Abstract
Recent studies indicate that reduced levels of certain tumor-suppressing microRNAs (miRNAs) circulating in the blood are linked to tumor progression and poor prognosis across various types of malignancies. Identified from a comprehensive analysis of the NCBI and miRNA databases, we tested tumor suppressor miR-3619-5p in esophageal squamous cell carcinoma (ESCC). Both test-scale and large-scale analyses demonstrated that plasma levels of miR-3619-5p were markedly lower in ESCC patients than in healthy volunteers. Lower plasma levels of miR-3619-5p showed a strong association with advanced pathological stages and were recognized as an independent prognostic marker. Overexpression of miR-3619-5p in ESCC cells inhibited cell proliferation, migration and invasion through the direct suppression of novel target protein, proviral insertion site in Moloney murine leukemia virus 1 (PIM1). PIM1 is overexpressed in various solid and hematological cancers including ESCC, and has proven to be a promising target of inhibitors in recent clinical trials. In vivo, increased plasma 3619-5p levels following subcutaneous injection in mice bearing ESCC tumors significantly inhibited tumor growth, with low expression of PIM1 in tumor. Until now, no study has demonstrated that the secretory-type miRNA such as miR- 3619-5p could contribute to nucleic acid therapy to PIM1. Reduced blood levels of miR-3619-5p are linked to ESCC progression and poor prognosis, suggesting that miR-3619-5p could act as a novel therapeutic focus for nucleic acid-based treatment targeting PIM1 in ESCC patients.
Similar content being viewed by others
Introduction
Globally, esophageal cancer ranks sixth in mortality rates, contributing significantly to cancer death statistics1. Esophageal cancer is typically classified into two histological types: adenocarcinoma and squamous cell carcinoma2. In Western countries, adenocarcinoma is the predominant form of esophageal cancer. In contrast, within the “esophageal cancer belt,” a region spanning from northern Iran through Central Asia to north-central China, esophageal squamous cell carcinoma (ESCC) accounts for roughly 90% of all cases1. In recent years, remarkable advancements have been seen in chemotherapy, radiotherapy, surgical procedures, and perioperative management for ESCC. Despite advancements in treatment, the 5-year survival rate for individuals with ESCC continues to hover around 20%3. To enhance patient outcomes, many studies have focused on elucidating the molecular mechanisms of tumorigenesis in ESCC, as well as identifying potential clinical biomarkers and therapeutic molecular targets. However, for ESCC there are still only a few early diagnostic biomarkers and therapeutic targets4,5.
Discovered in 1993, microRNAs (miRNAs) are small, non-coding RNA molecules that regulate gene expression by destabilizing mRNA and inhibiting its translation. Numerous studies have revealed altered miRNA expression associated with the onset and development of diverse diseases, including cancer6. In particular, several miRNAs have been shown to maintain their stability and detectability in plasma and serum by being encapsulated within exosomes and binding to proteins such as Argonaute2 and HDL. These interactions provide protection, preventing the miRNAs from being degraded by endogenous ribonucleases7,8,9,10. Furthermore, some of these blood miRNAs have been reported not only to be derived from tumor cell lysis but also actively secreted as mediators of intercellular communication8,11,12,13. Over the past decade, numerous blood-derived miRNAs, including those examined in this study, have emerged as potential biomarkers for detecting cancer, monitoring tumor progression, and predicting prognosis as well as chemoresistance14,15,16,17.
Kosaka and collaborators proposed a new theory based on findings from their in vitro research18. Over the past decade, numerous blood-derived miRNAs, including those examined in this study, have emerged as potential biomarkers for detecting cancer, monitoring tumor progression, and predicting prognosis as well as chemoresistance. However, as the disease advances, the compensatory mechanism of healthy cells weakens, contributing to cancer progression.
Our previous research revealed a decline in specific tumor-suppressor miRNAs in the blood as cancer advances14,15,17,19. We suggested that the levels of these miRNAs in the blood might indicate the capacity of adjacent healthy cells to transfer tumor-suppressor miRNAs to cancer cells. As a result, specific tumor-suppressor miRNAs in the bloodstream may serve as valuable biomarkers for detecting malignant cancers, evaluating prognosis and chemosensitivity, and acting as potential targets for nucleic acid-based therapies in digestive tract cancers17,20,21,22.
As part of this research, we concentrated on the tumor-suppressor miR-3619-5p, identified through a comprehensive analysis of the NCBI and miRNA databases. This miRNA, which targets the proviral insertion site in Moloney murine leukemia virus 1 (PIM1), was observed to be diminished in the plasma of ESCC patients. In conclusion, our results showed that lower plasma levels of miR-3619-5p were strongly linked to advanced pathological stages and recognized as an independent prognostic indicator. Enhanced expression of miR-3619-5p in ESCC cells suppressed cell proliferation by inducing apoptosis or causing G1/S arrest in a TP53-dependent manner, while also impairing cell migration and invasion. Overexpression of miR-3619-5p in ESCC cells directly suppressed PIM1 in vitro. In mice, increased plasma 3619-5p levels following subcutaneous injection significantly inhibited tumor growth, with downregulation of PIM1. Overall, our findings indicated that reduced blood levels of miR-3619-5p are linked to ESCC progression and poor prognosis, highlighting the possibility that miR-3619-5p could be a potential target for nucleic acid therapies targeting PIM1 in ESCC patients (Fig. 1).
Graphical abstract. In healthy persons, the plasma level of miR-3619-5p are sufficiently supplied. However, as esophageal squamous cell carcinoma (ESCC) progresses, the plasma level of miR-3619‐5p is decreasing. This dynamics of miR-3619‐5p could be the promising biomarker to detect cancer and predict prognoses of ESCC patients. Moreover, the overexpression of miR-3619‐5p suppresses proviral insertion site in Moloney murine leukemia virus 1 (PIM1) expression by inhibiting stability and translation of mRNA. Overexpression of miR-3619‐5p enhances the tumor-killing activity by suppressing PIM1 expression. Low blood levels of miR-3619‐5p are associated with ESCC progression and poor prognoses and could be a target of nucleic acid therapy in ESCC patients.
Results
Selection of plasma MiRNA candidates based on a systematic review of the NCBI database
From the 2,600 miRNA candidates available in miRBase, we identified 25 miRNAs that exhibit low expression levels in ESCC tissues, possess tumor-suppressive properties, and have not yet been reported as biomarkers in body fluids. Following the application of exclusion criteria, five miRNAs (miR-3619-5p, −3178, −1182, −637, and − 564) were selected based on their detectable signals in samples from healthy individuals (Supplementary Fig. 1). Among the five candidate miRNAs, miR-3619-5p was chosen in this study, based on its tumor-suppressive function, detectable levels in healthy plasma, and lack of prior reports as a circulating biomarker in ESCC23,24.
