Abstract
Circular RNAs (circRNAs) play a critical role in hepatocellular carcinoma (HCC) progression. Although circ_0008043 is predicted to be highly expressed in HCC tissues, its functional role and molecular mechanisms remain poorly understood. In this study, we investigated the effects of circ_0008043 on HCC metastasis and its regulation of the miR-661/PLEKHG4B axis. Functional assays revealed that circ_0008043 knockdown suppressed HCC cell viability, migration, and epithelial-mesenchymal transition (EMT). Mechanistically, circ_0008043 acted as a sponge for miR-661, which directly targeted PLEKHG4B. Rescue experiments demonstrated that miR-661 inhibition or PLEKHG4B overexpression counteracted the effects of circ_0008043 silencing or miR-661 overexpression. Furthermore, we identified ESRP1 as a key regulator promoting circ_0008043 biogenesis. In vivo experiments confirmed that circ_0008043 knockdown significantly inhibited lung metastasis. Collectively, our findings reveal that ESRP1-derived circ_0008043 facilitates HCC cell migration and EMT by modulating the miR-661/PLEKHG4B axis, thereby promoting tumor metastasis. This study provides novel insights into the molecular mechanisms of HCC progression and suggests a potential therapeutic target for HCC treatment.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC), the predominant histological subtype of liver cancer, accounts for over 50% of cases globally, with China bearing the highest disease burden. Current epidemiological projections indicate that liver cancer incidence will surpass 1 million cases annually by 20251. While viral hepatitis remains the primary etiological factor, the proportion of HCC cases attributable to obesity and non-alcoholic fatty liver disease (NAFLD) has risen dramatically in recent years2. The insidious nature of early-stage HCC often leads to delayed diagnosis, with most patients presenting at advanced stages where median survival is limited to 1–2 months3. Although hepatic resection remains the first-line therapeutic intervention, postoperative recurrence rates exceed 50% within five years4. Recent advances in targeted therapies and immune checkpoint inhibitors have expanded treatment options for advanced HCC and improved patient outcomes5,6; however, their efficacy in preventing metastasis and recurrence remains uncertain. These clinical challenges underscore the urgent need to elucidate the molecular mechanisms underlying HCC progression to identify novel therapeutic targets.
Emerging evidence highlights the crucial role of epigenetic regulation, particularly non-coding RNAs, in HCC pathogenesis, with many serving as promising diagnostic biomarkers7. Among these, circular RNAs (circRNAs) are single-stranded, covalently closed RNA molecules generated through back-splicing that have attracted increasing research attention8. The biogenesis of most circRNAs involves back-splicing of pre-mRNA, a process tightly regulated by splicing factors that determine the equilibrium between circRNA formation and canonical splicing9. Functionally, circRNAs predominantly exert their biological effects through the competitive endogenous RNA (ceRNA) mechanism, where they sequester microRNAs (miRNAs) to modulate expression of downstream target genes10,11. Growing evidence implicates specific circRNA-miRNA-mRNA regulatory axes in oncogenesis. For instance, circ_100367 promotes esophageal cancer progression by enhancing cell migration and radioresistance via the miR-217/Wnt3 pathway12. In HCC, hsa_circ_104348 has been shown to accelerate tumor proliferation and suppress apoptosis through the miR-187-3p/RTKN2 axis13. Similarly, circ_102002 facilitates papillary thyroid cancer metastasis by acting as a miR-488-3p sponge to upregulate HAS2 expression14. Despite these advances, the functional characterization of circRNAs remains largely incomplete, with the majority awaiting comprehensive annotation and mechanistic investigation in HCC.
Recent profiling studies have identified circ_0008043 as significantly upregulated in HCC tissues15,16. Moreover, our research group previously found that circ_0008043 promotes cellular proliferation, migration, invasion, and tumor growth by regulating the miR-326/RAB21 axis16. However, the underlying mechanism of circ_0008043 remains largely unknown. In this study, we investigated the role of circ_0008043 in HCC malignant progression, with particular focus on its regulation of the miR-661/PLEKHG4B axis. Our findings not only elucidate a novel molecular pathway in HCC metastasis but also provide potential therapeutic targets for clinical intervention.
Materials and methods
Microarray analysis
The GSE155949 dataset was retrieved from the Gene Expression Omnibus (GEO) database. All analyses were performed using the R programming language. To identify and visualize differentially expressed circRNAs between tumor and adjacent normal tissues, a volcano plot was generated. This graphical representation facilitates the identification of circRNAs that are significantly upregulated or downregulated in tumor samples compared to normal controls.
Specimen collection and ethical approval
Sixty patients diagnosed with hepatocellular carcinoma (HCC) at The Third People’s Hospital of Shenzhen were enrolled in this study. During surgical procedures, paired samples of tumor tissues and adjacent non-tumor tissues were collected from each patient. All collected tissue samples were immediately frozen in liquid nitrogen and stored at -80 °C until further processing. Written informed consent was obtained from every participant prior to their inclusion in the study. The study protocol was reviewed and approved by the Ethics Committee of The Third People’s Hospital of Shenzhen (Approval Number: 2023-108-02), ensuring compliance with the Declaration of Helsinki guidelines. The clinicopathological characteristics of the patients, categorized based on high or low expression levels of circ_0008043, are summarized in Table 1.
Cell culture
Human HCC cell lines (Focus and HA22T/VGH) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA), supplemented with 10% fetal bovine serum (FBS; Gibco). Human liver epithelial cells (THLE3), also sourced from the Chinese Academy of Sciences Cell Bank, were maintained in complete Bronchial Epithelial Cell Growth Medium (BEGM; Lonza, Basel, Switzerland). HEK293T cells (ATCC, Manassas, VA, USA) were grown in high-glucose Dulbecco’s Modified Eagle Medium (DMEM; Gibco), containing 10% FBS. All cell cultures were incubated at 37 °C in a humidified atmosphere containing 95% air and 5% CO2.
