Abstract
The biological process of fracture repair involves inflammation, vascularization, endochondral ossification, and cartilage production. Long non-coding RNAs (lncRNAs) are essential for the growth and maintenance of the skeleton. In this study, the role of the long non-coding RNA H19 in endochondral ossification during fracture repair was investigated. We examined H19 expression in the contexts of skeletal development and fracture repair and found that hypertrophic chondrocytes, growing endochondral bone spongiosa, and cortical bone exhibit H19 expression. H19 expression in cortical bone was similar to that in skeletal muscle, suggesting a potential role in bone development regulation. Inhibiting H19 near fractures in a mouse model resulted in disruption of vascularization and delayed endochondral ossification, ultimately leading to delayed fracture union. Importantly, H19 inhibition had no impact on the formation of the cartilaginous callus, indicating its specific role in vascularization and ossification. Further analysis revealed that H19 reduced miR-21a-5p’s inhibition of smad7 by functioning as a sponge for miR-21a-5p. H19 silencing led to a decrease in Smad7 expression and a rise in Smad2/3 phosphorylation, and delayed both ossification and vascularization. In conclusion, H19 influences bone remodeling and fracture healing by modulating the miR-21a-5p-smad7-TGFβ-smad2/3 axis, presenting therapeutic potential for fracture repair.
Similar content being viewed by others
Introduction
The goal of fracture repair aims to restore the structural integrity and functionality of the damaged bone tissue through a complex series of cellular events that are part of a highly coordinated biological process. Numerous cellular and molecular cues, including those related to inflammation, angiogenesis, cartilage formation, and endochondral ossification, strictly regulate this intricate process1. Understanding the molecular processes that govern these events is essential for enhancing our comprehension of bone regeneration and identifying potential therapeutic targets to improve fracture healing outcomes1,2.
Long non-coding RNAs (lncRNAs) have emerged as key regulators of numerous biological processes, such as bone repair and skeletal development, in recent years3. Initially, these RNA molecules, composed of over 200 nucleotides, were thought to be mere transcriptional noise4,5. However, increasing research has demonstrated their crucial roles in a range of biological processes, often serving as versatile regulators of gene expression through various mechanisms6. One such lncRNA, H19, has gained attention for its diverse roles in various cellular contexts, including skeletal development7,8.
Common molecular and cellular pathways, particularly those regulating processes like endochondral ossification and vascularization, underlie both skeletal development and fracture repair1,9,10. Endochondral ossification, the process of bone formation by cartilage replacement, mirrors the developmental pathways that generate long bones during embryogenesis11,12. Notably, the expression of several genes essential for the embryonic development of long bones also occurs in the fracture callus, with expression patterns and histological features resembling those found in the growth plate12,13. These observations suggest that the regulatory processes governing bone growth may also be critical for fracture healing.
H19, initially identified as an imprinted gene with preferential maternal expression, exhibits dynamic expression patterns during skeletal growth and fracture healing8,14,15. Recent investigations have illuminated its potential role in regulating chondrogenesis and influencing osteogenic differentiation8,16,17,18. H19 expression in growth plate cartilage and metaphyseal bone varied with age, as described by Anenisia et al.19, implying a role in long bone formation and endochondral ossification. Furthermore, H19 expression increased in chondrocytes exposed to hypoxia, alongside collagen type 2 and miR-675, further indicating its involvement in cartilage-related processes20. These intriguing findings underscore the need for further investigation into H19’s functional role in fracture healing and skeletal growth.
Angiogenesis plays a key role in fracture recovery by providing the healing region with essential nutrients, oxygen, and osteogenic cells through newly formed blood vessels. One major risk factor for impaired healing is ischemia, or lack of vascularization21. In addition to providing nutrients and oxygen, blood vessels are crucial for endochondral ossification and play a signaling role in promoting cell differentiation22,23,24. The hypoxic conditions during fracture repair activate hypoxia-inducible factor (HIF), a key regulator of angiogenesis. HIF enhances vascularization by transcriptionally upregulating vascular endothelial growth factor (VEGF), which promotes blood vessel formation25,26,27. Thus, the interplay between HIF, VEGF, and angiogenesis is critical during bone regeneration.
Recent studies have begun to uncover how lncRNAs influence angiogenesis during fracture healing28. Long noncoding RNAs, such as H19, may regulate angiogenesis by modulating pro-angiogenic protein expression, thereby impacting vascularization at the fracture site29,30,31. Nevertheless, further research is required to fully elucidate H19’s specific role in vascularization and its effects on the molecular pathways associated with angiogenesis.
Taking these factors into account, the goal of our research is to determine the role of H19 in the intricate processes of vascularization and endochondral ossification during fracture repair. By employing both in vitro and in vivo methods, we aim to elucidate the molecular mechanisms by which H19 influences these critical aspects of bone repair. Our research may provide novel insights into the broader roles of lncRNAs in coordinating complex biological processes and offer important understandings into the molecular mechanisms driving bone growth and repair. Moreover, understanding the regulatory role of H19 in fracture repair could lead to the development of novel therapeutic strategies to promote bone healing and enhance clinical outcomes.
Results
Expression of H19 during postnatal skeletal development
With the mechanisms controlling endochondral ossification and those essential for embryonic skeletal development sharing a great deal of overlap, we used a FISH test to investigate the expression of H19 genes in mouse forelimbs on the day of birth. Different patterns of H19 expression were found by our analysis. Significantly, H19 was detected within hypertrophic chondrocytes that without expressing COL1α1 (indicated by the red arrow). Furthermore, at later phases of bone development, H19 expressed itself in the spongiosa of growing cortical and endochondral bones (Fig. 1A). Notably, it was discovered that the cortical bone’s expression of H19 was comparable to that of the surrounding skeletal muscle (Fig. 1A). Furthermore, the major location of H19 expression within the cytoplasm was revealed by our FISH results (Fig. 1B). Taken together, our results point to a strong relationship between H19 expression and the complex process of endochondral ossification.
Expression of H19 during postnatal skeletal development. (A) H19 exhibited expression within the spongiosa of developing endochondral bones and cortical bone. The expression of H19 gene (red) in the forelimb metaphyseal to diaphyseal region was detected by fluorescence in situ hybridization at postnatal day 0. H19 detected in hypertrophic chondrocytes that do not express COL1α1 (green) (shown by red arrow). (B) H19 expression localization in cells, H19 (red) expression is mainly located in the cytoplasm. DAPI, 4′, 6-diaminyl-2-phenylindole, COL1α1, Collagen 1α1.
Impact of H19 loss on fracture healing delay
To assess the involvement of H19 in fracture repair, we established a mouse model targeting loss-of-function (LOF) in fracture sites. This was accomplished by injecting an Adenovirus that expressed simH19 into the surrounding muscle, periosteum, and bone marrow at the fracture sites. A decrease in H19 expression in the fracture sites of 8–10 weeks old mice was successfully validated by quantitative PCR tests carried out on the surrounding tissues (Fig. 2B). To discern the specific cell lineages targeted by simH19 and contributing to callus formation during fracture repair, we evaluated red fluorescent protein (RFP) expression derived from adenovirus simH19 in tibia fractures of AdsimH19 mice at 2 weeks post-operation (PO-2 W). RFP-positive cells were seen in the periosteum, external cartilaginous callus, and internal cartilaginous callus when fracture calluses were stained with immunohistochemistry (Fig. 2A). These results demonstrated that all skeletal-related cells in the callus were effectively transfected by adenovirus simH19, with chondrocytes in the cartilaginous callus being the primary target.