Study design to find novel biomarker MiRNAs in the plasma of ESCC patients
This study was structured as outlined below. (1) Plasma miR-3619-5p levels were analyzed to examine their correlations with clinicopathological characteristics and prognostic outcomes in ESCC patients; (2) an evaluation of the potential anti-tumor effects of miR-3619-5p overexpression in ESCC cells was undertaken in vitro; and (3) an assessment of its tumor-suppressive role was carried out in an in vivo tumor model (Fig. 2a).
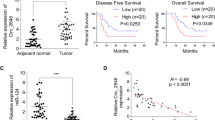
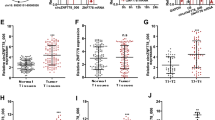
Study design and identification of plasma miRNA candidates. (a) Study design to identify novel candidate miRNAs with reduced levels in patient plasma as potential therapeutic targets for ESCC. The workflow included: (1) systematic in silico screening and experimental selection of candidate miRNAs; (2) large-scale qRT-PCR validation of plasma miR-3619-5p levels in ESCC patients (n = 94) and healthy volunteers (n = 81); (3) assessment of associations between plasma levels and clinicopathological features or survival; and (4) functional validation through in vitro and in vivo experiments. (b) The expression levels of miR-3619-5p were analyzed in normal human organs and ESCC cell lines. High expression of miR-3619-5p was observed in the skeletal muscle, heart, stomach, and lung. In contrast, 75% of ESCC cell lines exhibited lower miR-3619-5p expression compared to normal esophageal mucosa. (c) Quantitative analysis of plasma miR-3619-5p levels in clinical samples. Plasma RNA was isolated from peripheral blood of ESCC patients and healthy volunteers. Levels of miR-3619-5p were measured by TaqMan qRT-PCR and normalized to cel-miR-39. Statistical comparison using the Mann–Whitney U test revealed significantly reduced plasma miR-3619-5p levels in ESCC patients (P = 0.0002), suggesting potential utility as a non-invasive biomarker. (d) Kaplan–Meier survival analysis based on plasma miR-3619-5p expression. Prognostic analysis demonstrated that low plasma levels of miR-3619-5p were significantly correlated with poorer overall survival rates (P = 0.0471). Additionally, low plasma levels of miR-3619-5p were significantly correlated with poorer recurrence-free survival rates in ESCC patients (P = 0.0378).
Levels of miR-3619-5p in normal tissues and ESCC cell lines
We examined miR-3619-5p expression levels in normal human tissues and ESCC cell lines to determine its tissue distribution and potential cancer-specific downregulation. Total RNA was extracted from commercially available normal human tissue samples (stomach, thymus, prostate, skeletal muscle, kidney, spleen, intestine, pancreas, heart, lung, colon, testis, liver, thyroid, brain, and esophagus) and sixteen ESCC cell lines (TE1, TE2, TE4, TE5, TE6, TE8, TE9, TE10, TE11, TE13, TE14, TE15, KYSE70, KYSE150, KYSE170, and KYSE790). miR-3619-5p levels were quantified using TaqMan-based quantitative reverse transcription PCR (qRT-PCR), and normalized to U6 small nuclear RNA (RNU6B) using the 2^−ΔΔCt method. All measurements were conducted in triplicate (n = 3 biological replicates per sample).
As depicted in Fig. 2b, miR-3619-5p exhibited relatively high expression in the skeletal muscle, heart, stomach, and lung among the tested normal tissues, suggesting its physiological role in multiple organs. In contrast, its expression was consistently downregulated in ESCC cell lines when compared to normal esophageal mucosa. Specifically, 12 out of 16 (75%) ESCC cell lines showed more than 50% reduction in miR-3619-5p levels.
Comprehensive validation of plasma miR-3619-5p levels in ESCC patients
We initiated our analysis by validating the findings in a large-scale study. Plasma miR-3619-5p was successfully detected in all 94 ESCC patients and 81 healthy volunteers. Notably, plasma miR-3619-5p levels were significantly reduced in ESCC patients compared to healthy controls (P < 0.001) (Fig. 2c).
Association of plasma miR-3619-5p levels with clinicopathological features in ESCC patients
We analyzed the associations between plasma miR-3619-5p levels and clinicopathological characteristics in a cohort of 94 ESCC patients (Table 1). The patients were divided into two groups based on the median relative expression of plasma miR-3619-5p. Lower plasma miR-3619-5p levels were significantly correlated with advanced T stage (P = 0.025), N stage (P = 0.046), pathological stage (P = 0.016), and disease recurrence (P = 0.022).
Plasma miR-3619-5p as a candidate prognostic biomarker in ESCC cases
Prognostic analysis indicated that lower plasma levels of miR-3619-5p were significantly linked to poorer overall survival (P = 0.047) and recurrence-free survival (P = 0.038) in ESCC patients (Fig. 2d). Using the Cox proportional hazards model, univariate and multivariate analyses demonstrated that, alongside T and N factors—recognized prognostic markers in ESCC—a low plasma level of miR-3619-5p was also identified as a significant prognostic indicator (P = 0.028; hazard ratio: 2.09; 95% confidence interval: 1.08–4.03), independently predicting poor outcomes in ESCC patients (Table 2).
Tumor suppressive function of miR-3619-5p in ESCC cells and miR-3619-5p targeting PIM1
To examine the tumor-suppressive function of miR-3619-5p in ESCC cells, we conducted a series of in vitro functional assays using two representative ESCC cell lines: KYSE70 (TP53 wild-type) and KYSE170 (TP53 mutant). Cells were transfected with either miR-3619-5p mimics or a negative control mimic (miR-NC) at a final concentration of 12 nM using Lipofectamine RNAiMAX. Cell proliferation was assessed at 24, 48, and 72 h post-transfection using a WST-8–based colorimetric assay. Absorbance was measured at 450 nm, and all assays were performed in triplicate (n = 3 independent experiments). Results showed a significant reduction in cell proliferation at 72 h in both KYSE70 and KYSE170 cells transfected with miR-3619-5p compared to miR-NC (P < 0.01, Student’s t-test) (Fig. 3a). Flow cytometry analysis demonstrated that miR-3619-5p expression induced apoptosis in KYSE70 cells (wild-type TP53) and caused G1/S phase cell-cycle arrest in KYSE170 cells (mutant TP53), indicating a TP53-dependent mechanism in ESCC cells (Fig. 3a). To elucidate the mechanisms of growth suppression, we performed flow cytometry-based cell cycle and apoptosis analyses. For cell cycle analysis, cells were fixed in 70% ethanol, stained with propidium iodide (PI), and analyzed using a Becton Dickinson Accuri™ C6 Flow Cytometer 72 h post-transfection. miR-3619-5p expression led to G1/S cell cycle arrest in KYSE170 cells and an increase in the sub-G1 population in KYSE70 cells. For apoptosis analysis, annexin V-FITC/PI double staining was used. The proportion of early apoptotic (annexin V+/PI−) and late apoptotic (annexin V+/PI+) cells significantly increased in KYSE70 cells transfected with miR-3619-5p mimics compared to controls (P < 0.01, one-way ANOVA followed by Tukey’s post hoc test). This apoptotic response was not observed in KYSE170 cells, suggesting that the induction of apoptosis by miR-3619-5p is TP53-dependent (Fig. 3b).