Cell transfection
Focus and HA22T cells were seeded in six-well plates at a density of 2 × 10^5 cells per well and cultured until they reached approximately 70% confluence. Cells were then transfected with various constructs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The constructs included shRNA targeting circ_0008043 (sh-circ_0008043), shRNA negative control (sh-nc), miR-661 mimic, negative control mimic (nc mimic), miR-661 inhibitor, negative control inhibitor (nc inhibitor), PLEKHG4B overexpression plasmid, and empty vector (all from GenePharma, Shanghai, China). Additionally, HA22T cells were transfected with shRNAs targeting splicing factors (ESRP1, ESRP2, NOVA1, NOVA2, MEX3A, MEX3B, QKI, and SRSF1; GenePharma) using Lipofectamine 2000. After an initial incubation in serum-free medium for 6 h, the medium was replaced with complete growth medium and cells were further cultured for 48 h. The sequences of shRNAs are shown in Table 2.
Fluorescence in situ hybridization (FISH)
The localization of circ_0008043 was assessed using a FISH kit (Ribobio, Guangzhou, China). Focus and HA22T cells were first fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 for 15 min. The fluorescence-labeled circ_0008043 probe was denatured at 37 °C for 5 min before being incubated with the cells overnight at 42 °C. After incubation, cells were washed with 2×SSC solution to remove unbound probe. Subsequently, cells were stained with 10 µL of DAPI for 5 min in the dark to visualize nuclei. Fluorescence signals were observed and captured using a laser scanning confocal microscope (model FV1000; Olympus, Tokyo, Japan).
Quantitative real-time PCR (qPCR)
Total RNA was extracted from HCC cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Additionally, cytoplasmic and nuclear RNA fractions were isolated using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, Canada). RNA concentration was quantified using a spectrophotometer, and 1 µg of total RNA was reverse transcribed into cDNA. For miRNA analysis, cDNA synthesis was performed using the miRcute miRNA First-Strand cDNA Synthesis Kit (TIANGEN, Beijing, China), followed by quantification using the miRcute miRNA qPCR Detection Kit (TIANGEN). For circRNA and mRNA analysis, reverse transcription and quantitative PCR were conducted using the FastKing One Step RT-qPCR Kit (SYBR Green; TIANGEN). qPCR reactions were run on a real-time PCR system (ABI7500; Applied Biosystems, Foster City, CA, USA) under standard thermal cycling conditions. The relative expression levels of circRNA and mRNA were normalized to GAPDH, while miRNA expression was normalized to U6. The data were analyzed using the 2−ΔΔCT method. Additionally, GAPDH and U6 served as controls for the cytoplasmic and nuclear fractions, respectively. Specific primer sequences used for qPCR are shown in Table 3.
Actinomycin D treatment
Focus and HA22T cells were treated with Actinomycin D (MedChemExpress, Monmouth Junction, NJ, USA) for designated time points: 0, 4, 8, 12, and 24 h. At each time point, total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). qPCR was subsequently performed to determine the levels of circ_0008043 and linear mRNA PTGR1. The qPCR analysis was conducted using the FastKing One Step RT-qPCR Kit (SYBR Green; TIANGEN, Beijing, China) on a real-time PCR system (ABI7500; Applied Biosystems, Foster City, CA, USA).
Determination of cell viability
Cell viability was assessed using the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). HCC cells were seeded in 96-well plates at a density of 2,000 cells per well and cultured for 48 h. Following this incubation period, 10 µL of CCK-8 solution was added to each well and the cells were further incubated for 2 h at 37 °C. The absorbance was then measured at 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
Measurement of cell migration
Cell migration was evaluated using both the Transwell assay and the scratch test. For the Transwell assay, a 200 µL cell suspension containing 1 × 105 cells in serum-free medium was added to the upper compartment of Transwell chambers (pore size 8 μm; Corning, NY, USA). The lower compartments were filled with complete medium to serve as a chemoattractant. After incubation for 24 h at 37 °C in a humidified atmosphere containing 5% CO2, non-migrated cells on the upper surface of the membrane were removed, and the cells that migrated to the lower surface were fixed with 4% paraformaldehyde for 15 min and stained with 0.2% crystal violet solution for 10 min. The stained cells were then observed and photographed under an inverted microscope (model IX83; Olympus, Tokyo, Japan). For the scratch test, cells were seeded in six-well plates and cultured until they reached approximately 100% confluence. A uniform wound was created across the center of the cell monolayer using a sterile 200 µL pipette tip. Cellular debris was carefully washed away with PBS, and fresh complete medium was added to each well. The cells were then incubated for 24 h. Images of the wound closure were captured at 0 and 24 h under a microscope to assess the migration rate.
Western blotting
Proteins were extracted using RIPA buffer supplemented with protease inhibitors. Protein concentration was determined using a BCA Protein Assay Kit (Beyotime, Shanghai, China). Equal amounts of protein (20 µg) were separated by 10% SDS-PAGE and subsequently transferred onto PVDF membranes. The membranes were blocked with 5% skimmed milk in TBST for 1 h at room temperature to prevent non-specific binding. Following blocking, the membranes were incubated overnight at 4 °C with primary antibodies (Abcam, Cambridge, UK) against the proteins of interest. After extensive washing with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Abcam) for 1 h at room temperature. Protein bands were visualized using an ECL detection reagent (Beyotime), and images were captured using a ChemiDoc imaging system (Bio-Rad, Hercules, CA, USA).
Flow cytometry
Cell apoptosis was assessed using the Annexin V-FITC Apoptosis Detection Kit (Beyotime, Shanghai, China). Briefly, HCC cells were washed with PBS and detached using trypsin-EDTA solution. After detachment, cells were collected by centrifugation, resuspended in PBS, and counted. Approximately 5 × 104 cells were then resuspended in 195 µL of binding buffer (included in the kit). Next, 5 µL of Annexin V-FITC and 10 µL of PI were added to each sample, mixed gently, and incubated for 20 min at room temperature in the dark. Apoptotic cells were detected using a flow cytometer (CytoFLEX; Beckman Coulter, Miami, FL, USA). Data analysis was performed using associated software.