To determine if H19 is necessary for fracture repair, including the formation of the cartilage callus and endochondral ossification, semi-stabilized tibia fractures were performed on mice injected with AdsimH19 and AdGFP at 8–10 weeks of age. Radiographs of AdGFP fractures showed the first indications of the creation of an external callus by post-operation week 2 (PO-2 W), and the cortical gap disappeared by post-operation week 4 (PO-4 W) due to a bridging internal and external bony callus, showing the unification of the fracture (Fig. 2C). On the other hand, callus development was clearly visible in AdsimH19 fractures along the periosteum, extending away from the fracture line at PO-2 W and PO-3 W. It was noted that there was no bridging callus between the cortices. Radiographic examination at PO-4 W revealed a marked delay in union for AdsimH19 tibia fractures (Fig. 2C). Micro-CT (µCT) examinations of mineralized calluses from AdGFP fractures showed virtually complete bridging of bony calluses by PO-2 W, with complete bridging by PO-4 W, consistent with radiographic findings. On the other hand, at PO-2 W and PO-4 W, AdsimH19 fractures showed a significant radiolucent gap between fractured cortices (Fig. 2D). Quantitative analysis of reconstructed µCT data also revealed dynamic trends (Fig. 2E). In AdGFP control fractures, mineralized callus volumes peaked at PO-2 W and declined by PO-4 W, indicating ongoing external callus remodeling. In contrast, AdsimH19 fractures displayed significant new external callus bone growth following H19 silencing, suggesting a robust periosteal and soft tissue response. However, no significant decrease in callus volume was observed until PO-4 W, indicating a delay in endochondral ossification and bone remodeling in AdsimH19 fractures (Fig. 2D).
Effect of H19 loss on the process of fracture repair. (A) Expression of red fluorescent protein (RFP) derived from adenovirus simH19 in tibial fracture of AdsimH19 mice. Immunohistochemical staining of fracture callus revealed RFP-positive cells (brown) in periosteum, external cartilage callus and internal cartilage callus. (B) Quantitative analysis of H19 in the tissues around the fracture site. Quantitative PCR analysis of the tissues around the fracture site showed that the expression of H19 in the AdsimH19 group was significantly lower than that in the AdGFP group, *p < 0.05. (C) The effect of H19 on the process of fracture repair. X-ray images of fractures showed initial signs of external callus formation in the AdGFP group at the 2nd week post operation (PO-2 W), and cortical space disappeared by bridging internal and external callus at the 4th week post operation (PO-4 W). In the AdsimH19 group, obvious callus was formed and extended outward from the fracture line at week 2 (PO-2 W) and week 3 (PO-3 W), and fracture healing was delayed at week 4 (PO-4 W). (D) Micro-CT (µCT) analysis showed that the callus of fractures in the AdGFP group was almost completely bridged with PO-2 W and completely bridged with PO-4 W. The AdsimH19 group showed a large amount of radiative space between the cortices of the PO-2 W fissure and continued to PO-4 W. (E) Quantitative analysis by micro-CT (µCT).BV, bone tissue volume; BV/TV, bone volume fraction; SMI, structure model index; Tb.N, trabecular number; Tb.Th, trabecular thickness; Conn.Dn, Connectivity Density; *p < 0.05, **p < 0.01, compared with AdGFP group.
Effects of H19 silencing on endochondral ossification and remodeling of fracture repair
At PO-1, 2, and 4 W, histological evaluations of fracture repair were carried out using alcian blue/hematoxylin/orange-g (ABH/OG) staining (Fig. 3A). Both the AdGFP and the AdsimH19 groups’ fracture calluses at PO-1 W showed similarities, with the exterior calluses showing signs of early mesenchymal cell recruitment and cartilage development. The AdGFP group showed more callus production by PO-2 W, with a noteworthy shift from cartilaginous to bony callus formation. By PO-4 W, the AdGFP group showed active new bone remodeling, bony union, and cortical unification through bridging calluses. AdsimH19 group, on the other hand, showed unique fracture repair kinetics. An indication of the delayed cartilage-to-bone replacement at PO-2 W was the comparatively strong external callus production along periosteal surfaces. Undifferentiated mesenchymal tissue was observed in both the intramedullary space and the fracture gap. By PO-4 W, these regions remained partially non-unified in both the periosteal and intramedullary areas, with delayed new bone remodeling, and filled with some fibrous tissue (Fig. 3A).
To further investigate the fracture healing process, quantitative analyses of the tissue composition surrounding the calluses were conducted. Histomorphometric assays revealed considerably higher ratios of cartilage to bone areas during fracture repair in the AdsimH19 group, consistent with histological observations (Fig. 3B). Moreover, it was found that in C3H10 T1/2 cells cultivated using the micromass approach, the absence of H19 did not prevent the development of chondrocytes but rather did not promote chondrogenic differentiation (data not shown).
Tartrate-resistant acid phosphatase (TRAP) staining, used to identify osteoclasts, was also performed to assess whether improper bone remodeling contributed to the delayed union. However, compared to the AdGFP controls, fewer TRAP-positive cells were observed in the AdsimH19 group (Fig. 3C, D). These results suggest that in AdsimH19-injected mice, endochondral ossification and remodeling during fracture repair were significantly delayed, ultimately leading to delayed bone unification.
Silencing H19 delays endochondral osteogenesis and remodeling during fracture repair. (A) Histological evaluation of fracture repair was performed with alcian blue/hematoxylin/Orange-g (ABH/OG) staining. External calluses in PO-1 W, AdsimH19 and AdGFP groups showed obvious signs of early recruitment of mesenchymal cells and formation of cartilage (blue). Cartilage callus (blue) in the AdsimH19 group was more than that in the AdGFP group at PO-2 W, and the AdGFP group showed a transition from cartilage (blue) to bone (red) callus formation. The fractures in the PO-4 W and AdGFP group showed cortical unity through the bridge callus, accompanied by bone healing and active new bone remodeling. Partial fractures of periosteal and intramedullary region were not uniform in AdsimH19 group. (B) quantitative analysis of the tissue composition of fracture callus. In the process of fracture repair, the ratio of cartilage area to bone area in AdsimH19 group was significantly increased compared with AdGFP group, *p < 0.05, **p < 0.01. (C) Tartrate-tolerant acid phosphatase (TRAP) staining was used to detect osteoclasts. The total number of trap positive cells (purplish red) in AdsimH19 group was less than that in AdGFP control group. (D) TRAP staining was used for quantitative analysis of osteoclasts, *p < 0.05, compared with AdGFP group.
The required role of H19 on vascularization during endochondral ossification process
We implanted cartilage fragments subcutaneously in nude mice to analyze the potential impact of H19 on the endochondral ossification process, aiming to understand how these changes affect altered fracture healing. These fragments were transfected with AdGFP, AdBMP2, AdH19, and/or AdsimH19. Fluorescence microscopy confirmed a transfection efficiency of 30–40% before implantation (Figure supplement 1). Significant morphological changes were seen three weeks post-implantation (Fig. 4A). From a visual standpoint, the mass in the AdGFP group exhibited the smallest size, while the masses in the AdGFP and AdBMP2+simH19 groups displayed a more pronounced cartilage phenotype compared to those in the AdBMP2 and AdBMP2+H19 groups. By using microCT analysis, higher bone volume and trabecular mass parameters demonstrated that AdBMP2 promoted endochondral ossification in comparison to AdGFP. AdsimH19, on the other hand, prevented BMP2-induced endochondral ossification, as seen by reduced trabecular mass parameters and bone volume fraction (Fig. 4B). Safranin O staining was used to quantify the ratios of mineral deposition, calcification, and cartilage hypertrophy (Fig. 4C). Throughout the endochondral ossification process, the AdsimH19 group consistently exhibited lower ratios of mineral deposition, calcification, and hypertrophy. These observations can be attributed to the delayed formation of endochondral bone, culminating in a delay in bone remodeling (Fig. 4C). Collectively, these data suggested that silencing H19 in cartilage fragments largely inhibited the endochondral ossification.
Then, the masses sections in the AdBMP2 and AdBMP2+simH19 groups were stained with CD31 immunofluorescence (IF). The results showed that compared with AdBMP2 group, CD31 fluorescence in cartilage area of AdBMP2+simH19 group was significantly reduced and vascularization was abnormal (Fig. 4D). CD31 immunofluorescence was also used to assess whether delayed fracture union in the AdsimH19 group was associated with altered vascularization. At PO-2 W, both AdGFP and AdsimH19 fractures exhibited similar vascularization patterns in the external callus regions, with avascular cartilaginous tissue invading newly formed bone. However, the AdsimH19 group displayed more extensive cartilaginous tissue compared to the AdGFP group, indicating aberrant vascularization in the exterior calluses from both groups (Fig. 4E). Furthermore, regions of all the trabecular bone marrow tissue of the AdGFP group showed high CD31 expression up to PO-4 W (Fig. 4E). Nevertheless, naked vascular infiltration across the fracture gap comprising cartilaginous-like fibrous tissue without indications of bone formation was seen in the AdsimH19 group in addition to high CD31 expression in the trabecular bone tissue. Since the tissues within the fracture gap were scarcely vascularized, it was likely that the fracture delayed-unions seen in the AdsimH19 group might be attributable to a disturbed vascular network that led to postpone endochondral ossification.