An exploration of the potential role of miR-3619-5p in inhibiting tumor progression in ESCC cells. (a) Cell proliferation and cell cycle analysis following miR-3619-5p overexpression. KYSE70 (TP53 wild-type) and KYSE170 (TP53 mutant) ESCC cells were transfected with 12 nM of miR-3619-5p mimic or negative control mimic (miR-NC) using Lipofectamine RNAiMAX. Cell proliferation was measured at 24, 48, and 72 h using a WST-8–based colorimetric assay (Cell Counting Kit-8; n = 3). Absorbance at 450 nm was recorded to assess metabolic activity. Overexpression of miR-3619-5p significantly inhibited cell proliferation at 72 h compared to miR-NC (P < 0.01, Student’s t-test). Data are presented as mean ± SD from three independent experiments. For cell cycle analysis, cells were fixed in 70% ethanol, stained with propidium iodide, and analyzed by flow cytometry 72 h post-transfection. miR-3619-5p induced G1/S cell cycle arrest in KYSE170 cells and an increase in the sub-G1 population in KYSE70 cells, indicating apoptosis induction. (b) Apoptotic cell analysis was conducted by annexin V-FITC/propidium iodide (PI) double staining and flow cytometry 72 h after transfection. In KYSE70 cells, overexpression of miR-3619-5p significantly increased the proportions of early apoptotic (Annexin V+/PI−) and late apoptotic (Annexin V+/PI+) cells compared to miR-NC controls (P < 0.01, ANOVA with Tukey’s post hoc test; n = 3). This apoptotic response was not observed in KYSE170 cells. (c) Transwell migration and invasion assays revealed that miR-3619-5p inhibited the migration and invasion capabilities of both types of ESCC cells. Data are presented as mean ± SD from three independent experiments. (d) Western blot analysis was performed to assess PIM1 protein expression 72 h after transfection. miR-3619-5p overexpression led to a marked reduction in PIM1 levels compared to controls in both cell lines. β-actin served as a loading control. To confirm direct binding of miR-3619-5p to PIM1 mRNA, a dual-luciferase reporter assay was conducted. The 3′ untranslated region (UTR) of PIM1 (wild-type or with mutated seed sequence) was cloned downstream of the luciferase gene. Co-transfection with miR-3619-5p mimic significantly decreased luciferase activity in the wild-type construct (P < 0.01, Student’s t-test; n = 3), but not in the mutant, confirming that PIM1 is a direct target of miR-3619-5p. Data are presented as mean ± SD from three independent experiments.
Next, we performed transwell migration and Matrigel invasion assays using 8-µm pore size Boyden chambers. Twenty-four hours post-transfection, 5 × 10⁵ cells were seeded into the upper chambers in serum-free medium, and the lower chambers contained 10% FBS as a chemoattractant. After 22 h of incubation, migrated or invaded cells on the bottom surface of the membrane were fixed, stained, and counted in five randomly selected fields under a microscope. Both migration and invasion were significantly suppressed in miR-3619-5p-transfected KYSE70 and KYSE170 cells compared to controls (P < 0.01, unpaired Student’s t-test; n = 3) (Fig. 3c).
We then explored potential targets of the tumor suppressor miR-3619-5p in cancers using the TargetScan database, identifying several putative target genes. Among them, we focused on a novel candidate, PIM1, which contains complementary sequences to miR-3619-5p. Western blot analysis was performed to examine PIM1 protein levels in miR-3619-5p transfectants, revealing a decrease in PIM1 expression in miR-3619-5p transfectants compared to miR-NC transfectants (Fig. 3d). A luciferase reporter assay was conducted with plasmids harboring either the wild-type or mutant 3′ UTR of PIM1. The results revealed a significant decrease in luciferase activity in the wild-type constructs, whereas no such reduction was observed in the mutant constructs (Fig. 3d). Transfection with miR-3619-5p, leading to PIM1 knockdown, suppressed AKT phosphorylation, increased p21 production, and induced phosphorylation-mediated inactivation of Rb in both TP53 wild-type and TP53 mutant cells. However, phosphorylation of caspase-3 and PARP was observed only in the TP53 wild-type cell line (Fig. 4a). The findings indicated that low expression of tumor suppressor miR-3619-5p induced overexpression of PIM1 with cell proliferation, migration and invasion through the activation of the AKT pathway in ESCC cells. Figure 4b shows a hypothetical model of the overexpression of PIM1 by the reduced levels of miR-3619-5p in ESCC cells.
The suppression of malignant behaviors in ESCC cells was observed following PIM1 knockdown mediated by miR-3619-5p. The knockdown effects varied depending on the TP53 mutation status, highlighting the potential influence of TP53 alterations on the functional impact of miR-3619-5p in targeting PIM1. (a) Effects of miR-3619-5p-mediated PIM1 knockdown on downstream signaling pathways. KYSE70 (TP53 wild-type) and KYSE170 (TP53 mutant) cells were transfected with miR-3619-5p or miR-NC mimics at 12 nM. At 72 h post-transfection, protein lysates were analyzed by western blot. In both cell lines, miR-3619-5p overexpression suppressed phosphorylation of AKT (p-AKT), upregulated p21, and enhanced phosphorylation of retinoblastoma protein (p-Rb), suggesting inhibition of cell cycle progression. Notably, cleaved caspase-3 and cleaved PARP—markers of apoptosis—were observed only in TP53 wild-type cells, indicating a TP53-dependent apoptotic response. β-actin was used as a loading control. (b) Schematic illustration of a proposed model. Loss of miR-3619-5p in ESCC results in de-repression of PIM1, promoting downstream oncogenic signaling via AKT activation and inhibition of p21-mediated cell cycle arrest. Restoration of miR-3619-5p expression reverses this phenotype by suppressing PIM1 and downstream pathways, particularly in TP53-competent cells. (c) Restoration of plasma miR-3619-5p levels was shown to inhibit tumor growth in vivo. BALB/c nude mice (n = 4 per group) were inoculated with 2 × 10⁶ KYSE170 cells into the left lower flank. Once tumors were established (day 7), mice received subcutaneous injections of either miR-3619-5p mimic or miR-NC mimic (each 5 nmol) mixed with 20 µL AteloGene Local Use Quick Gelation into the right lower flank, away from the tumor site. Treatments were conducted weekly for three weeks following the initial injection. (d) Treatment with the miR-3619-5p mimic led to a marked reduction in tumor growth relative to the negative control mimic (P < 0.01, Student’s t-test). PIM1 expression in tumor tissues was evaluated using western blotting and immunohistochemistry. Tumors from mice treated with the miR-3619-5p mimic exhibited significantly lower PIM1 expression levels than those treated with the negative control mimic. β-actin was used as a loading control. Data are presented as mean ± SD from four independent experiments. (e) Plasma miR-3619-5p levels were compared between mice receiving the miR-3619-5p mimic and those treated with the negative control mimic. Mice treated with the miR-3619-5p mimic showed significantly higher plasma levels of miR-3619-5p as opposed to those treated with the negative control mimic (P < 0.01). Data are presented as mean ± SD from four independent experiments. (f) Analysis of blood parameters was executed to investigate potential side effects of miR-3619-5p treatment. No signs of organ dysfunction were observed, as indicated by normal levels of creatinine (Cre), aspartate aminotransferase (AST), alanine aminotransferase (ALT), amylase (AMY), and total bilirubin (T-Bil). Data are presented as mean ± SD from four independent experiments.
