Abstract
Dipeptidyl peptidase 4 (DPP-4) inhibitors (DPP-4i) are widely used to treat Type 2 diabetes (T2D) and are known for their cardiovascular and renal safety profiles. Systematic reviews have also shown that DPP-4i are associated with reduced dementia risk via unknown mechanisms. To examine vildagliptin (DPP-4i) effects on the intestinal microbiota in T2D patients, plasma metabolomics were conducted and inflammatory profiles collected to investigate correlations with potential neuroprotective effects. We examined 29 patients with T2D (not well controlled with metformin) before, and at 30 and 60 days after vildagliptin was introduced, and investigated intestinal microbiota, plasma metabolomic, and inflammatory profiles. In patients after 2 months, vildagliptin induced mild microbiota changes, represented by significant increases in Bariatricus and Butyricimonas genera and the Marinifilaceae family (short-chain fatty acids producers), reduced insulin, HOMA-IR, MCP1, and interferon (IFN)-γ levels, and elevated interleukin (IL)-4 and IL-10 levels, all of which represented an anti-inflammatory profile. Metabolomics results showed that leucine, 2-oxoisocaproate (branched-chain amino acid metabolite), and inosine were significantly reduced after vildagliptin was introduced. Additionally, choline, dimethylamine, and betaine levels were significantly higher, which may contribute to explain DPP-4i protective effects against dementia, as these metabolites are neuroprotective. In our T2D patient cohort (not well controlled with metformin), vildagliptin, in addition to improved glucose control and improved insulin resistance, modulated the intestinal microbiota, anti-inflammatory cytokine profiles, and metabolomics, and when combined, may contribute to explain DPP-4i’s neuroprotective effects.
Similar content being viewed by others
Introduction
Dipeptidyl peptidase 4 (DPP-4) inhibitors (DPP-4i) are widely used to treat Type 2 Diabetes (T2D) and are known for their cardiovascular and renal safety profiles1. Efficacious toward diabetes, these inhibitors improve glucose metabolism and pancreatic islet function2,3,4,5. Incretin-based DPP-4i therapies are based on the insulinotropic actions of glucagon-like peptide 1 (GLP-1)2. By increasing endogenous GLP-1 and insulin levels and reducing glucagon secretion6, DPP-4i effectively lower postprandial blood glucose levels by inhibiting incretin degradation. A previous systematic review and network meta-analysis comparing cognitive outcomes associated with antidiabetic agents demonstrated that DPP‐4i were associated with reduced dementia risk (Odds ratio = 0.78, 95% confidence interval (CI) 0.61–0.99)7. In a Bayesian network meta-analysis examining the impact of antidiabetic agents on dementia risk, DPP-4i similarly showed protective effects against Alzheimer’s disease when compared with no antidiabetic treatments (Hazard ratio = 0.48, 95% CI 0.25–0.92)8. Furthermore, animal studies also supported potential DPP-4i neuroprotective effects9,10,11,12,13,14,15,16,17.
While some studies have suggested links between DPP-4i and the intestinal microbiota18,19,20,21,22,23,24,25,26,27,28,29,30, few have been performed in humans, and without unclear correlation with microbiota and metabolic effect28,29. Moreover, in previous studies, correlations between DPP-4i-modulated microbiota, plasma metabolomics, insulin resistance, and inflammatory profiles were not investigated. To address this, we investigated vildagliptin (DPP-4i) effects on the intestinal microbiota, plasma metabolomics, and inflammatory profiles in T2D patients (not well controlled with metformin) to identify correlations with potential neuroprotective effects.
Methods
Study design and the patient population
The anthropometric and metabolic characteristics of T2D patients before and after 60 days of vildagliptin treatment are presented in Table 1. This longitudinal, paired, interventional study evaluated patients before, during, and after the intervention. Each participant served as their own control; therefore, a double-blind, placebo-controlled group was not included. Participants were recruited from the Public Health System. The protocol was approved by Research Ethics Committees at the University of Campinas (CAAE: 84087617.0.0000.5404) and was conducted in accordance with relevant guidelines and regulations. All patients involved in this study provided written informed consent.
Participants were 32–70 years old, of both sexes and diagnosed with T2DM (Table 1). HbA1c levels were 6.5–10%, body mass index 25–35 kg/m2, and a glomerular filtration rate was > 30 ml/min/1.75 m2. All participants had no prior DPP-4i use.