Silencing H19 obstructs endochondral osteogenesis leading to delayed bone remodeling. (A) The same proportion of AdBMP2, AdBMP2+simH19, AdGFP, AdBMP2+H19-infected cartilage fragments were subcutaneously implanted into the lateral abdomen of nude mice without thymus (nu/nu) for 3 weeks. In appearance, the tumor size of AdGFP group was the smallest. The masses in AdGFP and AdBMP2+simH19 groups showed a more distinct cartilage phenotype than those in AdBMP2 and AdBMP2+H19 groups. (B) Micro-CT quantitative analysis showed that compared with AdGFP, bone mass and trabecular mass parameters in AdBMP2 group were higher, while bone volume fraction and trabecular mass parameters in AdBMP2+simH19 group were lower than AdBMP2 group. “*” means p < 0.01 as compared with AdBMP2 group; “#” means p < 0.01 as compared with AdBMP2+simH19 group. BV, bone tissue volume; BV/TV, bone volume fraction; SMI, structure model index; Tb.N, trabecular number; Tb.Th, trabecular thickness; Conn.Dn, Connectivity Density. (C) The proportion analysis of cartilage hypertrophy, calcification and mineral deposition determined by saffranine O staining of cartilage mass showed that compared with AdGFP, chondrocytes in AdBMP2 group were significantly hypertrophic; Compared with AdBMP2, hypertrophic chondrocytes decreased in AdBMP2+simH19 group. Compared with AdBMP2+simH19, hypertrophic chondrocytes increased in AdBMP2+H19 group. (D) Immunofluorescence (IF) detection of CD31 in cartilage mass showed that compared with AdBMP2, the expression of CD31 (red) in cartilage ossification area in AdBMP2+simH19 group was significantly reduced. (E) Immunofluorescence (IF) detection of CD31 in mouse fracture sections showed that in the PO-2 W and PO-4 W, the expression of CD31 (red) of AdsimH19 groups was significantly lower than that in the AdGFP group in the cartilage ossification region, and the silence of H19 hindered vascularization, resulting in delayed endochondral osteogenesis.
H19’s regulation of smad7-TGFβ-smad2/3 pathway through sponging MiRNA 21a-5p
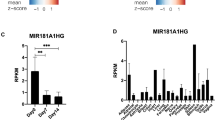
Considering that LncRNA is reported to primarily function as a competition endogenous RNA (ceRNA) and a primary microRNA precursor, our focus shifted to the examination of altered miRNA expression using microarray assays in C3H10T1/2 cells. These cells were transfected with AdBMP2, AdBMP2+AdsimH19, and cultured in micromass conditions for 12 days. Microarray results revealed significant changes in miRNA expression (Fig. 5A, Figure supplement 2B). Eight miRNAs (miR-21a-5p, miR-3963, miR-199a-3p, miR-322-5p, miR-214-3p, miR-27a-3p, miR-22-3p, and miR-15b-5p) were shown to be highly up regulated in the AdBMP2+simH19 group. On the other hand, five miRNAs (miR-5126, miR-210-3p, miR-1224-5p, let-7b-5p, and miR-8110) exhibited a notable down-regulation in the AdBMP2 group (Fig. 5B). MiR-21a-5p, one of the up-regulated miRNAs, showed the greatest increase in the AdBMP2+simH19 group, more than tripling in comparison to the control (Fig. 5B). These results suggest that miR-21a-5p may play a key role in mediating the effect of H19 on the Smad7-TGFβ-Smad2/3 pathway.
RNAHybrid investigations, which support these conclusions, showed that there is an ideal binding free energy between H19 and miR-21a-5p (Figure supplement 2C). Moreover, a significant level of conservation was seen at the H19 binding sites in a few species (Figure supplement 3A,B). In addition, qPCR assays indicated that AdsimH19 could elevate miR-21a-5p expression in mouse chondrocytes in vitro from day 3 to day 12 (Fig. 5C). Considerably, the reporter gene containing the wild-type sequence of H19, but not the mutant, had less luciferase activity in HEK 293 cells when miR-21a-5p was overexpressed (Fig. 5D, E and Figure supplement 2D). After that, it was examined how miR-21a-5p affected the expression of target genes in mouse chondrocytes. MiR-21a-5p may target 63 possible candidate genes in the skeletal system, according to bioinformatic analysis (Figure supplement 2A). Our team and others have previously emphasized the significance of smad7 as a crucial regulator in chondrogenesis and endochondral ossification32. Interestingly, in C3H10T1/2 cells stimulated by BMP2, a noteworthy association was found between the expression of smad7 mRNA and H19 (Fig. 5F). Moreover, smad7 mRNA levels were significantly decreased by transfection with the miR-21a-5p mimic (data not shown). All of these findings point to the ability of miR-21a-5p to bind to smad7′s 3′-UTR and inhibit its production (Figure supplement 4). Our attention turned to comprehending the possible influence of miR-21a-5p on the TGFβ-smad2/3 signaling pathway in mouse chondrocytes stimulated by BMP2, since smad7 functions as an inhibitor of this pathway by preventing the phosphorylation of smad2/3. Notably, endochondral ossification followed by a substantial upsurge in miR-21a-5p expression was caused by the exogenous production of BMP2 (Figs. 5C and 6A). In this setting, there was an increase in smad7 and p-smad2/3 expression (Fig. 6A). Intriguingly, H19 silencing reduced smad7 expression, while antmiR21a-5p treatment unexpectedly led to concurrent increases in both smad7 and p-smad2/3 levels (Fig. 6B). This phenomenon could be explained by two compensatory mechanisms: (1) AntmiR21a-5p alleviated miRNA-21a-5p-mediated suppression of H19 (Figure supplement 5), allowing sustained BMP2-driven p-smad2/3 activation that secondarily upregulated smad7 expression; (2) The neutralization of miRNA-21a-5p simultaneously relieved its direct inhibitory effect on smad7 transcription, resulting in amplified smad7 accumulation. These dual effects may be responsible for the co-elevation of both pathway components, despite their typical antagonistic relationship. Moreover, H19 silencing in the callus environment led to a rise in Smad2/3 phosphorylation and a decrease in Smad7 expression (Fig. 6C, D). These findings suggest that the inhibition of H19 may regulate the suppressive impact of miR-21a-5p to activate the TGFβ-smad2/3 signaling pathway by inhibiting Smad7 in mouse chondrocytes.
LncRNA H19 binds miR-21a-5p to regulate the expression of samd7. (A) BMP2, BMP2 + simH19 induced changes in miRNA expression. C3H10T1/2 cells were infected with AdBMP2 or AdBMP2+AdsimH19 and micro cultured. The changes of miRNA expression in C3H10T1/2 cells were detected by microarray at 12 days after infection, and the results showed significant changes in miRNA expression. (B) microarray results showed that eight miRNAs were significantly up-regulated in the AdBMP2+simH19 group, among which miR-21a-5p was increased threefold compared with the control group. (C) AdsimH19 increased the expression of miR-21a-5p in chondrocytes. qPCR was used to detect the expression of miR-21a-5p in cultured mouse chondrocytes infected with AdBMP2 or AdBMP2+AdsimH19 in vitro. Expression of miR-21a-5p at day 3, day 7 and day12 showed increased in the AdBMP2+AdsimH19 group. D, H19 wild type, miR-21a-5p simulator and H19 mutant double luciferase gene sequence. (E) H19 and miR-21a-5p have binding sites. Luciferase activity of HEK293 cells transfected with NC mimics + H19 wt, mmu-miR-21a-5p + H19 wt, NC mimics + H19 mu and mmu-miR-21a-5p + H19 mu was detected. The results showed that compared with the NC mimics + H19 wt group, the luciferase activity of the mmu-miR-21a-5p + H19 wt group was decreased, and compared with the mmu-miR-21a-5p + H19 wt group, the luciferase activity of the mmu-miR-21a-5p + H19 mu group was increased. “*” means p < 0.05, “**” means p < 0.01. (F) BMP2-induced mRNA expression of H19 and smad7. Stimulated by AdBMP2, the relative expression levels of H19 and smad7 were measured by reverse transcription and quantitative real-time PCR (RT-qPCR) at a series of times (days 1, 2, 3, 5, 7, 9, and 12). AdGFP was used as a control. Values for AdBMP2 / AdGFP are shown (each assay was triplicate and/or performed in at least three separate experiments).