Restoration and maintenance of miR-3619-5p levels in plasma could inhibit tumor growth in vivo
To investigate the systemic secretion of miR-3619-5p from healthy cells, its expression patterns were analyzed across multiple human organs. Quantitative RT-PCR was used to assess miR-3619-5p expression levels in human tissues (Fig. 2b). miR-3619-5p expression was observed to be relatively high in the skeletal muscle, heart, stomach, and lung, indicating its potential physiological relevance in multiple organ systems. To investigate its tumor-suppressive role in vivo, we established a subcutaneous xenograft model using BALB/c nude mice. KYSE170 cells (2.0 × 10⁶ cells in 100 µL PBS) were injected into the left lower flank of 4-week-old female mice (n = 4 per group). Tumor formation was visually confirmed 7 days post-implantation. Thereafter, mice received subcutaneous injections of either synthetic miR-3619-5p mimic or negative control (miR-NC) mimic, each mixed with atelocollagen (20 µL total volume), into the right lower flank once weekly for three consecutive weeks. This injection site was anatomically separated from the tumor site to mimic systemic delivery while minimizing direct contact (Fig. 4c).
Tumor volumes were measured with calipers twice a week and calculated using the formula: volume = (length × width²)/2. At the end of the observation period, mice were sacrificed, and tumors were excised, weighed, and analyzed. Compared with the miR-NC group, miR-3619-5p-treated mice exhibited significantly smaller and lighter tumors (P < 0.01, unpaired Student’s t-test; Fig. 4d), suggesting that miR-3619-5p exerts potent anti-tumor effects in vivo.
To explore the underlying molecular mechanism, PIM1 protein expression in tumor tissues was evaluated by both western blotting and immunohistochemistry. As shown in Fig. 4d, tumors from the miR-3619-5p-treated group exhibited a marked reduction in PIM1 expression, confirming in vivo suppression of the target gene by miR-3619-5p. Furthermore, qRT-PCR analysis of plasma samples revealed a significant elevation in circulating miR-3619-5p levels in the treatment group compared to controls (P < 0.01, Fig. 4e), confirming systemic delivery and stabilization of the miRNA mimic in circulation.
To assess treatment safety, serum biochemical analyses were performed to measure markers of renal, hepatic, and pancreatic function, including creatinine, AST, ALT, amylase, and total bilirubin. No significant abnormalities were observed in the miR-3619-5p group, indicating that the treatment did not cause organ toxicity (Fig. 4f). These findings strongly indicated that restoring miR-3619-5p in plasma could effectively suppress ESCC tumor growth.
Discussion
The clinical application of miRNAs has faced significant challenges over the past few decades25,26,27. Our findings also suggest that the blood levels of certain tumor-suppressor miRNAs could serve as valuable biomarkers for detecting malignancies, predicting poor prognosis, assessing chemosensitivity, and potentially functioning as therapeutic targets in nucleic acid-based treatments for various digestive tract cancers17,20,21,22. This study revealed that reduced plasma levels of miR-3619-5p were closely linked to advanced pathological stages and served as an independent prognostic marker in ESCC. In ESCC cells, miR-3619-5p overexpression suppressed cell proliferation, migration, and invasion by directly targeting and downregulating the novel protein PIM1.
We performed a systematic review of the NCBI and miRNA databases, focusing on the tumor-suppressor miR-3619-5p, which was a putative regulator of PIM1 and exhibits low plasma levels in ESCC patients. Our study demonstrated that reduced plasma miR-3619-5p levels were strongly associated with advanced pathological stages and identified as an independent prognostic marker. In ESCC cells, miR-3619-5p overexpression suppressed cell proliferation via apoptosis or G1/S arrest in a TP53-dependent manner and inhibited migration and invasion independently of TP53. Furthermore, miR-3619-5p overexpression directly downregulated the novel target protein PIM1 in vitro. In vivo, subcutaneous injections that elevated plasma miR-3619-5p levels significantly suppressed tumor growth in mice, accompanied by a reduction in PIM1 expression. This study is the first to demonstrate that miR-3619-5p may serve as a novel blood biomarker for cancer detection and prognosis prediction in ESCC. Additionally, miR-3619-5p shows potential as a nucleic acid-based therapeutic agent targeting the oncogene PIM1 in ESCC patients. Other candidate miRNAs identified during our initial screening will be investigated in future studies to further elucidate their potential roles in ESCC pathogenesis and therapy.
Various molecular functions of miR-3619-5p have been reported in a variety of cancers, including gastric cancer24, breast cancer28, prostate cancer29, gallbladder cancer30, colorectal cancer31, and hepatocellular carcinoma32. miR-3619-5p is known to target multiple oncogenes, suppressing their functions. For instance, it targets CTNNB1 in colorectal and hepatocellular carcinoma and HDGF in tongue squamous cell carcinoma, hepatocellular carcinoma, and infantile hemangiomas. Both CTNNB1 and HDGF play critical roles in the caspase activation pathway. These prior studies align with our findings, highlighting the tumor-suppressive role of miR-3619-5p in ESCC.
The most striking finding of the present study was that overexpression of miR-3619-5p induced the downregulation of PIM1. PIM1 is known to be overexpressed in numerous solid tumors and hematological malignancies. Its overexpression is linked to poor prognosis in a variety of cancers, including acute myeloid leukemia, acute lymphoblastic leukemia, multiple myeloma, breast cancer, colorectal cancer, papillary thyroid carcinoma, liver cancer, prostate cancer, and ESCC33,34,35,36,37,38,39,40. Therefore, clinical trials involving various PIM1 inhibitors have been performed, including AZD1208, PIM447, TP-3654, CTT1403, SGI-1776, and ETH-155,008, to evaluate the efficacy of therapeutic agents targeting PIM1, reflecting the growing research interest in exploiting this kinase for cancer treatment41,42,43,44,45. However, in some clinical trials, gastrointestinal disturbances, especially nausea and diarrhea, were commonly reported, and more than 70% of patients reported experiencing adverse events of grade 3 or higher, including febrile neutropenia, hypotension, and pneumonia41,43,44. These studies highlighted the significant adverse effects associated with conventional treatments. In contrast, our mouse model showed no evidence of blood-based organ damage or gastrointestinal issues following miR-3619-5p administration. This suggests that restoring miR-3619-5p to its normal levels in human blood may provide a cancer treatment option with fewer side effects in clinical practice. Similarly, our previous research demonstrated that replenishing depleted tumor-suppressor miRNAs in plasma to levels observed in healthy individuals effectively inhibited tumor progression in cancer-bearing mice without inducing notable adverse effects20,21,22.