Bioinformatic analysis
The targeted miRNAs of circ_0008043 were predicted using the Circular RNA Interactome database. The targeted mRNAs of miR-661 were predicted using the TargetScan database.
Dual-luciferase reporter assay
Wild-type (WT) sequences of circ_0008043 and PLEKHG4B containing the miR-661 binding sites were amplified and cloned into pmirGLO vectors (Promega, Madison, WI, USA). Similarly, mutant (MUT) sequences of circ_0008043 and PLEKHG4B were generated and also cloned into pmirGLO vectors. HEK293T cells were seeded in 24-well plates and co-transfected with either WT or MUT plasmids along with miR-661 mimic or negative control mimic (nc mimic) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h of transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. The relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity.
RNA pull-down assay
HEK293T cells were lysed using lysis buffer supplemented with RNase inhibitors and protease inhibitors. Biotin-labeled miR-661 and its negative control (biotin-nc) probes were used to pre-coat streptavidin magnetic beads (Beyotime, Shanghai, China) for 3 h at room temperature. The cell lysate was then incubated with the probe-coated beads overnight at 4 °C with gentle rotation. After incubation, the beads were washed five times with cold lysis buffer to remove unbound material. Total RNA was extracted from the bound fraction using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The expression levels of circ_0008043 and PLEKHG4B in the pull-down samples were analyzed by qPCR using the FastKing One Step RT-qPCR Kit (SYBR Green; TIANGEN, Beijing, China).
RNA-binding protein Immunoprecipitation (RIP)
RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (GENESEED, Guangzhou, China) according to the manufacturer’s protocol. Briefly, HA22T cells were lysed in RIP lysis buffer supplemented with RNase inhibitors and protease inhibitors. Anti-ESRP1 antibody and control anti-IgG antibody (both from GENESEED) were incubated with protein A/G magnetic beads at 4 °C for 2 h to form antibody-bead complexes. Cell lysates were then incubated with these antibody-bead complexes overnight at 4 °C with gentle rotation. After extensive washing with RIP wash buffer, the immunoprecipitated RNA was isolated using the provided protocol and reagents. The expression levels of circ_0008043 in the precipitated RNA samples were analyzed by qPCR using the FastKing One Step RT-qPCR Kit (SYBR Green; TIANGEN, Beijing, China).
In vivo metastasis model
The animal study was approved by the Ethics Committee of The Third People’s Hospital of Shenzhen (Approval Number: 2024-069-01) and conducted in accordance with the ARRIVE guidelines. Short hairpin RNA targeting circ_0008043 (sh-circ_0008043) and a negative control shRNA (sh-nc) were cloned into adenovirus (Ad) vectors and transfected into HEK293T cells to produce adenovirus particles. Focus cells were subsequently infected with either Ad-sh-circ_0008043 or Ad-sh-nc. BALB/c nude mice (male, six-week-old) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). The mice were randomly divided into two groups (n = 6 per group). Each mouse was injected with 1 × 106 infected Focus cells via the tail vein. Eight weeks post-injection, the mice were euthanized by intraperitoneal injection of sodium pentobarbital at a dose of 160 mg/kg. Lungs were harvested from all animals, and the number of metastatic nodules on the lung surface was counted.
Immunohistochemistry
The metastatic nodules were collected to made paraffin sections. The sections were incubated with primary antibodies against PLEKHG4B, E-cadherin, N-cadherin, and Vimentin at 4 °C overnight, and then incubated with secondary antibody (Abcam) at 37 °C for 1 h. The sections were incubated with 3,3′-diaminobenzidine for color developing. Results were observed under a light microscope.
Hematoxylin and Eosin (H&E) staining
Paraffin-embedded sections of metastatic nodules were deparaffinized and rehydrated through a graded ethanol series. The sections were then stained with hematoxylin (MedChemExpress, Monmouth Junction, NJ, USA) for 10 min to highlight nuclei, followed by a brief wash in running tap water to enhance color development. Subsequently, the sections were counterstained with eosin (MedChemExpress) for 3 min to visualize cytoplasmic structures. After staining, the sections were dehydrated through an ascending ethanol series, cleared in xylene, and mounted with neutral balsam. The stained sections were observed and imaged under a light microscope (Olympus, Tokyo, Japan).
Statistical analysis
All experiments were independently repeated at least three times. Data analysis was conducted using GraphPad Prism 7 software (GraphPad Software, San Diego, CA, USA). Results are presented as mean ± SD. Comparisons between two groups were analyzed using Student’s t-test, while one-way ANOVA followed by Tukey’s post hoc test was used for comparisons among multiple groups. Kaplan–Meier curves were employed to analyze the overall survival rates of HCC patients, and the log-rank test was used to assess statistical significance. A P-value < 0.05 was considered statistically significant.