Silencing of H19 resulted in increased miR-21a-5p, decreased smad7 expression, and increased smad2/3 phosphorylation. (A) BMP2 induced increased expression of smad7 protein. Blot of smad7 expression was performed on day 7 after transfection of AdBMP2 into mouse chondrocytes. Using beta-actin as a control, the relative protein expression was analyzed by Image Lab software. Compared with the AdGFP group, “*” indicates p < 0.05. Each assay was triplicate and/or performed in at three separate experiments; Representative results are shown. Full-length/ uncropped gels and blots were included in a Supplementary Information file. (B) Decreased miR-21a-5p increases the phosphorylation of smad2/3. On the 7th day after transfection of AdBMP2+simH19 + miRNC, AdBMP2+simH19+AntmiR-21a-5p, the expression of smad7, p-smad2/3 and samd2/3 were imprinted. Using beta-actin as a control, the relative protein expression was analyzed by Image Lab software. Compared with AdBMP2+simH19 + miRNC group, “*” indicates p < 0.05. Each assay was triplicate and/or performed in at three separate experiments; Representative results are shown. Full-length/ uncropped gels and blots were included in a Supplementary Information file. (C) silencing H19 leads to decreased smad7 expression. The expression of smad7 in the callus transfected with AdGFP and AdsimH19 was detected by immunohistochemistry. Compared with the AdGFP group, the expression of smad7 (brown) in the AdsimH19 group was significantly reduced. (D) silencing H19 promotes smad2/3 phosphorylation. The expression of p-smad2/3 in callus transfected with AdGFP and AdsimH19 was detected by immunofluorescence. Compared with AdGFP group, the expression of p-smad2/3 (red) in AdsimH19 group was significantly increased.
Reversal of delayed endochondral ossification by AntagomiR-21a-5p
Finally, we investigated the impact of antagomiR-21a-5p on the repair of delayed unions induced by silencing H19. The normal production of calluses along the periosteum, extending away from the fracture line at PO-2 W and PO-3 W, was efficiently restored by antagomiR-21a-5p. Radiographs of fractures treated with AntagomiR-21a-5p showed that PO-3 W and 4 W successfully united the fracture by bridging the cortical gap via internal and exterior bony calluses (Fig. 7A). Furthermore, in AntagomiR-21a-5p treated fractures, µCT investigations on mineralized calluses showed near-total bridging of bony calluses by PO-2 W, followed by complete bridging at PO-4 W. However, similar to the delayed-union seen in the simH19 group, fractures treated with AntagomiR-NC still showed a significant radiolucent area between fractured cortices at PO-4 W (Fig. 7B). In antagomiR-21a-5p treated fractures, quantitative µCT data showed ongoing external callus remodeling until PO-4 W, reversing the delayed bone remodeling (Supplemental Table 1). Histological evaluations corroborated these results by showing that AntagomiR-21a-5p could successfully stop the delayed osseous restoration of cartilage in AdsimH19 fracture repairs at PO-2 W. Furthermore, the fibrous tissue in the surrounding intramedullary area and fracture gap in AdsimH19 fractures vanished after receiving treatment with AntagomiR-21a-5p. The fractures were fully bridged by PO-4 W, demonstrating ongoing bone remodeling (Fig. 7C and Supplemental Table 2). These persuasive results imply that the insertion of AntagomiR-21a-5p can significantly reverse the delayed endochondral ossification and remodeling seen throughout the fracture repair process caused by AdsimH19 (Figure supplement 6).
Effect of miR-21a-5p inhibitors on silencing H19-induced delayed fracture healing. (A) The effect of Ant-miR-21a-5p on the process of silent-H19-induced fracture repair, fracture radiographs showed that obvious callus was formed in the AdsimH19 + miRNC group at week 2 (PO-2 W) and week 3 (PO-3 W) after surgery, and fracture healing was delayed at week 4 (PO-4 W). AdsimH19 + Ant miR-21a-5p group restored the normal formation of periosteum along the periosteum and extended outward from the fracture line at PO-2 W and PO-3 W. The cortical space disappeared through internal and external callus bridging at the 4th week after surgery (PO-4 W), and the fracture was successfully unified. (B) micro-CT (µCT) analysis showed that the AdsimH19 + miRNC group showed a large amount of radiation-permeable space between the crack cortex of PO-2 W and continued to PO-4 W, and the callus of the fracture of AdsimH19 + Ant-miR-21a-5p group was almost completely bridged with PO-4 W. (C) Histological evaluation of fracture repair was performed by alcian blue/hematoxylin/Orange g (ABH/OG) staining. At PO-1 W, the external calluses of AdsimH19 + miRNC group and AdsimH19 + Ant miR-21a-5p group showed signs of early recruitment of mesenchymal cells and formation of cartilage (blue). At PO-2 W, Cartilage callus(blue) in AdsimH19 + miRNC group was more than that in AdsimH19 + Ant-miR-21a-5p group, and the transition from cartilage (blue) to bone (red) callus formation in AdsimH19 + Ant-miR-21a-5p group was observed. At PO-4 W, partial fractures in periosteum and intramedullary region in AdsimH19 + miRNC group were not uniform, and fractures in AdsimH19 + Ant miR-21a-5p group showed cortical unity through the bridge callus, accompanied by bone healing and active new bone remodeling.
Discussion
The emerging role of long non-coding RNA (lncRNA) H19 as a crucial regulator in both skeletal development and diseases has garnered significant attention7,8,14,16,33. The possible role of H19 in fracture recovery has drawn attention since the process bears similarities to the complex spatiotemporal cell events and signaling controls observed during skeletal development1,9,10,13. While earlier research has suggested that H19 is important for osteogenesis and skeletal growth, its precise function in fracture repair at lesion sites is still unknown1,7,8,9,13. Our study presents novel genetic evidence demonstrating that inhibiting H19 signaling at fracture sites disrupts vascularization, delays endochondral ossification, and postpones bone union. Interestingly, H19 inhibition does not affect cartilaginous callus development. Through comprehensive approaches, including semi-stabilized fractures, endochondral ossification models with cartilage fragments in vivo, and loss-of-function gene targeting, we underscore the critical requirement of H19 signaling for proper vascularization and endochondral ossification during fracture repair. Notably, this is unrelated to its function in the development of cartilaginous calluses. Overall, our data demonstrates that H19 interacts with miR21a-5p and the smad7-TGFβ-psmad2/3 signaling pathway to regulate vascularization and endochondral ossification. These findings imply that H19 targeting may be a useful treatment approach to improve bone fracture repair in clinical situations. The interesting finding that faulty H19 can promote the production of cartilaginous callus while also causing improper or delayed endochondral ossification and vascularization, which can lead to fracture delayed unions, is particularly remarkable. Moreover, the fact that antagomiRNA21a-5p may restore defective endochondral ossification in the absence of H19 suggests that the H19-miRNA21a-5p axis may be essential for the processes involved in bone healing.
The intricate procedures that control endochondral ossification and remodeling are likewise similar to those needed for the formation of the embryonic skeleton1,2. Many genes essential for the embryonic development of long bones (through endochondral ossification) are also expressed in the fracture callus, with both gene expression patterns and tissue histology resembling those in the growth plate12,34. H19 expression is noteworthy for showing both geographical and temporal trends. H19 expression in growth plate cartilage and metaphyseal bone was previously discovered to decrease with age by Anenisia et al.19. They also observed higher levels of H19 expression in chondrocytes of resting and proliferative zones as opposed to the hypertrophic zone and metaphyseal bone. On the contrary hand, our FISH investigation shows increased H19 expression in hypertrophic chondrocytes and the spongiosa of growing endochondral bones. It is remarkable that cortical bone and the surrounding skeletal muscle have similar levels of H19 expression. These results suggested that H19 might have specific functions related to the creation of long bones (endochondral ossification).