This is the first study to demonstrate that the tumor suppressor miR-3619-5p could be a therapeutic agent targeting PIM1 as well as a novel plasma biomarker in cases of ESCC. Therefore, Clinical applications using this promising powerful suppressor of tumors might be possible in cancers associated with PIM1 overexpression. We also revealed the novel potential functions of miR-3619-5p in modulating the PIM1 pathway, which plays a crucial role in tumorigenesis. However, there remains a limitation to this work. Further investigation into the physiological effects of miR-3619-5p is essential to ensure its safe application in clinical settings in vivo. Additionally, more research is needed to elucidate the mechanisms governing the cellular uptake and secretion of tumor-suppressor miRNAs. Such studies will be instrumental in the advancement of miRNA delivery systems for both therapeutic and diagnostic applications in the future.
Materials and methods
Individuals and associated samples
The institutional review boards of Kyoto Prefectural University of Medicine (ERB-C-319-1) approved this study, and all experimental procedures adhered to corresponding guidelines and directives. Informed written consent was obtained from all participants for the collection and use of their tissue and blood samples. From June 2010 to December 2012, plasma samples were gathered from 94 ESCC patients in succession and 81 healthy individuals, excluding individuals with a history of cancer. All tissue and plasma samples were collected at Kyoto Prefectural University of Medicine. Tumor staging was conducted based on the Union for International Cancer Control (UICC) classification system46. Peripheral blood samples of 7 mL were collected from each patient before surgery and from healthy volunteers. Resected specimens were formalin-fixed and paraffin-embedded for pathological analysis. Histological evaluation of the adjacent tissues was carried out following the World Health Organization criteria. In all cases, the pathological findings were reviewed and confirmed by at least two pathologists. The group of 81 healthy volunteers included medical personnel as well as individuals with non-malignant conditions, such as gallstones or inguinal hernias. They underwent comprehensive medical evaluations, including blood tests, endoscopy, and CT scans, to confirm the absence of esophageal or other cancerous diseases. Blood samples were collected in sodium heparin tubes (BD Vacutainer, Franklin Lakes, NJ) and processed using a three-step centrifugation process (1,500 rpm for 30 min, followed by 3,000 rpm and 4,500 rpm for 5 min each) to reduce contamination from cellular nucleic acids. The resulting plasma was preserved at −80 °C for future analysis.
RNA extraction
Using the mirVana PARIS Kit (Ambion, Austin, TX) and following the protocol provided by the manufacturer, total RNA was isolated from 400 µL of plasma. The RNA was finally collected in 100 µL of preheated elution buffer for optimal recovery at 95 °C. A consistent volume of 400 µL of plasma was used, as previous studies have highlighted the lack of a universally accepted internal control for plasma miRNA analysis14,15. Total RNA was obtained from tissue samples preserved in formalin and embedded in paraffin, consisting of four 15 μm-thick slices (total thickness of 60 μm), using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Austin, TX) following the manufacturer’s protocol. The RNA was then eluted into 60 µL of Elution Solution.
Systematic review of the NCBI database to select candidate MiRNAs
We conducted a study aimed at identifying innovative plasma microRNA indicators in patients with ESCC (Supplementary Fig. 1). From the pool of 2,600 miRNA candidates listed in the human miRNA miRBase (Version 18) (http://microrna.sanger.ac.uk/), we conducted a PubMed search for all studies related to these miRNAs published up to February 2019. Abstracts without corresponding full-text articles were deemed ineligible for inclusion, as were other incomplete articles and articles not published in English. The selection of candidate miRNAs was guided by these criteria: (I) miRNAs identified as circulating or oncogenic miRNAs, and (II) miRNAs not previously described as tumor-suppressive. Two authors independently screened all retrieved articles according to predefined criteria, with any disagreements resolved through consultation with a third author.
MiRNA levels were quantified using qRT-PCR
The levels of miRNA were measured using qRT-PCR with the Human TaqMan MicroRNA Assay Kit (Applied Biosystems, Foster City, CA) on a StepOnePlus PCR system (Applied Biosystems). The qPCR conditions comprised an initial denaturation at 95 °C for 10 min, followed by 40 amplification cycles: 95 °C for 15 s for denaturation and 60 °C for 1 min for annealing and extension. Cycle threshold (Ct) values were analyzed using StepOne Software v2.0 (Applied Biosystems). Consistent with previous studies, data normalization was performed by spiking samples with cel-miR-39, a synthetic RNA oligonucleotide absents within the human genome7. C. elegans cel-miR-39 was obtained as a custom-synthesized RNA oligonucleotide (Qiagen, Valencia, CA). Data normalization across samples was performed employing the 2-ΔΔCt method with cel-miR-39 as the reference. For miRNA expression analysis in human tissue samples and cultured cells, U6 small nuclear RNA (RNU6B) acted as the control for normalization via the same 2-ΔΔCt method. Gene expression changes were analyzed using the 2-ΔΔCt method47,48. Reverse transcription was carried out with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) in a 5 µL reaction volume. The reaction mixture consisted of 1.67 µL of extracted RNA, 0.05 µL of 100 mM dNTPs, 0.33 µL of Multiscribe Reverse Transcriptase (50 U/µL), 0.5 µL of 10× reverse transcription buffer, 0.06 µL of RNase inhibitor (20 U/µL), 1 µL of a gene-specific primer (hsa-miR-3619-5p, Assay ID: 479689), and 1.39 µL of nuclease-free water. To protect against degradation by endogenous plasma RNases, 25 fmol of oligo in a total volume of 5 µL was added after treating the plasma sample with 2× denaturing solution (Ambion). Cel-miR-39 served as the control for each RNA sample in TaqMan qRT-PCR assays (Applied Biosystems), adhering to established protocols. ΔCt values were obtained by normalizing the Ct values of target miRNAs against those of RNU6B or cel-miR-39. ΔΔCt values were subsequently calculated by subtracting the mean ΔCt of control plasma (healthy volunteers) or normal esophageal tissue from the ΔCt of plasma or tissue samples from ESCC patients, enabling comparative analysis.