Results
Circ_0008043 expression is upregulated in HCC tissues and cells
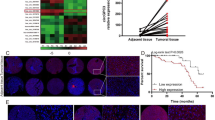
Numerous circRNAs are differentially expressed in cancers and play crucial roles in regulating cancer progression. In this study, we analyzed the GSE155949 dataset and identified several circRNAs that were either upregulated or downregulated in HCC tissues compared to normal tissues (Fig. 1A). To further investigate the role of circ_0008043 in HCC, we examined its expression in 60 paired HCC and adjacent non-tumor tissues using qPCR. The results demonstrated that circ_0008043 expression was significantly higher in HCC tissues than in normal tissues (Fig. 1B). The association between circ_0008043 expression and clinicopathological characteristics of HCC patients is summarized in Table 1. We found that elevated circ_0008043 expression was significantly associated with AFP levels, tumor size, tumor differentiation, and distant metastasis, but not with age, sex, or tumor number. Kaplan–Meier survival analysis revealed that patients with high circ_0008043 expression exhibited poorer overall survival compared to those with low expression (Fig. 1C). Next, we evaluated circ_0008043 expression in HCC cell lines. qPCR analysis showed that circ_0008043 was highly expressed in Focus and HA22T cells compared to normal liver epithelial THLE3 cells (Fig. 1D). Structural analysis indicated that circ_0008043 is derived from exons 5–9 of the PTGR1 gene (Fig. 1E). To determine its subcellular localization, fluorescence in situ hybridization (FISH) was performed, revealing that circ_0008043 was predominantly localized in the cytoplasm of Focus and HA22T cells rather than in the nucleus (Fig. 1F). This observation was further validated by isolating cytoplasmic and nuclear RNA fractions, followed by qPCR, which confirmed the cytoplasmic localization of circ_0008043 in HCC cells (Fig. 1G). To assess the stability of circ_0008043, HCC cells were treated with actinomycin D, and the levels of circ_0008043 and linear PTGR1 mRNA were measured using qPCR. The results demonstrated that linear PTGR1 mRNA had a shorter half-life compared to circ_0008043, indicating the enhanced stability of the circular RNA (Fig. 1H). In summary, our findings demonstrate that circ_0008043 is significantly upregulated in both HCC tissues and cell lines, exhibits cytoplasmic localization, and possesses greater stability compared to its linear counterpart.
Circ_0008043 expression is upregulated in HCC tissues and cells. (A) Differentially expressed circRNAs in HCC tumor and normal tissues were predicted using the GSE155949 dataset. Red: upregulated circRNAs; blue: downregulated circRNAs. (B) Circ_0008043 expression in tumor tissues and paired adjacent normal tissues from patients with HCC (n = 60). (C) The percentage of surviving patients with high or low circ_00088043 expression. (D) Circ_0008043 expression in HCC cell lines (Focus and HA22T) and liver epithelial cells (THLE3). (E) Diagram of the formation of circ_0008043 from the exon of PTGR1 transcripts. (F) The location of circ_0008043 was measured in HCC cells was meas8red using FISH. Scale bar = 50 μm. (G) qPCR was used to measure the expression of circ_0008043 in the cytoplasm and nucleus in HCC cells. U6 and GAPDH were the internal controls in the nucleus and cytoplasm. (H) HCC cells were treated with actinomycin D, and the expression of circ_0008043 and linear RNA PTGR1 was measured by qPCR. **P < 0.01.
Silencing of circ_0008043 suppresses HCC cell migration and epithelial-mesenchymal transition (EMT), and induces apoptosis
To investigate the biological functions of circ_0008043 in HCC cells, we first established stable circ_0008043 knockdown cell lines. As shown in Fig. 2A, transfection with sh-circ_0008043 significantly reduced the expression of circ_0008043 in both Focus and HA22T cells. Next, we evaluated the effects of circ_0008043 knockdown on cell viability using the CCK-8 assay. The results demonstrated that silencing circ_0008043 markedly inhibited the viability of Focus and HA22T cells (Fig. 2B). Cell migration was assessed using both Transwell assays and wound healing assays. The data revealed that knockdown of circ_0008043 significantly suppressed the migratory capacity of HCC cells (Fig. 2C–F). To further explore the role of circ_0008043 in EMT, we measured the protein levels of key EMT markers, including E-cadherin, N-cadherin, and Vimentin, using western blotting. As illustrated in Fig. 2G, silencing circ_0008043 increased the expression of E-cadherin, a marker of epithelial phenotype, while downregulating the mesenchymal markers N-cadherin and Vimentin. Additionally, apoptosis was analyzed using flow cytometry, and the results indicated that knockdown of circ_0008043 significantly promoted apoptosis in both Focus and HA22T cells (Supplementary Fig. 1). Collectively, these findings demonstrate that silencing circ_0008043 suppresses HCC cell viability, migration, and EMT, while inducing apoptosis.
Silencing of circ_0008043 suppresses HCC cell migration and EMT. (A) circ_0008043 expression in sh-nc and sh-circ_0008043 transfected cells. (B) CCK-8 was conducted to assess cell viability. (C, D) Transwell assay was used to determine cell migration, and migration rate was quantified. Scale bar = 50 μm. (E, F) Scratch test was used to evaluate cell migration, and wound healing percentage was quantified. Scale bar = 200 μm. (G) Protein levels of EMT markers (E-cadherin, N-cadherin, and Vimentin) were examined by western blotting. Original bands are presented in supplementary Fig. 2. **P < 0.01.
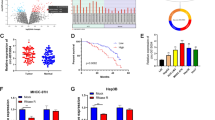
Circ_0008043 functions as an effective miR-661 sponge
It is well established that circRNAs can act as miRNA sponges by binding to miRNA response elements (MREs)17. To explore the downstream miRNAs of circ_0008043, we utilized the Circular RNA Interactome database to predict potential targets. Our analysis identified miR-661 as a putative target of circ_0008043 (Fig. 3A). To validate this targeting relationship, we performed dual-luciferase reporter assays and RIP experiments. In the dual-luciferase reporter assay, co-transfection of WT circ_0008043 with miR-661 significantly reduced luciferase activity, indicating direct binding between miR-661 and circ_0008043 (Fig. 3B). Furthermore, RIP experiments using biotin-labeled miR-661 demonstrated increased enrichment of circ_0008043 compared to control conditions (Fig. 3C), confirming the interaction between circ_0008043 and miR-661. Next, we assessed the expression levels of miR-661 in HCC cells. qPCR analysis revealed that miR-661 expression was significantly lower in HCC cell lines (Focus and HA22T) compared to normal liver epithelial THLE3 cells (Fig. 3D). Additionally, silencing circ_0008043 led to an upregulation of miR-661 expression in HCC cells (Fig. 3E). Similarly, miR-661 expression was found to be downregulated in HCC tissues compared to adjacent non-tumor tissues (Fig. 3F). Collectively, these results indicate that circ_0008043 acts as a sponge for miR-661, thereby negatively regulating its expression in HCC cells and tissues.