Indeed, chondrogenesis and endochondral ossification play a critical role in the repair of bone fractures1,2. It is commonly known that defects in the production of cartilage calluses can seriously impair bone healing35,36. Nevertheless, there isn’t much study available right now on H19’s function in chondrogenic differentiation7,8,15. According to Eric et al., H19 expression rose in chondrocytes exposed to hypoxia coupled with collagen type 2 and miR-675, but it declined in response to IL-1β and TNFα-induced inflammation20. In a similar vein, Dudek et al. showed that miR-675 might alter the expression of collagen type 2 in human articular chondrocytes37. Their research showed that collagen type 2 expression was inhibited when H19 and/or miR-675 expression was suppressed. Interestingly, our investigation found that faulty H19 at fracture sites greatly increased the ratios of cartilage areas in the external callus, which runs counter to these findings. Furthermore, C3H10 T1/2 cells’ chondrogenic development in micromass culture seems to be improved by blocking H19. This surprising disparity thus calls for more research into the precise pathways through which H19 affects chondrogenic differentiation.
Research on H19’s function in boosting osteogenic differentiation is still underway, and it appears that the underlying mechanisms are context-dependent and complex38. Increased expression patterns of H19 have been documented during the osteogenic differentiation process of human mesenchymal stem cells (hMSCs)39. There has been discussion on the specific processes by which H19 stimulates osteogenic differentiation. Two mature miRNAs, miR-675-5p and miR-675-3p, are produced from the main transcript of miR-675, which is encoded by H1940. H19 and miR-675 have both been reported to be elevated during MSC osteogenic differentiation in certain investigations41,42. MiR-675 has been demonstrated to block Smad3 phosphorylation, downregulate TGF-β1, and decrease HDAC4/5, which results in a reduction in HDAC recruitment to the promoter of the osteogenic transcription factor Runx242. However, conflicting findings have also been reported. Some studies have shown that miR-675-5p can suppress osteogenic differentiation and negatively regulate H19 through direct binding in a feedback loop mechanism39. Furthermore, it has been proposed that H19 functions as a sponge and a ceRNA for miR-141 and miR-22, which both target the Wnt/β-catenin pathway and are negative regulators of osteogenic differentiation39. It’s interesting to note that different studies have different roles for H19 in reaction to different stimuli, such as BMP2, a strong endochondral ossification factor32,43. Other studies have revealed that the expression of H19 in BMP9-induced osteogenic differentiation exhibits a bidirectional homeostasis with early elevation and later reduction33,44, despite the fact that no discernible change in miR-675 expression was detected during BMP2-induced osteogenic differentiation in C2C12 cells. Moreover, miRNAs that target the notch-signaling pathway may be modulated in relation to H19’s effect on osteogenic differentiation44.In order to fully comprehend how H19 affects osteogenic development, it is crucial to investigate putative miRNA binding sites and related pathways, given the intricacy and context-dependent character of H19’s regulatory activities. To reconcile these conflicting results and determine the precise processes behind H19’s role in either promoting or inhibiting osteogenic differentiation, more research is required.
Without a doubt, vascularization is essential to the healing of bone fractures properly1,2. The creation of new blood vessels during the angiogenesis process is crucial for bringing nutrients and oxygen to the healing site21. In addition to supplying oxygen, vascularization aids in the inflow of osteoblasts and stimulates signaling that promotes cell proliferation and differentiation, both of which are essential for effective bone repair1,2. Moreover, endochondral ossification—the process by which bone replaces cartilage during fracture healing—requires the growth of new blood vessels. The migration and multiplication of mesenchymal stem cells (MSCs) can be adversely affected by hypoxic conditions, or low oxygen levels, however the function of hypoxia in fracture healing is complicated45. The healing process can be positively impacted by hypoxia-inducible factor (HIF) activity, which is induced by low oxygen levels34. Transcription factors known as HIFs are essential for controlling how cells react to variations in oxygen availability. HIFs have the ability to promote angiogenesis and other tissue-repairing processes in the context of fracture healing34. At the same time, HIF-1α signal transduction can combine angiogenesis with bone formation. However, our study only confirmed that silencing H19 can inhibit angiogenic differentiation, and its specific mechanism can be further studied in the future.
In summary, elucidating the role of H19 in fracture repair is essential for understanding the mechanisms underlying fracture healing. Although several studies have been conducted, the details of how H19 regulates the fracture repair process remain unclear. In this study, we established that H19 plays a crucial role in regulating angiogenesis and endochondral osteogenic differentiation during fracture repair. Furthermore, we identified that the endochondral osteogenic differentiation and bone remodeling processes in fracture healing are mediated by the miR-21a-5p-Smad7-TGFβ-Smad2/3 axis. Our findings provide a novel perspective on the mechanisms of fracture healing and lay the groundwork for developing drugs targeting delayed or non-union fractures. However, further research is required to delineate the specific pathways through which H19 influences chondrogenic differentiation and to clarify how H19 inhibits angiogenic differentiation.
Methods and materials
Ethical approval and consent to participate
The present study received approval from the Animal Ethics Committees of Chongqing Medical University (Project: The role and molecular mechanism of LncRNA H19/miRNA-21a-5p signaling pathway in BMP2 regulation of acetabular endochondral ossification in developmental hip dysplasia, Ethical batch number: IACUC-CQMU-2024-0449, Date: 2024.05.27), adhering to the principles of animal protection and in compliance with the pertinent regulations of national animal welfare and ethics. All surgical procedures were carried out under the influence of sodium pentobarbital anesthesia, and diligent measures were taken to mitigate the discomfort experienced by the animals enrolled in the investigation. The euthanasia method used in this study was to select high concentrations of carbon dioxide and suffocate the animals to death. We confirmed the study is reported in accordance with ARRIVE guidelines.
Chemicals and cell culture
The American Type Culture Collection (ATCC, Manassas, VA, USA) provided the mouse bone marrow mesenchymal stem cell C3H10T1/2 and human embryonic kidney (HEK) 293 cell lines. Complete Dulbecco’s modified Eagle’s medium (SH30023.01, DMEM, Hyclone, China) was used to cultivate these cell lines. It was supplemented with 10% fetal bovine serum (10099141, FBS, Gibco, Australia), 100 U/ml penicillin, and 100 mg/ml streptomycin. The culture was kept at 37 °C in a humidified, 5% carbon dioxide (CO2) environment. All chemical reagents, unless otherwise noted, came from Corning or Sigma-Aldrich.
Isolation and culture of cartilage fragments
Following ethical guidelines, the six 10-week-old C57BL/6 mice (n = 3 males and n = 3 females, all the mice were purchased from GemPharmatech Co., Ltd) were euthanized and sterilized using 70% ethanol. Subsequently, the designated joint (such as knee or hip) was meticulously dissected to expose the cartilage, employing sterile forceps and scissors. The dissected tissue was collected in a container containing Hank’s Balanced Salt Solution (HBSS) or Phosphate Buffered Saline (PBS) to ensure tissue hydration. Carefully remove the intact cartilage from the joint and cut the cartilage into small pieces with sterile scissors. After that, sterilized scissors were used to carefully cut the cartilage into tiny pieces. These pieces of cartilage were then cultivated in α-minimum essential medium (12571063, αMEM, Gibco, Australia) with 10% fetal bovine serum (10099141 C, FBS, Gibco, Australia) added, along with 100 U/ml of penicillin and 100 mg/ml of streptomycin. The culture was kept in a humidified atmosphere with 5% carbon dioxide (CO2) at 37 °C.
Construction and generation of recombinant adenoviral vectors (AdBMP2, adgfp, AdH19, and AdsimH19)
AdEasy technology, a previously disclosed approach, was used to create recombinant adenoviruses46. AdGFP, used as a control mock virus, and AdBMP2, described in previous research16,47,48, were essential components of the investigation. In short, PCR amplification and subcloning into an adenoviral shuttle vector were performed on the full-length transcript of mouse lncRNA H19 and the coding area of human BMP2. Then, recombinant adenoviral vectors were created using these designs. Subsequent development includes creating recombinant adenoviruses with either lncRNA H19 or BMP2 in HEK-293 cells. For AdsimH19, a special strategy was used. Using the Gibson Assembly technique as outlined49, three siRNAs that target mouse lncRNA H19 were assembled to an adenoviral shuttle vector at the same time.