mRNA levels were quantified using qRT-PCR
Single-stranded complementary DNA (cDNA) synthesized from total RNA was amplified using gene-specific primers, including those for E-cadherin and vimentin. mRNA levels were quantified through qRT-PCR using the Human TaqMan Gene Expression Assay Kit (Applied Biosystems). The qPCR was performed on a StepOnePlus PCR system (Applied Biosystems), and Ct values were determined using StepOne Software v2.0 (Applied Biosystems). Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) in a reaction volume of 20 µL. The mixture included 10 µL of extracted RNA, 0.8 µL of 100 mM dNTPs, 1.0 µL of Multiscribe Reverse Transcriptase (50 U/µL), 2.0 µL of 10× Reverse Transcription Buffer, 1.0 µL of RNase inhibitor (20 U/µL), 2.0 µL of 10× RT random primer, and 3.2 µL of nuclease-free water. The reaction conditions for cDNA synthesis were set as follows: incubation at 25 °C for 10 min, 37 °C for 120 min, and 85 °C for 5 min, followed by a hold at 4 °C. For amplification, 1.4 µL of cDNA was mixed with 10 µL of TaqMan 2× Universal PCR Master Mix (without AmpErase UNG; Applied Biosystems), 1.0 µL of gene-specific primers/probes (Hs00170423_m1 for E-cadherin and Hs00185584_m1 for vimentin; Applied Biosystems), and 7.6 µL of nuclease-free water, resulting in a total reaction volume of 20 µL. The qPCR was performed on a StepOnePlus PCR system (Applied Biosystems) under the following conditions: initial incubation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Ct values were determined using StepOne Software v2.0 (Applied Biosystems). For cDNA synthesis, the reaction mixtures were incubated in a sequential manner: 16 °C for 30 min, 42 °C for 30 min, 85 °C for 5 min, and then held at 4 °C. For amplification, 0.67 µL of cDNA was mixed with 5 µL of TaqMan 2× Universal PCR Master Mix (without AmpErase UNG; Applied Biosystems), 0.5 µL of gene-specific primers/probes, and 3.83 µL of nuclease-free water, achieving a final reaction volume of 10 µL. The qPCR was carried out on a StepOnePlus PCR system (Applied Biosystems) under the following conditions: initial incubation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Ct values were calculated using StepOne Software v2.0 (Applied Biosystems).
Culture of ESCC cell lines
ESCC cell lines, including TE2 (CVCL 4455), TE5 (CVCL 1764), TE8 (CVCL 1766), TE9 (CVCL 1767), TE15 (CVCL 1763), KYSE150 (CVCL 1348), KYSE170 (CVCL 1358), and KYSE790 (CVCL 8510), were obtained from RIKEN Cell Bank (Tsukuba, Japan) and cultured in Roswell Park Memorial Institute 1640 medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (Trace Scientific, Melbourne, Australia). All cells were maintained in a humidified chamber with 5% carbon dioxide at 37 °C. Cell line authentication was confirmed through short tandem repeat profiling conducted by the RIKEN Cell Bank (Tsukuba, Japan), and no mycoplasma contamination was observed in any cultures.
Transfection of ESCC cells with MiRNA mimics
To achieve overexpression of miR-3619-5p, KYSE70 and KYSE170 cells were transfected with either the miR-3619-5p mimic (Assay ID: MC20221) or a negative control mimic (mirVana miRNA mimic Negative Control #1), both obtained from the mirVana miRNA mimic panel (Ambion). Transfection was performed using Lipofectamine RNAiMAX (Invitrogen) at a final concentration of 12 nM, following the manufacturer’s instructions. After 72 h, miR-3619-5p overexpression was validated through qRT-PCR with the Human TaqMan MicroRNA Assay Kit (Applied Biosystems).
Proliferation assay and cell cycle analysis
Cell growth was measured at various time points following transfection using a WST-8–based colorimetric assay (Cell Counting Kit 8; Dojindo Laboratories, Kumamoto, Japan) to determine the number of viable cells. The cell cycle was analyzed 72 h post-transfection using flow cytometry, following previously established protocols49. The assessment of cell viability was conducted by measuring the optical density at 450 nm, indicative of metabolic activity. For flow cytometry analysis, collected cells were fixed in 70% chilled ethanol and subsequently treated with RNase A and propidium iodide. The samples were then analyzed using a Becton Dickinson Accuri™ C6 Flow Cytometer (Becton Dickinson, San Jose, CA).
Apoptotic cell analysis
Non-transfected cells exposed to staurosporine for 24 h were used as a control. At 72 h following transfection with the miRNA mimic, cells were collected and subjected to staining with fluorescein isothiocyanate-conjugated annexin V and propidium iodide, following the guidelines provided by the Annexin V Kit manufacturer (Beckman Coulter, Brea, CA). The percentage of apoptotic cells was analyzed with a Becton Dickinson Accuri™ C6 Flow Cytometer.
Transwell migration and invasion assays
Migration and invasion assays were performed using 24-well Boyden chambers (Transwell chambers, BD Transduction, Franklin Lakes, NJ). For invasion assays, 8-µm pore filters were coated with Matrigel to mimic the extracellular matrix, while uncoated filters served as controls for migration assays (BD Transduction). Cells transfected with the miRNA mimic (5 × 10⁵ per well) were plated in the upper chamber. After a 22-hour incubation, cells that migrated to the underside of the filters were fixed and stained using Diff-Quik stain (Sysmex, Kobe, Japan). The nuclei of stained cells were counted in triplicate.
Luciferase activity assay
Luciferase reporter constructs were created by cloning the 3’-untranslated region (UTR) of PIM1 downstream of the luciferase gene in the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA). Site-specific mutations were introduced using the GeneTailor Site-Directed Mutagenesis System (Thermo Fisher Scientific). KYSE70 and KYSE170 cells were transfected with either the luciferase reporter or control plasmids (pmirGLO) via Lipofectamine RNAiMAX (Thermo Fisher Scientific). After 24 h, 10 nmol/L of miRNA (either a negative control mimic (miR-NC) or miR-3619-5p) was added to the cells. Two days post-treatment, Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). The relative luciferase activity was calculated by calculating the ratio of Firefly luciferase luminescence to that of the Renilla luciferase, serving as an internal normalization control.
Western blot analysis
Mouse monoclonal anti-PIM1 antibodies (Cat. No. 13513) were purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology, CA). Antibodies against p21, caspase-3, cleaved caspase-3, AKT, phospho-AKT, PARP, and ACTB were obtained from Cell Signaling Technology (Cat. No., 2946, 9662, 9664,4691, 4060, 5732, 9532 and 4970, respectively; Cell Signaling Technology, USA). Cells were lysed using a Tris buffer (50 mmol/L, pH 7.5) containing 150 mmol/L NaCl, 1 mmol/L EDTA, 0.5% NP-40, 10% glycerol, 100 mmol/L NaF, 10 mmol/L sodium pyrophosphate, 2 mmol/L Na2VO3, and a protease inhibitor cocktail (Roche, Tokyo, Japan). Proteins were extracted using the M-PER Mammalian® Protein Extraction Reagent (Thermo Scientific, USA), and 20 µg of protein per lane was loaded onto the gel for electrophoresis. To ensure equal protein loading, β-actin (ACTB) was used as an internal loading control.