Circ_0008043 functions as an effective miR-661 sponge. (A) The potential binding sites of circ_0008043 and miR-661. The mutant circ_0008043 sequences were designed. (B) Dual-luciferase reporter assay and (C) RNA pull-down were performed to affirm the targeting relationship. (D) miR-661 expression in HCC cells (Focus and HA22T) and normal cells (THLE3). (E) miR-661 expression in sh-nc and sh-circ_0008043 transfected cells. (F) miR-661 expression in 60 paired HCC tissues and adjacent non-tumor tissues. **P < 0.01. NS: no significance.
Circ_0008043 sponges miR-661 to regulate HCC cell migration and EMT
To further elucidate the functional interplay between circ_0008043 and miR-661 in HCC cells, we transfected HCC cells with miR-661 inhibitor and assessed its impact on miR-661 expression. As shown in Fig. 4A, transfection with the miR-661 inhibitor significantly reduced miR-661 levels in Focus and HA22T cells. We then evaluated the effects of circ_0008043 knockdown and miR-661 inhibition on cell viability, migration, and EMT. The results demonstrated that silencing circ_0008043 significantly inhibited cell viability and migration (Fig. 4B–E). However, downregulation of miR-661 partially reversed these inhibitory effects, indicating that circ_0008043 exerts its regulatory functions, at least in part, through sponging miR-661. To investigate the role of circ_0008043 and miR-661 in EMT, we examined the protein levels of key EMT markers using western blotting. Knockdown of circ_0008043 increased the expression of the epithelial marker E-cadherin while decreasing the mesenchymal markers N-cadherin and Vimentin (Fig. 4F). Importantly, reducing miR-661 expression reversed these changes, restoring N-cadherin and Vimentin levels and diminishing the upregulation of E-cadherin induced by circ_0008043 knockdown. In summary, our findings indicate that circ_0008043 suppresses HCC cell migration and EMT by acting as a sponge for miR-661. This interaction plays a critical role in regulating cellular phenotypes associated with HCC progression.
Circ_0008043 sponges miR-661 to regulate HCC cell migration and EMT. (A) miR-661 expression in HCC cells following inhibitor and nc inhibitor transfection. (B) Cell viability was measured using CCK-8. (C) Transwell assay and (D, E) wound healing assay were carried out to determine cell migration. Scale bar for Transwell assay = 50 μm. Scale bar for wound healing assay = 200 μm. (F) Protein levels of EMT markers (E-cadherin, N-cadherin, and Vimentin) were examined by western blotting. Original bands are presented in supplementary Fig. 3. **P < 0.01.
PLEKHG4B is a downstream target of miR-661
Accumulating evidence has demonstrated that miRNAs regulate pathophysiological processes by modulating mRNA translation and degradation18. To identify potential targets of miR-661, we utilized the TargetScan database for prediction analysis. Our results revealed that PLEKHG4B was a putative target of miR-661 (Fig. 5A). Dual-luciferase reporter assays confirmed that co-transfection of miR-661 significantly reduced the luciferase activity associated with the PLEKHG4B 3′-UTR (Fig. 5B). Furthermore, RNA pull-down experiments using biotinylated miR-661 demonstrated a significant enrichment of PLEKHG4B mRNA (Fig. 5C), indicating a direct interaction between miR-661 and PLEKHG4B. These findings collectively suggest that miR-661 directly targets PLEKHG4B. Next, we examined the expression levels of PLEKHG4B in HCC cells. Our data showed that PLEKHG4B expression was significantly higher in HCC cell lines compared to the normal liver cell line THLE3 (Fig. 5D). Next, we found that PLEKHG4B expression was elevated by transfection of miR-661 inhibitor (Fig. 5E and F). Notably, overexpression of circ_0008043 led to a reduction in PLEKHG4B levels, while this effect was reversed by miR-661 inhibitor (Fig. 5G and H). Additionally, PLEKHG4B expression was markedly upregulated in HCC tissues compared to adjacent non-tumor tissues (Fig. 5I). Taken together, these results demonstrate that miR-661 directly targets PLEKHG4B and negatively regulates its expression.
PLEKHG4B is a downstream target of miR-661. (A) The potential binding sites of PLEKHG4B and miR-661 and the mutant PLEKHG4B sequences. (B) Dual-luciferase reporter assay and (C) RNA pull-down were used to verify the targeting relationship. (D) PLEKHG4B expression in HCC cells (Focus and HA22T) and normal cells (THLE3). (E) PLEKHG4B mRNA and (F) protein levels mediated by miR-661 inhibitor were detected using qPCR and western blotting, respectively. Original bands are presented in supplementary Fig. 4. (G) PLEKHG4B mRNA and (H) protein levels in HCC cells after circ_0008043 knockdown and miR-661 inhibition were detected using qPCR and western blotting, respectively. Original bands are presented in supplementary Fig. 5. (I) PLEKHG4B expression in 60 paired HCC tissues and adjacent non-tumor tissues. **P < 0.01. NS: no significance.
miR-661 targets PLEKHG4B to regulate cell migration and EMT
To elucidate the effects of miR-661 and PLEKHG4B on cellular behaviors, we conducted a series of functional assays. Overexpression of miR-661 was achieved by transfecting cells with a miR-661 mimic, resulting in significantly increased miR-661 expression (Fig. 6A). Similarly, transfection with a PLEKHG4B overexpression vector led to a marked elevation in PLEKHG4B levels (Fig. 6B). Functional assays, including CCK-8, Transwell migration, and wound healing (scratch) tests, were performed to assess the impact of these manipulations on cell viability and migration. The results indicated that overexpression of miR-661 inhibited both cell viability and migration. Notably, enforced expression of PLEKHG4B partially reversed this inhibitory effect (Fig. 6C-F). Furthermore, Western blot analysis demonstrated that miR-661 upregulated E-cadherin levels while downregulating N-cadherin and Vimentin, indicative of its role in suppressing EMT. However, the overexpression of PLEKHG4B counteracted these effects mediated by miR-661, restoring N-cadherin and Vimentin levels and reducing E-cadherin expression (Fig. 6G). In summary, our findings suggest that miR-661 exerts its inhibitory effects on cell viability, migration, and EMT in HCC cells by targeting PLEKHG4B.
