether immune cell content could distinguish between tumor and normal tissues. A clear separation in the PCA plot indicates systematic differences in the immune microenvironment between the two groups. Differential expression of immune cells was assessed using the “Wilcox. Test”46with P < 0.05 indicating statistically significant distinctions between groups.
Analysis of shared DEGs features
The intersection of DEGs from the seven sampled locations was identified and visualized using a Venn diagram. Hub genes associated with CRC were determined, and their specific chromosomal locations were mapped using a chromosome-gene auxiliary file. The “RCircos” package47 was used to visually depict the chromosomal distribution of these genes. Functional enrichment analysis was performed using the STRING online tool (version 12, https://cn.string-db.org/)48 to establish protein-protein interaction (PPI) networks.
Profiling the specific characteristics of DEGs in each region
To identify unique genes from the Venn diagram of DEGs from different locations, we performed the Wilcoxon rank-sum test49. The GEPIA database was used to test whether gene expression differed statistically between normal and tumor groups. Using TCGA normal and GTEx data from COAD patients, significant differences were determined with a cutoff of |log2FC| > 1 and P < 0.05. To analyze the drug sensitivity and response of different tumor cells, we utilized the Genomics of Drug Sensitivity in Cancer (GDSC) database50 (https://www.cancerrxgene.org/), which integrates and reanalyzes data from multiple studies and resources.
Cell lines source and culture
Human colon and rectal tissue CCD-841CoN cells were purchased from Guangzhou Jennio Biotech Co., Ltd (Guangzhou, China), while human colorectal cancer cell lines SW480 and HCT116 were acquired from Starfish Biotechnology Co., Ltd (Suzhou, China). Cells were cultured in DMEM medium supplemented with 10% fetal bovine serum and 1% streptomycin/penicillin.
SiRNA transfection
siRNA transfection was conducted following the Lipofectamine 2000 protocol, as per the manufacturer’s instructions (Thermo, United States). Cells were harvested 48 h post-transfection for subsequent functional assays. The sequences of the siRNAs used are listed in Table 3.
RT-qPCR and Western blot assay
Total RNA was extracted from cells using the RNA pure Tissue & Cell Kit (CWBIO, China), followed by reverse transcription utilizing the HiFi Script cDNA Synthesis Kit (CWBIO, China). Quantitative real-time PCR (qRT-PCR) was conducted using SYBR Green Mix (CW3008H, CWBIO). Relative mRNA expression levels were calculated using the 2−ΔΔCt method51. Primer sequences are detailed in Table 4. Western blotting analysis was performed as described previously52.
Cell proliferation assay
SW480 cells were pre-cultured at a density of 8 × 103 cells per well in a 96-well plate for 24 h. Cells were then treated with either negative control (NC) or siRNA for 24, 48, 72, and 96 h. Subsequently, 10 µL of CCK-8 solution was added to each well, and absorbance at 450 nm was measured using a microplate reader after a 2-hour incubation53,. EdU staining was conducted on transfected cells according to the manufacturer’s instructions.
Transwell assays
For Transwell assays, 8 × 104 cells/well were seeded in the upper chamber with 100 µL of serum-free medium, while the lower chamber contained 500 µL of medium supplemented with 20% fetal bovine serum. After a 24-hour incubation, crystal violet staining was performed to visualize the cells that migrated. Images were captured using a 100x optical microscope. Additionally, SW480 cells (1.4 × 106 cells/well) were seeded in a 6-well plate, transfected with NC or siRNA for 6 hours, and three scratches were made per well using a 200 µL pipette tip. Scratch widths were imaged at 0 and 24 h post-scratching for analysis.
Statistical analysis
Statistical analysis and data visualization were conducted using Perl language (Strawberry Perl 64-bit) and R language (Version 4.1.3). The Wilcoxon test was employed for inter-group differential analysis, while the Kruskal-Wallis test54 was employed for comparisons involving more than two groups. For cellular experimental data, statistical analysis was conducted utilizing GraphPad Prism 7 (IBM, USA). Multiple group comparisons were assessed through one-way analysis of variance (ANOVA). Results were considered statistically significant at a p-value < 0.0555.