Immunohistochemistry
Mouse monoclonal anti-PIM1 antibodies (Cat. No. 13513) were provided by Santa Cruz Biotechnology (Santa Cruz Biotechnology, CA). Tumor tissues were preserved in 10% formaldehyde prepared in PBS and subsequently embedded in paraffin. The paraffin blocks were kept at room temperature in a dark environment, and section staining via the HRP method was completed within two weeks. After deparaffinization, antigen retrieval was achieved by heating the sections in 10 mmol/L citrate buffer (pH 9.0) at 95 °C for 60 min. Endogenous peroxidase activity was inhibited by treating the slides with 3% H2O2 for 20 min. The sections were then blocked with Block Ace (Dainippon Sumitomo Pharmaceutical) for 30 min at room temperature and incubated with anti-PIM1 antibodies (1:100) for 1 h at room temperature. PBS was used for all washing steps and dilutions. The EnVision + HRP System (EnVision + Dual Link System-HRP; Dako North America) was employed to detect the primary antibody binding. HRP activity was detected using diaminobenzidine tetrahydrochloride, and the slides were counterstained with Mayer’s hematoxylin.
Protocol for animal experiments
This study aimed to investigate the tumor-suppressive effects of microRNA in an in vivo model of esophageal cancer using BALB/c nude mice. A subcutaneous transplantation model was established to evaluate tumor growth and treatment efficacy. BALB/c nude female mice, four weeks of age, were used, with a total of eight mice divided into two groups: a control group and a treatment group, each consisting of four mice. Randomization and blinding were not performed, as direct tumor observation by the experimenter was necessary to ensure appropriate intervention. The sample size was determined without a power analysis. Under isoflurane anesthesia, esophageal cancer cells (KYSE170, 2.0 × 10⁶ cells) and tumor-suppressive synthetic microRNA were subcutaneously injected to establish a tumor xenograft model. Tumor engraftment was visually confirmed, and after three weeks, blood samples were collected from the inferior vena cava or portal vein, and primary tumor tissues along with peritumoral tissues were harvested under anesthesia to minimize distress. The collected samples underwent immunohistochemical staining and molecular analysis, including RNA and protein extraction followed by RT-PCR, Western blot, and microarray analysis, to assess tumor-associated protein expression and genetic changes. The primary outcome of this study was tumor size, measured in terms of tumor diameter and tumor weight. Secondary outcomes included the assessment of circulating nucleic acid levels in the blood, immunohistochemical evaluation of tumor-associated proteins, and changes in RNA and protein expression. Statistical analysis was conducted using a t-test, with a significance threshold set at p < 0.05, and multiple comparison corrections were not applied. To minimize suffering, humane endpoints were established in accordance with UKCCCR guidelines. Mice were euthanized if the tumor exceeded 17 mm in diameter, if tumor weight surpassed 10% of body weight, or if body weight decreased by more than 20%. Euthanasia was performed through an overdose of barbiturate anesthesia. Throughout the study, appropriate measures were taken to prevent animal escape, and carcasses were incinerated under proper infection control protocols. This study was approved by the Kyoto Prefectural University of Medicine Animal Experimentation Committee (Approval No.: M2023-506) and followed ARRIVE guidelines to ensure ethical and rigorous experimental conduct.
Analysis of statistical data
The Mann–Whitney U test and Student’s t-test were used for comparing plasma and tissue sample data from independent groups, while the Wilcoxon test was applied to analyze paired tumor and normal tissue samples. The chi-squared test or Fisher’s exact test was used to assess the relationship between plasma miR-3619-5p levels and clinicopathological characteristics. A threshold of P < 0.05 was used to define statistical significance in this study. For survival analysis, Kaplan–Meier curves were generated based on univariate predictors, and group differences were evaluated using the log-rank test. Both univariate and multivariate survival analyses were conducted using the likelihood ratio test within the stratified Cox proportional hazards model, with a P-value < 0.05 considered statistically significant.
Data availability
The datasets produced and/or analyzed in this study are registered with the Research Registry. The unique identifying number for this study is researchregistry10758. The data can be accessed via the following hyperlink: https://www.researchregistry.com/browse-the-registry#home/.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Siewert, J. R. & Ott, K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat. Oncol. 17, 38–44. https://doi.org/10.1016/j.semradonc.2006.09.007 (2007).
Lagergren, J., Smyth, E., Cunningham, D. & Lagergren, P. Oesophageal cancer. Lancet 390, 2383–2396. https://doi.org/10.1016/s0140-6736(17)31462-9 (2017).
Lin, D. C., Du, X. L. & Wang, M. R. Protein alterations in ESCC and clinical implications: A review. Dis. Esophagus 22, 9–20. https://doi.org/10.1111/j.1442-2050.2008.00845.x (2009).
Ohashi, S. et al. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology 149, 1700–1715. https://doi.org/10.1053/j.gastro.2015.08.054 (2015).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. https://doi.org/10.1016/0092-8674(93)90529-y (1993).
Mitchell, P. S. et al. Circulating micrornas as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 105, 10513–10518. https://doi.org/10.1073/pnas.0804549105 (2008).
Kosaka, N. et al. Secretory mechanisms and intercellular transfer of MicroRNAs in living cells. J. Biol. Chem. 285, 17442–17452. https://doi.org/10.1074/jbc.M110.107821 (2010).
Arroyo, J. D. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 108, 5003–5008. https://doi.org/10.1073/pnas.1019055108 (2011).
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D. & Remaley, A. T. Micrornas are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433. https://doi.org/10.1038/ncb2210 (2011).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. https://doi.org/10.1038/ncb1596 (2007).
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. https://doi.org/10.1038/ncb1800 (2008).
Ichikawa, D., Komatsu, S., Konishi, H. & Otsuji, E. Circulating MicroRNA in digestive tract cancers. Gastroenterology 142, 1074–1078e1071. https://doi.org/10.1053/j.gastro.2012.03.008 (2012).
Tsujiura, M. et al. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer 102, 1174–1179. https://doi.org/10.1038/sj.bjc.6605608 (2010).
Komatsu, S. et al. Circulating micrornas in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer 105, 104–111. https://doi.org/10.1038/bjc.2011.198 (2011).
Kawaguchi, T. et al. Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers. Int. J. Mol. Sci. https://doi.org/10.3390/ijms17091459 (2016).
Imamura, T. et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci. Rep. 7, 5708. https://doi.org/10.1038/s41598-017-06137-8 (2017).
Kosaka, N. et al. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J. Biol. Chem. 287, 1397–1405. https://doi.org/10.1074/jbc.M111.288662 (2012).
Komatsu, S. et al. Prognostic impact of Circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin. Biol. Ther. 12 (Suppl 1), 53–59. https://doi.org/10.1517/14712598.2012.681373 (2012).
Kiuchi, J. et al. Low levels of tumour suppressor miR-655 in plasma contribute to lymphatic progression and poor outcomes in oesophageal squamous cell carcinoma. Mol. Cancer. 18, 2. https://doi.org/10.1186/s12943-018-0929-3 (2019).
Takashima, Y. et al. Plasma miR-1254 as a predictive biomarker of chemosensitivity and a target of nucleic acid therapy in esophageal cancer. Cancer Sci. 114, 3027–3040. https://doi.org/10.1111/cas.15830 (2023).
Kamiya, H. et al. Low blood level of tumour suppressor miR-5193 as a target of immunotherapy to PD-L1 in gastric cancer. Br. J. Cancer 130, 671–681. https://doi.org/10.1038/s41416-023-02532-3 (2024).