miR-661 targets PLEKHG4B to regulate cell migration and EMT. (A) miR-661 expression in mimic and nc mimic transfected cells. (B) PLEKHG4B expression in HCC cells after PLEKHG4B overexpression vector and empty vector transfection. (C) Cell viability was measured using CCK-8. (D) Transwell assay and (E, F) wound healing assay were carried out to determine cell migration. Scale bar for Transwell assay = 50 μm. Scale bar for wound healing assay = 200 μm. (G) Protein levels of EMT markers (E-cadherin, N-cadherin, and Vimentin) were examined by western blotting. Original bands are presented in supplementary Fig. 6. **P < 0.01.
ESRP1 promotes the biogenesis of circ_0008043
circRNAs are generated through back-splicing of precursor mRNAs, a process regulated by various splicing factors depending on the biological context19. To identify which splicing factors regulate the biogenesis of circ_0008043, we transfected HA22T cells with shRNAs targeting ESRP1, ESRP2, NOVA1, NOVA2, MEX3A, MEX3B, QKI, and SRSF1. qPCR analysis confirmed the effective knockdown of each targeted splicing factor compared to the sh-nc group (Fig. 7A). Subsequent qPCR analysis revealed that only the knockdown of ESRP1 significantly reduced the expression of circ_0008043, whereas the other splicing factors had no effect (Fig. 7B). These findings suggest that ESRP1 may promote the biogenesis of circ_0008043. Further investigation identified five potential ESRP1 binding sites within the flanking introns of the circ_0008043 formed by PTGR1 (Fig. 7C). To determine which of these sites interacted with ESRP1, RIP assays were performed. The results indicated that ESRP1 bound specifically to sites b, c, d, and e within the PTGR1 pre-mRNA (Fig. 7D). Mutagenesis studies showed that disruption of all four motifs (b, c, d, and e) or any two motifs within intron 4 (b/c) or intron 9 (d/e) abolished ESRP1’s ability to modulate circ_0008043 expression. Interestingly, mutation of a single motif was sufficient to reduce circ_0008043 levels (Fig. 7E). Additionally, qPCR analysis demonstrated that knockdown of ESRP1 not only decreased circPTGR1 (circ_0008043) expression but also increased the expression of linear PTGR1 mRNA (Fig. 7F). Taken together, these results indicate that ESRP1 facilitates the biogenesis of circ_0008043 by binding to specific motifs within its flanking introns.
ESRP1 promotes the biogenesis of circ_0008043. (A) After shRNAs targeting these splicing factors were transfected into HA22T cells, their expression was measured using qPCR. (B) Effect of splicing factor knockdown on the expression of circ_0008043. (C) The binding sites of ESRP1 in the flanking of circ_0008043. (D) The binding relationship between circ_0008043 and ESRP1 motif was analyzed using RIP. (E) Circ_0008043 expression was measured using qPCR after ESRP1 knockdown when the motif was wild-type or mutated. (F) Effect of ESRP1 knockdown on the expression of circ_0008043 (circPTGR1) and linear PTGR1. **P < 0.01. NS: no significance.
Interfering with circ_0008043 suppresses tumor metastasis in vivo
To evaluate the effect of circ_0008043 on tumor metastasis in vivo, we conducted animal experiments using a tail vein injection model. Focus cells infected with either Ad-sh-circ_0008043 or Ad-sh-nc (negative control) were injected into the tail veins of mice. After a defined experimental period, the lungs of the mice were harvested for analysis. As shown in Fig. 8A, knockdown of circ_0008043 significantly suppressed tumor metastasis to the lungs, as evidenced by a marked reduction in the number of metastatic nodules. Furthermore, H&E staining confirmed that silencing circ_0008043 effectively inhibited lung metastasis in HCC (Fig. 8B). To further explore the underlying mechanisms, IHC was performed to measure the expression levels of PLEKHG4B and EMT-related markers in the metastatic nodules. The results demonstrated that silencing circ_0008043 led to a significant decrease in the expression of PLEKHG4B, N-cadherin, and Vimentin, while E-cadherin levels were markedly increased (Fig. 8C). These findings suggest that circ_0008043 knockdown suppresses tumor metastasis by downregulating PLEKHG4B and reversing EMT. In summary, our data indicate that circ_0008043 plays a critical role in promoting tumor metastasis in vivo, and its knockdown inhibits metastasis by reducing PLEKHG4B expression and modulating EMT-related pathways.
Interfering with circ_0008043 suppresses HCC metastasis in vivo. (A) Images of the lungs to evaluate tumor metastasis, and the number of metastatic nodules was counted. The arrows indicated the metastatic nodules. (B) Represent images of H&E staining results of pulmonary metastatic focis. (C) The levels of PLEKHG4B and ETM markers (E-cadherin, N-cadherin, and Vimentin) were measured in these metastatic nodules using immunohistochemistry. **P < 0.01.
Discussion
In the present study, we demonstrated for the first time that knockdown of circ_0008043 modulates the miR-661/PLEKHG4B axis to suppress the migration and EMT of HCC cells, based on the ceRNA regulatory mechanism. Furthermore, we revealed that the splicing factor ESRP1 facilitates the biogenesis of circ_0008043.