Adenovirus and AntagomiRNA transfection
In vitro, chondrogenic differentiation and endochondral ossification were induced in C3H10 T1/2 cells using a micromass culture approach, which closely resembles the condensation of mesenchymal stem cells (MSCs) as described in earlier research. After being seeded at 60% confluence, C3H10T1/2 cells were infected with AdGFP, AdBMP2, or AdsimH19. The cells were extracted and suspended at a high density (about 105 cells per 25ul of media) following a 24-hour incubation period following infection. These cell suspensions were then incubated in a CO2 incubator after being positioned in the middle of each well in 12-well plates. Two to three milliliters of full DMEM were applied to each well following a one-hour incubation period. Every three days after that, half of the medium in each well was changed.
Regarding cartilage fragments in ex vivo: AdGFP, AdBMP2, or AdsimH19 were subsequently infected onto 50% densely seeded cartilage fragments. Before infection, 10 ug/ml of these adenoviruses were introduced to a polybrene solution. Fluorescence microscopy verified an infection effectiveness of roughly 30–40% following a 24-hour incubation period. After that, the cartilage pieces were gathered and injected subcutaneously into the athymic nude mice’s flanks (all the mice were purchased from GemPharmatech Co., Ltd).
For in vivo cells in fracture sites: AdGFP, AdsimH19, or AntagomiRNA 21a-5p were pre-incubated with Lipofectamine-3000(1.5 µl per well) for 15 min after tibia fracture surgery, and then they were injected into the surrounding muscle, periosteum, and bone. The tibia was removed for additional examination five days after the procedure. Red fluorescent protein (RFP)-targeting immunohistochemistry assay and qPCR assay to measure H19 and miRNA21a-5p expression levels were used to confirm transfection.
Fluorescence in situ hybridization (FISH)
On the first postnatal day, fetal mouse limbs were taken (the mouse was purchased from GemPharmatech Co., Ltd), preserved for the night in 4% paraformaldehyde treated with diethylpyrocarbonate (G3004, DEPC, Servicebio, Wuhan, China), and then embedded in paraffin. After obtaining 5 μm-thick serial sections, they were deparaffinized using xylene and rehydrated using graded ethanol and RNase-free deionized Millipore water (A57775, Invitrogen). The hybridization process was carried out as previously described50. Tissue sections were boiled for 20 min at 95–100 °C in citrate buffer (pH6.0) before being blocked with regular goat serum before hybridization. A primary antibody specific to collagen 1α1 (ab270993, Abcam, Cambridge, US) at a dilution of 1:200 was subsequently applied to these sections and incubated for the entire night at 4 °C. After that, a second antibody tagged with GFP was administered for 30 min. Sections were then pretreated for 15 min using the boiled target retrieval buffer from the RNAscope kit (320700, Invitrogen). After that, a hybridization process was conducted in a hybridization oven for three hours at 50 °C. This involved combining the hybridization buffer included in the kit with Cy3-tagged mouse lncRNA H19 probes that were made by Ribobio (Guangzhou, China). In accordance with the normal methodology, a sequence of successive incubations and washes came next. Lastly, an antifade mounting medium containing 4′, 6-diamidino-2-phenylindole (H-1200-10, DAPI, Vector Lab, Inc. Burlingame, CA) was used to mount the plates. A fluorescence microscope (Olympus, USA) was used to take microscopy pictures of the sections. At 561, 475, and 405 nm, respectively, Cy3 (lncRNA H19), GFP (collagen 1α1), and DAPI (nuclei) were activated. The lncRNA H19 probe has the sequence 5′-Cy3/cagttgccctcagacggagatggacg/Cy3-3′.
RNA isolation and RT-qPCR
Total RNA extraction was performed using TRIZOL regent (15596026CN, Invitrogen, CA, USA) according to the manufacturer’s instructions, subjected to reverse transcription reactions for mRNA using PrimeScript RT reagent kit (RR047A, Takara, Dalian, China) and for miRNA using reagent kit (RR047A, Takara, Dalian, China). Quantitative PCR analysis was carried out utilizing the CFX96 Real time PCR Detection System (Bio-Rad, California, USA), along with the SYBR premix Ex Taq II kit (RR820A, Takara, Dalian, China), following the manufacturer’s instructions. Programs for real-time PCR are as follows: an initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 60 °C for 5 s and extension at 72 °C for 45 s. GAPDH and U6 were used as a reference gene. The primer sequences for PCR are listed in Table 1.
Western blot analysis
Protein extraction was carried out using a 2% SDS lysis buffer containing 100mM Tris-HCL and 100mM β-Mercaptoethanol. The total protein content was denatured by boiling and quantified using a Bicinchoninic Acid (BCA) Protein Assay kit (P0012, Beyotime, Beijing, China). Equivalent amounts of protein were separated via electrophoresis on Bis-Tris gels containing 5% concentrate glue and 10% separation glue (WG1403BOX, Life Technologies, MA, USA), and subsequently transferred onto polyvinylidene fluoride membranes (C3117, PVDF, Millipore, MA, USA). Afterward, the membranes were blocked with 5% skimmed milk for 1 h at room temperature and then incubated with primary antibodies targeting samd7 (sc-365846, Santa Cruz Biotechnology Inc., Texas, USA), Smad2/3 (3102 S, Cell signaling Technology, MA, USA), and phosphor-Smad2/3 (8828 S, Cell signaling Technology, MA, USA) at a 1:1000dilution overnight. Subsequent to this primary antibody incubation, the membranes were incubated with the corresponding secondary antibody conjugated with horseradish peroxidase (HRP). Detection of protein bands on the blots was achieved using Immobilon Western Chemiluminescent HRP Substrate (WBKLS0500, Millipore, MA, USA).
Dual-luciferase reporter gene assay
The full sequence of H19 was obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/gene). Subsequently, The sequence containing 250 base pairs upstream and downstream of the predicted site was cloned into the downstream polyclonal site of the pmirGLO Luciferase gene vector (Promega, WI, USA) to construct the H19-wild type (WT) vector. Employing site-directed mutagenesis via PCR, the H19-mutant (Mut) vector was generated. For the assay, the renilla luciferase expression vector pRLTK (Takara Biotechnology Ltd., Dalian, China) was utilized as an internal control. The targeting relationship between miR-21a-5p and H19 was verified through a dual-luciferase reporter gene assay. The miRNA 21a-5p mimics or a negative control (NC) were co-transfected with the respective luciferase reporter vectors (H19-WT or Mut) into HEK-293T cells. Subsequently, the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was employed to measure the dual luciferase activity. The experiment was repeated three times.
Subcutaneous implantation of cartilage fragments
The Institutional Animal Care and Use Committee approved the study’s use of animals and their care. Every experimental protocol was conducted strictly in accordance with the approved criteria. Ex vivo culturing of cartilage fragments was used to simulate the previously documented endochondral ossification in order to induce it. In summary, AdGFP, AdBMP2, or AdsimH19-infected cartilage fragments were gathered and suspended in DMEM at a density of roughly 2 × 105 cells/ul (100ul per injection). Following that, these pieces were injected subcutaneously into the flanks of three female athymic nude mice (n = 3/group) that were five to six weeks old (all the mice were purchased from GemPharmatech Co., Ltd). The animals were mercifully put down when they reached the designated time points (subcutaneously implanted cartilage fragments were harvested at 3 W). For additional analysis, the ectopic lumps at the injection sites were removed. This entailed micro-CT scanning after the masses were exposed to X-ray imaging using an automatic exposure system set to 45 kv and 500 mA. After these imaging techniques, the masses were fixed for 24 h at room temperature in 4% paraformaldehyde (P0099, Beyotime, Beijing, China). After that, they were decalcified in 0.5 M ethylene diamine tetraacetic acid (EDTA) for 14 days at 4 °C before being embedded in paraffin. After obtaining 5 μm-thick serial sections, histological examinations and other specialized staining evaluations were conducted.
Semi-stabilized fracture model establishment and grouping
Fracture-healing studies were performed using 8–10 weeks old mice (Species: C57, all the mice were purchased from GemPharmatech Co., Ltd), employing a semi-stabilized tibia fracture model as previously described35. Briefly, mice under anesthesia through intraperitoneal administration of sodium pentobarbital were prepared for surgery. The procedure involved shaving the fur around the right knee region and disinfecting the skin with Betadine. Then, A midline anterior knee incision was made to displace the patella, and access to the intramedullary canal was achieved by creating an opening at the midpoint of the tibia plateau. A guidewire was inserted into the tibia to traverse the intramedullary needle channel, subsequently being removed. Following this, a 32G-gauge needle (0.23 mm in diameter) was carefully inserted into the intramedullary canal of the right tibia to provide stabilization. To induce fracture, a bone scissor was applied perpendicular to the bone’s long axis at the metaphysis of the pinned tibia. The surgical wound was sutured and closed, and the animals were administered Buprenex to alleviate any postoperative discomfort. Mice were humanely euthanized at 1 W, 2 W, and 4 W post-fracture (n = 5–7/group/timepoint). X-rays were obtained on day 0, week 1, week 2, week 3, and week 4 after surgery to confirm fracture occurrence and fracture healing using the KUBTEC NC X-ray system, which had an automatic exposure setting of 45 KV, 500 mA.