Data availability
“The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Siegel, R. L., Wagle, N. S., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 73, 233–254. https://doi.org/10.3322/caac.21772 (2023).
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48. https://doi.org/10.3322/caac.21763 (2023).
Kratzer, T. B. et al. Cancer statistics for American Indian and Alaska native individuals, 2022: including increasing disparities in early onset colorectal cancer. CA Cancer J. Clin. 73, 120–146. https://doi.org/10.3322/caac.21757 (2023).
Huang, J. et al. Updated epidemiology of Gastrointestinal cancers in East Asia. Nat. Rev. Gastroenterol. Hepatol. 20, 271–287. https://doi.org/10.1038/s41575-022-00726-3 (2023).
Spaander, M. C. W. et al. Young-onset colorectal cancer. Nat. Rev. Dis. Primers. 9 https://doi.org/10.1038/s41572-023-00432-7 (2023).
Sinicrope, F. A. Increasing incidence of Early-Onset colorectal Cancer. N Engl. J. Med. 386, 1547–1558. https://doi.org/10.1056/NEJMra2200869 (2022).
Patel, S. G., Karlitz, J. J., Yen, T., Lieu, C. H. & Boland, C. R. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol. Hepatol. 7, 262–274. https://doi.org/10.1016/S2468-1253(21)00426-X (2022).
Feng, R. et al. Cancer situation in china: what does the China cancer map indicate from the first National death survey to the latest cancer registration? Cancer Commun. (Lond). 43, 75–86. https://doi.org/10.1002/cac2.12393 (2023).
Rebuzzi, F., Ulivi, P. & Tedaldi, G. Genetic predisposition to colorectal cancer: how many and which genes to test?? Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms24032137 (2023).
Qiu, M. Z. et al. Comparison of survival between right-sided and left-sided colon cancer in different situations. Cancer Med. 7, 1141–1150. https://doi.org/10.1002/cam4.1401 (2018).
Wismayer, R., Kiwanuka, J., Wabinga, H. & Odida, M. Colorectal adenocarcinoma in uganda: are right-sided and left-sided colon cancers two distinct disease entities? World J. Surg. Oncol. 21, 215. https://doi.org/10.1186/s12957-023-03094-7 (2023).
Gholamalizadeh, H. et al. Prognostic value of primary tumor location in colorectal cancer: an updated meta-analysis. Clin. Exp. Med. https://doi.org/10.1007/s10238-023-01120-2 (2023).
Kneis, B. et al. Colon cancer Microbiome landscaping: differences in Right- and Left-Sided Colon cancer and a tumor Microbiome-Ileal Microbiome association. Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms24043265 (2023).
Hong, Z. et al. Proteomic profiling and tumor microenvironment characterization reveal molecular and immunological hallmarks of Left-Sided and Right-Sided Colon cancer tumorigenesis. J. Proteome Res. https://doi.org/10.1021/acs.jproteome.3c00302 (2023).
Peng, Q. et al. Identification of genomic expression differences between right-sided and left-sided colon cancer based on bioinformatics analysis. Onco Targets Ther. 11, 609–618. https://doi.org/10.2147/OTT.S154207 (2018).
Mao, Y. et al. Prediction values of tertiary lymphoid structures in the prognosis of patients with left- and right-sided colon cancer: a multicenter propensity score-matched study. Int. J. Surg. 109, 2344–2358. https://doi.org/10.1097/JS9.0000000000000483 (2023).
Cao, L. et al. Comparative analyses of the prognosis, tumor immune microenvironment, and drug treatment response between left-sided and right-sided colon cancer by integrating scRNA-seq and bulk RNA-seq data. Aging (Albany NY). 15, 7098–7123. https://doi.org/10.18632/aging.204894 (2023).