With advancements in high-throughput sequencing technology and bioinformatics analysis, increasing attention has been directed toward the biological roles of circRNAs. Compared with linear mRNAs, circRNAs are more stable due to their closed-loop structure, making them promising biomarkers for disease diagnosis and prognosis20. Accumulating evidence has shown that numerous circRNAs are dysregulated in HCC and play crucial roles in tumor progression by regulating processes such as cell proliferation, drug resistance, invasion, migration, angiogenesis, and apoptosis21,22,23. However, the functional roles of many circRNAs in HCC remain poorly understood. Previous studies have reported that circ_0008043 is upregulated in HCC15,16. Consistent with these findings, our results showed that circ_0008043 expression was significantly elevated in both HCC tissues and cell lines. Functional experiments further demonstrated that silencing circ_0008043 suppressed HCC cell migration and EMT while promoting apoptosis. Moreover, in vivo experiments confirmed that knockdown of circ_0008043 inhibited tumor metastasis. These findings reinforce the notion that circ_0008043 acts as a tumor promoter in HCC, aligning with prior research16. Nevertheless, there are limitations to our study. We analyzed only 60 paired HCC and adjacent normal tissue samples, which represents a relatively small cohort. Future studies with larger sample sizes are warranted to validate our findings and provide more robust insights into the role of circ_0008043 in HCC.
CircRNAs function as miRNA sponges to participate in HCC progression. Previous studies have shown that circ_0008043 negatively regulates the miR-326/RAB21 axis in HCC16. Generally, a single circRNA can simultaneously target multiple miRNAs, forming a complex regulatory network. In this study, we confirmed that circ_0008043 acts as an effective sponge for miR-661. miR-661 has been identified as a key regulator in various malignancies, including non-small cell lung cancer, glioma, melanoma, and ovarian cancer24,25,26,27. Moreover, miR-661 has been proposed as a potential biomarker for HCC28with its expression increased in the serum of HCC patients. Additionally, circ_0000221 has been reported to inhibit HCC progression by suppressing miR-661 29. In our study, we observed that miR-661 expression was reduced in HCC tissues and cells compared to their normal counterparts. Furthermore, overexpression of miR-661 reversed the effects on cellular behaviors induced by circ_0008043, indicating that miR-661 functions as a tumor suppressor in HCC. Notably, these findings are inconsistent with previous reports29which suggest that miR-661 promotes HCC progression. This discrepancy may be attributed to the complex regulatory mechanisms involving miR-661, where it can act as both a tumor promoter and suppressor depending on the specific context and the presence of other regulatory factors.
The downstream targets of miR-661 were subsequently identified, with a particular focus on the role of PLEKHG4B in HCC. PLEKHG4B is known to be a key driver in cell junction formation, promoting actin remodeling30. Importantly, it has also been implicated as a carcinogenic factor in HCC31. In our study, we explored the functional impact of PLEKHG4B on HCC cellular processes. Our results demonstrated that miR-661 inhibits the migration and EMT of HCC cells by targeting PLEKHG4B, suggesting that PLEKHG4B functions as an oncogene in HCC.
Additionally, we investigated the upstream regulatory mechanisms of circ_0008043. To the best of our knowledge, splicing factors play a critical role in modulating circRNA biogenesis by splicing the flanking long introns of pre-mRNAs during back-splicing32. In this study, we identified ESRP1 as the only splicing factor that significantly affected circ_0008043 expression. Several studies have reported that ESRP1 promotes the biogenesis and circularization of various circRNAs, including circUHRF1, circTNPO3, and circANKS1B33,34,35. Our findings align with these reports, as we demonstrated that ESRP1 facilitates the splicing of introns 4 and 9, thereby promoting the circularization of exons 5–9 in circ_0008043. These results suggest that ESRP1 is a key regulator of circ_0008043 biogenesis. Nevertheless, several questions remain unanswered. For instance, whether other splicing factors are involved in regulating circ_0008043 biogenesis and what mechanisms regulate ESRP1 expression warrant further investigation in future studies. Addressing these gaps will provide deeper insights into the complex regulatory network governing circ_0008043 and its role in HCC progression.
In summary, this study reveals that ESRP1-mediated biogenesis of circ_0008043 modulates the miR-661/PLEKHG4B axis to accelerate HCC progression by promoting cell migration and EMT. Furthermore, our in vivo experiments demonstrate that knockdown of circ_0008043 effectively inhibits tumor metastasis. These findings suggest that circ_0008043 may serve as a novel therapeutic target for HCC treatment. However, several important questions remain unanswered. For instance, whether other circRNAs or alternative pathways might compensate for the loss of circ_0008043 in HCC remains largely unknown. Additionally, translating these findings into clinical applications, particularly developing strategies to target circ_0008043 therapeutically, requires further investigation. Future studies should focus on elucidating the broader regulatory networks involving circ_0008043 and exploring its potential as a biomarker and therapeutic target in HCC.
Data availability
The dataset (GSE155949) analysed during the current study are available in the GEO database. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE155949).
References
Torimura, T. & Iwamoto, H. Treatment and the prognosis of hepatocellular carcinoma in Asia. LIVER INT 42 (2022). (2042).
Zhang, C. H., Cheng, Y., Zhang, S., Fan, J. & Gao, Q. Changing epidemiology of hepatocellular carcinoma in Asia. LIVER INT 42 (2022). (2029).
Ren, Z., Ma, X., Duan, Z. & Chen, X. Diagnosis, Therapy, and Prognosis for Hepatocellular Carcinoma. Anal Cell Pathol (Amst) 8157406 (2020). (2020).
Zhang, W., Zhang, B. & Chen, X. P. Adjuvant treatment strategy after curative resection for hepatocellular carcinoma. Front. Med. 15, 155 (2021).
Galle, P. R., Dufour, J. F., Peck-Radosavljevic, M., Trojan, J. & Vogel, A. Systemic therapy of advanced hepatocellular carcinoma. FUTURE ONCOL. 17, 1237 (2021).
Vogel, A., Meyer, T., Sapisochin, G., Salem, R. & Saborowski, A. Hepatocellular carcinoma. LANCET 400, 1345 (2022).