Sun, B., Han, Y., Cai, H., Huang, H. & Xuan, Y. Retraction note to: long non-coding RNA SNHG3, induced by IL-6/STAT3 transactivation, promotes stem cell-like properties of gastric cancer cells by regulating the miR-3619-5p/ARL2 axis. Cell. Oncol. (Dordr). 45, 199. https://doi.org/10.1007/s13402-022-00662-z (2022).
Wu, H. et al. HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell. Death Dis. 11, 233. https://doi.org/10.1038/s41419-020-2426-z (2020).
Lanford, R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201. https://doi.org/10.1126/science.1178178 (2010).
Gebert, L. F. et al. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 42, 609–621. https://doi.org/10.1093/nar/gkt852 (2014).
Gallant-Behm, C. L. et al. A MicroRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J. Invest. Dermatol. 139, 1073–1081. https://doi.org/10.1016/j.jid.2018.11.007 (2019).
Chen, C. et al. Chloroprocaine antagonizes progression of breast cancer by regulating LINC00494/miR-3619-5p/MED19 axis. J. Biochem. Mol. Toxicol. https://doi.org/10.1002/jbt.23524 (2023).
Li, S. et al. miR-3619-5p inhibits prostate cancer cell growth by activating CDKN1A expression. Oncol. Rep. 37, 241–248. https://doi.org/10.3892/or.2016.5250 (2017).
Liu, S. et al. DGCR5 promotes gallbladder cancer by sponging miR-3619-5p via MEK/ERK1/2 and JNK/p38 MAPK pathways. J. Cancer. 11, 5466–5477. https://doi.org/10.7150/jca.46351 (2020).
Song, B. et al. LINC00882 plays a tumor-promoter role in colorectal cancer by targeting miR-3619-5p to up-regulate CTNNB1. Arch. Med. Res. 53, 29–36. https://doi.org/10.1016/j.arcmed.2021.06.001 (2022).
Zuo, X. et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. https://doi.org/10.1186/s13045-019-0839-x (2020).
Cheng, H. et al. PIM-1 mRNA expression is a potential prognostic biomarker in acute myeloid leukemia. J. Transl. Med. 15, 179. https://doi.org/10.1186/s12967-017-1287-4 (2017).
La Starza, R. et al. High PIM1 expression is a biomarker of T-cell acute lymphoblastic leukemia with JAK/STAT activation or t(6;7)(p21;q34)/TRB@-PIM1 rearrangement. Leukemia 32, 1807–1810. https://doi.org/10.1038/s41375-018-0031-2 (2018).
Keane, N. A., Reidy, M., Natoni, A. & Raab, M. S. O’Dwyer, M. Targeting the Pim kinases in multiple myeloma. Blood Cancer J. 5, e325. https://doi.org/10.1038/bcj.2015.46 (2015).
Chen, J. & Tang, G. PIM-1 kinase: A potential biomarker of triple-negative breast cancer. Onco Targets Ther. 12, 6267–6273. https://doi.org/10.2147/ott.S212752 (2019).
Wen, Q. L. et al. Role of oncogene PIM-1 in the development and progression of papillary thyroid carcinoma: Involvement of oxidative stress. Mol. Cell. Endocrinol. 523, 111144. https://doi.org/10.1016/j.mce.2020.111144 (2021).
Liu, H. T., Wang, N., Wang, X. & Li, S. L. Overexpression of Pim-1 is associated with poor prognosis in patients with esophageal squamous cell carcinoma. J. Surg. Oncol. 102, 683–688. https://doi.org/10.1002/jso.21627 (2010).
Wu, M. et al. Double mutant P53 (N340Q/L344R) promotes hepatocarcinogenesis through upregulation of Pim1 mediated by PKM2 and LncRNA CUDR. Oncotarget 7, 66525–66539. https://doi.org/10.18632/oncotarget.9089 (2016).
Kirschner, A. N. et al. PIM kinase inhibitor AZD1208 for treatment of MYC-driven prostate cancer. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/dju407 (2015).
Cortes, J. et al. Phase I studies of AZD1208, a proviral integration moloney virus kinase inhibitor in solid and haematological cancers. Br. J. Cancer 118, 1425–1433. https://doi.org/10.1038/s41416-018-0082-1 (2018).
Wu, J., Chu, E. & Kang, Y. PIM kinases in multiple myeloma. Cancers (Basel)https://doi.org/10.3390/cancers13174304 (2021).
Dutta, A. et al. Genetic ablation of Pim1 or pharmacologic inhibition with TP-3654 ameliorates myelofibrosis in murine models. Leukemia 36, 746–759. https://doi.org/10.1038/s41375-021-01464-2 (2022).
Sharma, A. et al. Assessment of structural and activity-related contributions of various PIM-1 kinase inhibitors in the treatment of leukemia and prostate cancer. Mol. Divers. https://doi.org/10.1007/s11030-023-10795-4 (2024).
Chen, S., Yang, Y., Yuan, Y. & Bo, L. Targeting PIM kinases in cancer therapy: an update on Pharmacological small-molecule inhibitors. Eur. J. Med. Chem. 264, 116016. https://doi.org/10.1016/j.ejmech.2023.116016 (2024).
Rice, T. W. et al. Recommendations for pathologic staging (pTNM) of cancer of the esophagus and esophagogastric junction for the 8th edition AJCC/UICC staging manuals. Dis. Esophagus 29, 897–905. https://doi.org/10.1111/dote.12533 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. https://doi.org/10.1093/nar/29.9.e45 (2001).
Komatsu, S. et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis 30, 1139–1146. https://doi.org/10.1093/carcin/bgp116 (2009).
Acknowledgements
Not applicable to this study.
Funding
The authors have no commercial or financial incentives related to the publication of this study.
Author information
Authors and Affiliations
Contributions
HA, SK, and TK designed the research, and SK and EO reviewed the research; HA, SK, TK, HK, RI, and YT performed the cell culture, molecular biology, and animal experiments; HA, SK, TK, KN, JK, TI, TO, HS, TA, HK, AS, TK, HF, and EO provided clinical specimens and performed the clinical data analyses. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Competing interests
The authors report no competing interest.
Ethics approval
Research Protocol Approval: This study received approval from the Institutional Ethics Review Board (ERB-C-319-1).
Informed consent
The study adhered to the Declaration of Helsinki, and written informed consent regarding the treatment and study participation was obtained from all patients prior to surgery. Animal Studies: The animal study protocol was approved by the Institutional Animal Care and Use Committee of Kyoto Prefectural University of Medicine (M2023-506). Accordance: We confirmed that all experiments in this study were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Arakawa, H., Komatsu, S., Kishimoto, T. et al. Low levels of tumor suppressor miR-3619 in plasma contribute to malignant outcomes and a target for nucleic acid therapy in esophageal cancer. Sci Rep 15, 27358 (2025). https://doi.org/10.1038/s41598-025-12246-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12246-6