Yang, G., Yu, X. R., Weisenberger, D. J., Lu, T. & Liang, G. A. Multi-Omics overview of colorectal Cancer to address mechanisms of disease, metastasis, patient disparities and outcomes. Cancers (Basel). 15. https://doi.org/10.3390/cancers15112934 (2023).
Francois, M., Donovan, P. & Fontaine, F. Modulating transcription factor activity: interfering with protein-protein interaction networks. Semin Cell. Dev. Biol. 99, 12–19. https://doi.org/10.1016/j.semcdb.2018.07.019 (2020).
Huang, Y., Che, X., Wang, P. W. & Qu, X. p53/MDM2 signaling pathway in aging, senescence and tumorigenesis. Semin Cancer Biol. 101, 44–57. https://doi.org/10.1016/j.semcancer.2024.05.001 (2024).
Aljama, S. et al. Dichotomous colorectal cancer behaviour. Crit. Rev. Oncol. Hematol. 189, 104067. https://doi.org/10.1016/j.critrevonc.2023.104067 (2023).
Cai, Y. M. et al. Genome-wide enhancer RNA profiling adds molecular links between genetic variation and human cancers. Mil Med. Res. 11, 36. https://doi.org/10.1186/s40779-024-00539-2 (2024).
Zhang, Y. & Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 17, 807–821. https://doi.org/10.1038/s41423-020-0488-6 (2020).
Noti, L. et al. A combined Spatial score of granzyme B and CD68 surpasses CD8 as an independent prognostic factor in TNM stage II colorectal cancer. BMC Cancer. 22, 987. https://doi.org/10.1186/s12885-022-10048-x (2022).
Paredes, J. et al. Immune-Related gene expression and cytokine secretion is reduced among African American Colon cancer patients. Front. Oncol. 10, 1498. https://doi.org/10.3389/fonc.2020.01498 (2020).
Meng, J., Su, R., Liao, Y., Li, Y. & Li, L. Identification of 10 hub genes related to the progression of colorectal cancer by co-expression analysis. PeerJ 8, e9633. https://doi.org/10.7717/peerj.9633 (2020).
Stachel, I. et al. Modulation of nuclear factor E2-related factor-2 (Nrf2) activation by the stress response gene immediate early response-3 (IER3) in colonic epithelial cells: a novel mechanism of cellular adaption to inflammatory stress. J. Biol. Chem. 289, 1917–1929. https://doi.org/10.1074/jbc.M113.490920 (2014).
Xiao, F., Dai, Y., Hu, Y., Lu, M. & Dai, Q. Expression profile analysis identifies IER3 to predict overall survival and promote lymph node metastasis in tongue cancer. Cancer Cell. Int. 19, 307. https://doi.org/10.1186/s12935-019-1028-2 (2019).
Kuang, T., Zhang, L., Chai, D., Chen, C. & Wang, W. Construction of a T-cell exhaustion-related gene signature for predicting prognosis and immune response in hepatocellular carcinoma. Aging (Albany NY). 15, 5751–5774. https://doi.org/10.18632/aging.204830 (2023).
Jin, H. et al. IER3 is a crucial mediator of TAp73β-induced apoptosis in cervical cancer and confers Etoposide sensitivity. Sci. Rep. 5, 8367. https://doi.org/10.1038/srep08367 (2015).
Tsutsumi, C. et al. Tumor-infiltrating monocytic myeloid-derived suppressor cells contribute to the development of an immunosuppressive tumor microenvironment in gastric cancer. Gastric Cancer. 27, 248–262. https://doi.org/10.1007/s10120-023-01456-4 (2024).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Zhang, Y., Parmigiani, G. & Johnson, W. E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform. 2, lqaa078. https://doi.org/10.1093/nargab/lqaa078 (2020).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The Sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. https://doi.org/10.1093/bioinformatics/bts034 (2012).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. https://doi.org/10.1093/nar/gkv007 (2015).