Nagaraju, G. P., Dariya, B., Kasa, P., Peela, S. & El-Rayes, B. F. Epigenetics in hepatocellular carcinoma. SEMIN CANCER BIOL. 86, 622 (2022).
Chen, L. & Shan, G. CircRNA in cancer: fundamental mechanism and clinical potential. CANCER LETT. 505, 49 (2021).
Ashwal-Fluss, R. et al. CircRNA biogenesis competes with pre-mRNA splicing. MOL. CELL. 56, 55 (2014).
Zhang, M. et al. circRNA-miRNA-mRNA in breast cancer. CLIN. CHIM. ACTA. 523, 120 (2021).
Gao, M., Zhang, Z., Sun, J., Li, B. & Li, Y. The roles of circRNA-miRNA-mRNA networks in the development and treatment of osteoporosis. Front. Endocrinol. (Lausanne). 13, 945310 (2022).
Liu, J. et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY). 11, 12412 (2019).
Huang, G. et al. CircRNA hsa_circrna_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/beta-catenin pathway. CELL. DEATH DIS. 11, 1065 (2020).
Zhang, W., Liu, T., Li, T. & Zhao, X. Hsa_circRNA_102002 facilitates metastasis of papillary thyroid cancer through regulating miR-488-3p/HAS2 axis. CANCER GENE THER. 28, 279 (2021).
Han, J. et al. Clinicopathological-Associated regulatory network of deregulated circrnas in hepatocellular carcinoma. Cancers (Basel) 13, 2772 (2021).
Zhang, K. et al. Circular RNA circ_0008043 promotes the proliferation and metastasis of hepatocellular carcinoma cells by regulating the MicroRNA (miR)-326/RAB21 axis. BIOENGINEERED 13, 6600 (2022).
Tong, K. L., Tan, K. E., Lim, Y. Y., Tien, X. Y. & Wong, P. F. CircRNA-miRNA interactions in atherogenesis. MOL. CELL. BIOCHEM. 477, 2703 (2022).
Fabian, M. R., Sonenberg, N. & Filipowicz, W. Regulation of mRNA translation and stability by MicroRNAs. ANNU. REV. BIOCHEM. 79, 351 (2010).
Li, X., Yang, L. & Chen, L. L. The biogenesis, functions, and challenges of circular RNAs. MOL. CELL. 71, 428 (2018).
Chen, L. et al. The bioinformatics toolbox for circrna discovery and analysis. BRIEF. BIOINFORM. 22, 1706 (2021).
Xu, J. et al. CircRNA-SORE mediates Sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal. Transduct. Target. Ther. 5, 298 (2020).
Su, Y. et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 11, 8183 (2019).
Huang, X. Y. et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 39, 20 (2020).
Bao, Y. et al. Hsa_Circ_0001947/MiR-661/DOK7 Axis restrains Non-Small cell lung Cancer development. J. Microbiol. Biotechnol. 31, 1508 (2021).
Li, Z., Liu, Y. H., Diao, H. Y., Ma, J. & Yao, Y. L. MiR-661 inhibits glioma cell proliferation, migration and invasion by targeting hTERT. Biochem. Biophys. Res. Commun. 468, 870 (2015).
Lin, X., Zhong, L., Wang, N., Chu, X. & Liu, B. Hsa_circ_0103232 promotes melanoma cells proliferation and invasion via targeting miR-661/RAB3D. CELL. CYCLE. 21, 1811 (2022).
Zhu, T. et al. MiR-661 contributed to cell proliferation of human ovarian cancer cells by repressing INPP5J expression. BIOMED. PHARMACOTHER. 75, 123 (2015).
Ali, M. A. et al. Investigating miRNA-661 and ATG4-B mRNA expression as potential biomarkers for hepatocellular carcinoma. BIOMARK. MED. 12, 245 (2018).
Matboli, M. et al. Impact of circ-0000221 in the Pathogenesis of Hepatocellular via Modulation of miR-661-PTPN11 mRNA Axis. PHARMACEUTICS 14 (2022).
Ninomiya, K., Ohta, K., Yamashita, K., Mizuno, K. & Ohashi, K. PLEKHG4B enables actin cytoskeletal remodeling during epithelial cell-cell junction formation. J. CELL. SCI. 134, jcs249078 (2021).
Lin, S. Y. et al. Recurrent HBV Integration Targets as Potential Drivers in Hepatocellular Carcinoma. CELLS-BASEL 10 (2021).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. NAT. REV. GENET. 20, 675 (2019).
Zhao, W. et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. CELL. DEATH DIFFER. 27, 919 (2020).
Pan, X. et al. Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin. Transl Med. 12, e994 (2022).
Zeng, K. et al. The pro-metastasis effect of circANKS1B in breast cancer. MOL. CANCER. 17, 160 (2018).
Funding
This work was supported in part by grants from the Shenzhen High-level Hospital Construction Fund (XKJS-PWK-001); Medical Science and Technology Research Foundation of Guangdong Province (No.B2023229); Shenzhen Key Medical Discipline Construction Fund (No.SZXK079); Youth Fund of The Third People’s Hospital of Shenzhen (No. G2021003); Shenzhen Fundamental Research Program (NO.JCYJ20210324131809027).
Author information
Authors and Affiliations
Contributions
KZ conceived the study; KZ conducted the experiments; TF, QC, XY, XJ and NM analyzed the data; KZ was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study was approved by the Ethics Committee of The Third People’s Hospital of Shenzhen. All experiments were performed in accordance with the ARRIVE guidelines. All experiments were performed in accordance with relevant guidelines and regulations.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, K., Fang, T., Chen, Q. et al. Circ_0008043 promotes the metastasis of hepatocellular carcinoma by regulating the miR-661/PLEKHG4B axis. Sci Rep 15, 24580 (2025). https://doi.org/10.1038/s41598-025-10618-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10618-6