Micro-computed tomography
At 1 W, 2 W, 4 W after the fracture model (5–7 mice per group per time point), we obtained isolated tibia specimens and fixed them with 4% paraformaldehyde for 24 h. Then we scanned the fracture site along the transverse tibia surface using a micro-computed tomography (µCT) system (VivaCT 40; Scanco Medical AG, Bruker, Germany), with a voltage of 70 kV, a current of 114 uA and a voxel size of 10.5 μm. µCT V.6.1 Software was used to construct and analyze parameters. This technique was supported by the Laboratory of Biomechanics and Oral Tissue Engineering of the affiliated Oral hospital of Chongqing Medical University.
Histologic evaluation: hematoxylin and Eosin (H&E), Alcian blue and hematoxylin/orange (ABH/O), Safranin O staining and TRAP
Sections were deparaffinized with xylene and rehydrated using graded ethanol. H&E, ABH/O and safranin O /fast green staining was performed using standard protocol as previously described5,6. Briefly, the deparaffinized samples were subjected to antigen retrieval and fixation. The sections were stained with hematoxylin and eosin (H&E), Alcian blue/Orange g and Safranin O/fast green staining. Histological evaluation was performed using a light microscope (Olympus, Tokyo, Japan).
Immunohistochemistry assay
In general, tissue sections underwent deparaffinization with using xylene, followed by rehydration through a series of graded ethanol solutions. A treatment with 3% hydrogen peroxide (H2O2) for 10 minutes was performed to suppress endogenous peroxidase activity. The sections were then subjected to boiling in citrate buffer (pH 6.0) at 95–100 °C for 20 minutes. Following this, they were blocked using normal goat serum. Subsequently, the sections were incubated with primary antibodies targeting Smad7 (sc-365846, Santa Cruz Biotechnology Inc., Texas, USA; 1:300 dilution), CD31 (ab9498, Abcam, Cambridge, US; 1ug/ml, 1:200 dilution), and phosphor-Smad2/3 (8828S, Cell Signaling Technology, MA, USA; 1:200 dilution) overnight at 4 °C. After washing, sections intended for smad7 detection were incubated with biotin-labeled secondary antibody for 30 minutes, followed by incubation with streptavidin-HRP conjugate for 20 minutes at room temperature. Sections intended for phosphor-Smad2/3 detection were incubated with Cy3-labeled secondary antibody and mounted using antifade mounting media containing 4’, 6-diamidino-2-phenylindole (H-1200-10, DAPI, Vector Lab, Inc., Burlingame, CA). Microscopic images of the sections were captured using a fluorescence microscope (Olympus, Tokyo, Japan). Cy3 (CD31) and DAPI (nuclei) were excited at wavelengths of 561 nm and 405 nm, respectively.
Microarray assay
Total RNAs were isolated from differentiating C3H10 T1/2 mesenchymal stem cells that were subjected to transfection using RNeasy Mini kit with DNase treatment (74104, Qiagen). Subsequently, the hybridization of the mouse miRNA (8*60K) V21.0 array (Agilent) was conducted. This array encompasses a total of 1900 gene transcripts. The procedure followed the instructions provided by the manufacturer, and signal acquisition was performed using Agilent Feature Extraction (AFE) software version 10.7.1.1.
Bioinformatics prediction
As described previously, the binding sites of miRNA21a-5p on H19 were predicted using a web-based program RNAhybrid (The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs, Molecular Cell (2013), https://doi.org/10.1016/j.molcel.2013.08.027). To examine the evolutionary conservation of the binding sites in human H19 lncRNA, we used conversation scores for each nucleotide of H19 lncRNA. For the NCBI human H19 lncRNA sequence (NR_002196, build 37.3), the nucleotide-specific conservation score files were downloaded from UCSC genome browser (http://genome.ucsc.edu/). The binding sites of miRNA21a-5p on Smad7 were predicted using a website of TargetScan (https://www.targetscan.org/mmu_72/) and Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis
All images were acquired using an Olympus microscope (Japan) and subsequently analyzed using Image J Pro software. The presented data are expressed as the mean ± standard deviation (SD) and were subjected to analysis utilizing SPSS software (version 21, IBM, USA). To assess inter- and intragroup variations, a one-way analysis of variance (ANOVA) was conducted. Additionally, a t-test was employed to compare differences between any two specific groups. Spearman’s correlation coefficients were computed to evaluate the associations between lncRNA H19 expression and smad7 mRNA levels. A significance level of P < 0.05 was utilized to determine statistical significance.
Data availability
The datasets analyzed during the study are available in the NCBI database (http://www.ncbi.nlm.nih.gov/gene) (NR_002196, build 37.3). Uncropped and unedited blot images have been included in the supplementary file (original western blot for H19 in bone fracture). All other data is provided within the manuscript or supplementary information files.
References
Saul, D. & Khosla, S. Fracture healing in the setting of endocrine diseases, aging, and cellular senescence. Endocr. Rev. 43, 984–1002 (2022).
Oranger, A. et al. Irisin modulates inflammatory, angiogenic, and osteogenic factors during fracture healing. Int. J. Mol. Sci. 24, 1809 (2023).
Razmara, E. et al. non-coding RNAs in cartilage development: an updated review. Int. J. Mol. Sci. 20, 4475 (2019).
Okuyan, H. M. & Begen, M. A. LncRNAs in osteoarthritis. Clin. Chim. Acta. 532, 145–163 (2022).
Nojima, T. & Proudfoot, N. J. Mechanisms of LncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell. Biol. 23, 389–406 (2022).
Ferrer, J. & Dimitrova, N. Transcription regulation by long non-coding rnas: mechanisms and disease relevance. Nat. Rev. Mol. Cell. Biol. 25, 396–415 (2024).
Zhou, Z., Hossain, M. S. & Liu, D. Involvement of the long noncoding RNA H19 in osteogenic differentiation and bone regeneration. Stem Cell. Res. Ther. 12, 74 (2021).
Wang, L. & Qi, L. The role and mechanism of long non-coding RNA H19 in stem cell osteogenic differentiation. Mol. Med. 27, 86 (2021).
Einhorn, T. A. & Gerstenfeld, L. C. Fracture healing: mechanisms and interventions. Nat. Rev. Rheumatol. 11, 45–54 (2015).
Ramasamy, S. K., Kusumbe, A. P., Wang, L. & Adams, R. H. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature 507, 376–380 (2014).
Long, F. & Ornitz, D. M. Development of the endochondral skeleton. Cold Spring Harb Perspect. Biol. 5, a008334 (2013).
He, J. et al. Dissecting human embryonic skeletal stem cell ontogeny by single-cell transcriptomic and functional analyses. Cell. Res. 31, 742–757 (2021).
Hadjiargyrou, M. & O’Keefe, R. J. The convergence of fracture repair and stem cells: interplay of genes, aging, environmental factors and disease. J. Bone Min. Res. 29, 2307–2322 (2014).
Liao, J. et al. Long noncoding RNA (lncRNA) H19: an essential developmental regulator with expanding roles in cancer, stem cell differentiation, and metabolic diseases. Genes Dis. 10, 1351–1366 (2023).
Zhou, Q. P., Zhang, F., Zhang, J. & Ma, D. H19 promotes the proliferation of osteocytes by inhibiting p53 during fracture healing. Eur. Rev. Med. Pharmacol. Sci. 22, 2226–2232 (2018).
Dai, G. et al. LncRNA H19 regulates BMP2-induced hypertrophic differentiation of mesenchymal stem cells by promoting Runx2 phosphorylation. Front. Cell. Dev. Biol. 8, 580 (2020).
Stefańska, K. et al. Expression profile of new marker genes involved in differentiation of human wharton’s jelly-derived mesenchymal stem cells into chondrocytes, osteoblasts, adipocytes and neural-like cells. Int. J. Mol. Sci. 24, 12939 (2023).