Ambroise, J. et al. Impact of the spotted microarray preprocessing method on fold-change compression and variance stability. BMC Bioinform. 12, 413. https://doi.org/10.1186/1471-2105-12-413 (2011).
Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. Journal of the Royal Statistical Society Series A: Statistics in Society 174, 245–246, (2011). https://doi.org/10.1111/j.1467-985X.2010.00676_9.x %J Journal of the Royal Statistical Society Series A: Statistics in Society.
Consortium, G. O. Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056. https://doi.org/10.1093/nar/gku1179 (2015).
M., C. org.Hs.eg.db: Genome wide annotation for Human. (2015).
Aut, A. K. ggpubr: ‘ggplot2’ Based Publication Ready Plots. (2017).
Walter, W., Sánchez-Cabo, F. & Ricote, M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914. https://doi.org/10.1093/bioinformatics/btv300 (2015).
Han, H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386. https://doi.org/10.1093/nar/gkx1013 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M. & Alizadeh, A. A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 1711, 243–259. https://doi.org/10.1007/978-1-4939-7493-1_12 (2018).
Ringnér, M. What is principal component analysis? Nat. Biotechnol. 26, 303–304. https://doi.org/10.1038/nbt0308-303 (2008).
Lachin, J. M., Nonparametric Statistical & Analysis JAMA 323, 2080–2081, doi:https://doi.org/10.1001/jama.2020.5874 (2020).
Zhang, H., Meltzer, P. & Davis, S. RCircos: an R package for circos 2D track plots. BMC Bioinform. 14, 244. https://doi.org/10.1186/1471-2105-14-244 (2013).
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. https://doi.org/10.1093/nar/gkac1000 (2023).
Fang, Z., Du, R. & Cui, X. Uniform approximation is more appropriate for Wilcoxon Rank-Sum test in gene set analysis. PLoS One. 7, e31505. https://doi.org/10.1371/journal.pone.0031505 (2012).
Yang, W. et al. Genomics of drug sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961. https://doi.org/10.1093/nar/gks1111 (2013).
Weng, H. et al. Aspartoacylase suppresses prostate cancer progression by blocking LYN activation. Mil Med. Res. 10 https://doi.org/10.1186/s40779-023-00460-0 (2023).
Xiong, H. J. et al. Elevated FBXL6 activates both wild-type KRAS and mutant KRAS(G12D) and drives HCC tumorigenesis via the ERK/mTOR/PRELID2/ROS axis in mice. Mil Med. Res. 10, 68. https://doi.org/10.1186/s40779-023-00501-8 (2023).
Xu, H. et al. ADP-dependent glucokinase controls metabolic fitness in prostate cancer progression. Mil Med. Res. 10, 64. https://doi.org/10.1186/s40779-023-00500-9 (2023).
Clark, J. S. C. et al. Empirical investigations into Kruskal-Wallis power studies utilizing bernstein fits, simulations and medical study datasets. Sci. Rep. 13, 2352. https://doi.org/10.1038/s41598-023-29308-2 (2023).
Kwak, S. Are only p-Values less than 0.05 significant?? A p-Value greater than 0.05 is also significant?! J. Lipid Atheroscler. 12, 89–95. https://doi.org/10.12997/jla.2023.12.2.89 (2023).
Funding
This work was supported by Guangdong Basic and Applied Basic Research Foundation (2022A1515220075), Shenzhen Science and Technology Program (JCYJ20220531092607017), and Guangzhou Basic and Applied Basic Research Foundation (202201020509).
Author information
Authors and Affiliations
Contributions
YQ.L. and YT. Y. conceived the work, performed the experiments, analyzed the data, and wrote the manuscript. YW.H, and YB.L. analyzed the data. P.W. edited the manuscript, and supervised the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, Y., Huang, Y., Luo, Y. et al. Identification of heterogeneity and common characteristics in colorectal carcinoma located in distinct sites. Sci Rep 15, 25496 (2025). https://doi.org/10.1038/s41598-025-10528-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10528-7