Exclusion criteria: type 1 diabetes, renal and hepatic insufficiency, pregnancy, and any intestinal pathology. Patients who had used DPP-4i, proton pump inhibitors, or antibiotics within 3 months before study commencement were also excluded. In total, 36 patients were selected; but three missed one fecal sample collection, two had compromised serum samples due to technical issues, and two experienced fecal and serum sample losses, leading to their exclusion. The remaining 29 patients completed the study.
Lipopolysaccharide (LPS) and interleukin serum levels
Serum was separated from bloods, diluted to 20% (v/v) in endotoxin-free water, and heated to 70 °C for 10 min to inactivate serum proteins. LPS was quantified using a commercial Limulus Amebocyte Assay kit (Cambrex, Walkersville, MD, USA) following the manufacturer’s instructions31. Samples were aliquoted for multiplex immunoassays (Bio-Plex 200; Bio-Rad Laboratories, Hercules, CA, USA), which used magnetic bead panels to analyze IFN-γ, IL-1-α, IL-1-α, IL-1RA, IL-4, IL-6, IL-8, IL-10, IL-13, IL-17A, IL-18, MCP-1, MCSF, and TNF-α (HCYTOMAG-60K and HMMP2MAG-55K-01, MILLIPLEX MAP Human; Millipore Sigma, Merck KGaA).
Metabolite quantification
Metabolites were processed and quantified using Nuclear Magnetic Resonance Suite software v.8.1 (Chenomx Inc™, Edmonton, AB, Canada). The processor module was used to adjust the spectral phase and perform baseline corrections. A 0.5 Hz line-broadening function was used to reduce signal noise and fit metabolite signals to spectral peaks. The water signal was suppressed, and spectra were calibrated using the TMSP-d4 reference signal at 0.5 mM. Spectra were then individually transferred to a Profiling module to determine metabolomic profiles. Metabolites were identified and concentrations measured. Metabolite concentration data were exported to Excel® (Microsoft Office™ 365) and normalized as required32.
Microbiota analysis
Stool samples were collected before, and at 30 and 60 days after vildagliptin was introduced, stored at − 80 °C, and processed in a controlled environment to minimize contamination. Genomic DNA was extracted from a 200 mg sample using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). A negative control (water from the mini kit) was used from extraction through to final sequencing steps, and a mock microbial DNA community standard was used as a positive control (ZymoBIOMICS, Irvine, CA, USA). For each sample the V3–V4 hyper-variable region (Primer Forward TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG; Reverse GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC) of the bacterial 16S rRNA gene was amplified followed by Illumina 16S Metagenomic Sequencing Library Preparation guide. The taxonomic composition of the bacterial communities was obtained by analyzing the V3–V4 region of the 16S rRNA gene using the Illumina® MiSeq platform. The constructions of the DNA sequencing libraries were performed according to the manufacturer’s instructions (Illumina, San Diego, CA, USA) and followed the same flow described by Caporaso et al.33. The fastq sequences were analyzed using the Illumina 16S Metagenomics software.
Statistical analysis
Quantitative variables were presented according to distribution patterns: mean and standard deviation, or median, minimum, and maximum values. Student’s t-tests were used to compare two independent samples, and non-parametric (Mann–Whitney) tests were used for non-normally distributed variables. Categorical variables were presented as proportions, and either Pearson χ2 or Fisher’s exact tests were used to compare two proportions from independent samples. The taxonomic classification was performed using the DADA2 pipeline and the GreenGenes2 database34,35. Paired abundance analyzes were performed using the IBM SPSS® 20.0 software (Wilcoxon Signed Ranks Test). The analysis of alpha (Metrics: Pielou’s evenness) and beta diversity (Metrics: Unweighted UniFrac, weighted UniFrac ) was performed using QIIME36. The graphs were generated by GraphPad Prism 7.0 and Graphical Software: R (version 4.3.1). A 5% significance level was adopted in statistical tests.
Results
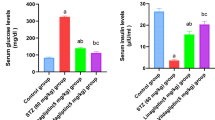
As expected, in T2D patients (not well controlled with metformin), vildagliptin use for 2 months improved metabolic control, as evidenced by significant decreases in fasting plasma glucose levels, and more importantly, HbA1c levels. Although fasting insulin levels did not change after administration, a clear and significant decrease in HOMA-IR was recorded, indicating improved insulin sensitivity (Fig. 1).