Yan, L., Liu, G. & Wu, X. Exosomes derived from umbilical cord mesenchymal stem cells in mechanical environment show improved osteochondral activity via upregulation of LncRNA H19. J. Orthop. Translat. 26, 111–120 (2021).
Andrade, A. C., Lui, J. C. & Nilsson, O. Temporal and Spatial expression of a growth-regulated network of imprinted genes in growth plate. Pediatr. Nephrol. 25, 617–623 (2010).
Steck, E. et al. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J. Mol. Med. (Berl). 90, 1185–1195 (2012).
Menger, M. M., Laschke, M. W., Nussler, A. K., Menger, M. D. & Histing, T. The vascularization paradox of non-union formation. Angiogenesis 25, 279–290 (2022).
Peng, Y., Wu, S., Li, Y. & Crane, J. L. Type H blood vessels in bone modeling and remodeling. Theranostics 10, 426–436 (2020).
Kusumbe, A. P., Ramasamy, S. K. & Adams, R. H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature 507, 323–328 (2014).
Tuckermann, J. & Adams, R. H. The endothelium-bone axis in development, homeostasis and bone and joint disease. Nat. Rev. Rheumatol. 17, 608–620 (2021).
Zhang, W. et al. Hypoxia inducible factor-1α related mechanism and TCM intervention in process of early fracture healing. Chin. Herb. Med. 16, 56–69 (2024).
Guo, Q. et al. Salidroside improves angiogenesis-osteogenesis coupling by regulating the HIF-1α/VEGF signalling pathway in the bone environment. Eur. J. Pharmacol. 884, 173394 (2020).
Towler, D. A. Vascular biology and bone formation: hints from HIF. J. Clin. Invest. 117, 1477–1480 (2007).
Bai, Y. et al. Long non-coding RNA HCAR promotes endochondral bone repair by upregulating VEGF and MMP13 in hypertrophic chondrocyte through sponging miR-15b-5p. Genes Dis. 9, 456–465 (2022).
Behera, J., Kumar, A., Voor, M. J. & Tyagi, N. Exosomal lncRNA-H19 promotes osteogenesis and angiogenesis through mediating Angpt1/Tie2-NO signaling in CBS-heterozygous mice. Theranostics 11, 7715–7734 (2021).
Fang, H. et al. Long noncoding RNA H19 overexpression protects against hypoxic-ischemic brain damage by inhibiting miR-107 and up-regulating vascular endothelial growth factor. Am. J. Pathol. 191, 503–514 (2021).
Hou, J. et al. Long noncoding RNA H19 upregulates vascular endothelial growth factor A to enhance mesenchymal stem cells survival and angiogenic capacity by inhibiting miR-199a-5p. Stem Cell. Res. Ther. 9, 109 (2018).
Zhao, C. et al. Sox9 augments BMP2-induced chondrogenic differentiation by downregulating Smad7 in mesenchymal stem cells (MSCs). Genes Dis. 4, 229–239 (2017).
Du, C. et al. LncRNA H19 mediates BMP9-induced angiogenesis in mesenchymal stem cells by promoting the p53-Notch1 angiogenic signaling axis. Genes Dis. 10, 1040–1054 (2023).
Jiang, W., Caruana, D. L., Back, J. & Lee, F. Y. Unique Spatial transcriptomic profiling of the murine femoral fracture callus: a preliminary report. Cells 13, 522 (2024).
Mi, M. et al. Chondrocyte BMP2 signaling plays an essential role in bone fracture healing. Gene 512, 211–218 (2013).
Wang, F. et al. Loss of Bcl-3 delays bone fracture healing through activating NF-κB signaling in mesenchymal stem cells. J. Orthop. Translat. 35, 72–80 (2022).
Martinez-Sanchez, A., Dudek, K. A. & Murphy, C. L. Regulation of human chondrocyte function through direct Inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145). J. Biol. Chem. 287, 916–924 (2012).
Moura, S. R. et al. Long non-coding RNA H19 regulates matrisome signature and impacts cell behavior on MSC-engineered extracellular matrices. Stem Cell. Res. Ther. 14, 37 (2023).
Liang, W. C. et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci. Rep. 6, 20121 (2016).
Dey, B. K., Pfeifer, K. & Dutta, A. The H19 long noncoding RNA gives rise to MicroRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 28, 491–501 (2014).
Ma, X. et al. Human amnion-derived mesenchymal stem cells promote osteogenic differentiation of human bone marrow mesenchymal stem cells via H19/miR-675/APC axis. Aging (Albany NY). 12, 10527–10543 (2020).
Huang, Y., Zheng, Y., Jia, L. & Li, W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells. 33, 3481–3492 (2015).
Zhou, N. et al. BMP2 induces chondrogenic differentiation, osteogenic differentiation and endochondral ossification in stem cells. Cell. Tissue Res. 366, 101–111 (2016).
Liao, J. et al. LncRNA H19 mediates BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) through Notch signaling. Oncotarget 8, 53581–53601 (2017).
Liu, J. et al. Mesenchymal stem cells and their microenvironment. Stem Cell. Res. Ther. 13, 429 (2022).
Liao, J., Xiao, H., Dai, G., He, T. & Huang, W. Recombinant adenovirus (AdEasy system) mediated exogenous expression of long non-coding RNA H19 (lncRNA H19) biphasic regulating osteogenic differentiation of mesenchymal stem cells (MSCs). Am. J. Transl Res. 12, 1700–1713 (2020).
Zeng, Y. et al. Sox9-increased miR-322-5p facilitates BMP2-induced chondrogenic differentiation by targeting Smad7 in mesenchymal stem cells. Stem Cells Int. 9778207 (2021). (2021).
Zhou, N. et al. HIF-1α as a regulator of BMP2-induced chondrogenic differentiation, osteogenic differentiation, and endochondral ossification in stem cells. Cell. Physiol. Biochem. 36, 44–60 (2015).
Deng, F. et al. A simplified and versatile system for the simultaneous expression of multiple SiRNAs in mammalian cells using Gibson DNA assembly. PLoS One. 9, e113064 (2014).
Li, D. Y. et al. H19 induces abdominal aortic aneurysm development and progression. Circulation 138, 1551–1568 (2018).
Acknowledgements
We extend our sincere gratitude to Professor Tong-chuan He and the Molecular Oncology Laboratory at the University of Chicago Medical Center for generously providing AdBMP2, AdH19, AdsimH19, and AdGFP. The authors declare that they have not use AI-generated work in this manuscript.
Funding
This research was made possible through the support of the National Natural Science Foundation of China (NSFC) under grants (#81972069 and #82102519). Additionally, we acknowledge the funding received from the China Postdoctoral Science Foundation under grant #2022M720600, the special project for Postdoctoral Science Foundation of Chongqing #2022CQBSGTB2012, the Youth Basic Research Project of the Ministry of Education Key Laboratory under grant #YBRP-202105, and the Program of Excellent Scientist Fund at the Children’s Hospital of Chongqing Medical University.
Author information
Authors and Affiliations
Contributions
Guangming Dai, Wei Huang and Dawei He designed and supervised the experiments and revised the manuscript. Bowen Chen, Junyi Liao, Hong Chen, Aowen Bao and Chen Zhao provided critical reading and revision of the manuscript. Qiuyue Sun and Chen Zhang wrote the manuscript and analyzed the data. Qiuyue Sun, Chen Zhang, Junyi Liao and Aowen Bao performed animal experiments. Qiuyue Sun, Chen Zhang and BoWen Chen performed immunohistochemical and immunofluorescence analyses and image acquisition. Qiuyue Sun and Bowen Chen performed cell and parameters culture studies and Western blotting. Chen Zhang performed RNA isolation and RT-qPCR. Guangming Dai, Qiuyue Sun and Chen Zhang performed Dual‐luciferase reporter gene assay and Bioinformatics prediction. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Written informed consent for publication was obtained from all participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, Q., Zhang, C., Chen, B. et al. Knockdown of LncRNA H19 inhibits vascularization and endochondral ossification via the MiRNA-21a-5p-Smad7/p-Smad2/3 pathway in fracture repair. Sci Rep 15, 26054 (2025). https://doi.org/10.1038/s41598-025-11300-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-11300-7