Vildagliptin effects on glycemia, HbA1c, and HOMA-IR over time (0 and 60 days). Data points represent mean values, with error bars indicating standard deviation. Statistical comparisons between time points are shown, with p values indicating significant differences: Glycemia: Significant reduction from day 0 to 60 (p = 0.0194). HbA1c: Significant decrease from day 0 to 60 (p = 2.85e−07). HOMA-IR: Significant difference from day 0 to 60 (p = 0.0492).
Lipopolysaccharide (LPS) levels showed no statistically significant differences between groups. To investigate inflammatory patterns, we examined expression of the following molecules: interferon (IFN)-γ, interleukin (IL)-1-α, IL-1-β, IL-1RA, IL-4, IL-6, IL-8, IL-10, IL-13, IL-17A, IL-18, MCP-1, MCSF, and TNF-α (Fig. S1). Most cytokines levels were similar between groups, but IL-4 and IL-10 levels (anti-inflammatory ILs) were higher at 2 months after vildagliptin administration, while MCP1 levels (inflammatory cytokine) were reduced (Fig. 2).
Cytokine (IL-10, IL-4, IFN-γ, and MCP-1) level changes over time (0 and 60 days). Data points represent mean values, with error bars indicating standard deviation. Statistical comparisons between time points are shown, with p values indicating significant differences: IL-10: Increase from day 0 to 60 (p = 0.0097), IL-4: Increase from day 0 to 60 (p = 0.00542). IFN-γ: Significant difference from day 0 to 60 (p = 0.0370). MCP-1: Significant difference from day 0 to 60 (p = 0.0202).
Metabolomics and microbiota analysis
We performed metabolomics analyses in patients before and 60 days after vildagliptin administration (Fig. S1). Levels of leucine, inosine, and the BCAA 2-oxoisocaproate were significantly reduced after vildagliptin was introduced (Fig. 3); however, choline, dimethylamine, and betaine levels were significantly higher (Fig. 3).
Changes in metabolite levels (leucine, 2-oxoisoacropate, choline, dimethylamine, betaine, aspartate, inosine, and serine) over time (0 and 60 days). Data points represent mean values, with error bars indicating standard deviation. Statistical comparisons between time points are shown, with p values indicating significant differences: Leucine: Significant decrease from day 0 to 60 (p = 0.0129). 2-Oxoisoacropate: Increase from day 0 to 60 (p = 0.0122). Choline: Significant increase from day 0 to 60 (p = 0.0077). Dimethylamine: Increase from day 0 to 60 (p = 0.0002). Betaine: Significant difference from day 0 to 60 (p = 0.0451). Aspartate: Significant increase from day 0 to 60 (p = 0.0408). Inosine: Significant increase from day 0 to 60 (p = 0.0386). Serine: Increased from day 0 to 60 (p = 0.0003).
We also performed microbiota compositional analyses on patients before and 60 days after vildagliptin was introduced. The Bray–Curtis index reflects microbial β-diversity, which assesses compositional dissimilarities between microbial communities, accounting for species presence and abundance. A pseudo-F value = 1.2655 indicated a slight difference between groups 0 and 60, with a p value = 0.053, suggesting a potential trend toward a significant difference. However, a q-value = 0.159 indicated that this difference was not statistically significant after correction for multiple testing (Fig. 4).
Gut microbiota analysis. (A) Beta diversity. The beta diversity based on Bray–Curtis dissimilarity is presented in a non-metric multidimensional scaling (NMDS) plot showing the microbiota dissimilarity between groups. UniFrac metric measures the phylogenetic distance between sets of microbial communities (A pseudo-F value of 1.2655 for 0 and 60, with a p value = 0.04 and q-value = 0.159). Box plots showing α-diversity metrics across different time points (0 and 60). Metrics included (B) Observed Features, (C) Inverse Simpson, (D) Shannon, (E) Pielou’s Evenness, and (F) Simpson’s Evenness. Box plots show the distribution of values for each time period, with 0 in red and 60 in blue. These metrics highlighted species richness, diversity, and evenness at different time points. Groups are represented by different colors: 0 is red and 60 days blue. (G) Linear Discriminant Analysis (LDA) scores of enriched microbial taxa in 60 days. Taxa include Bariatricus and Butyricimonas genera and the Marinifilaceae family. Higher LDA scores (log10) indicated a greater taxa contribution to the unique microbial profile at this period, suggesting significant enrichment when compared to day 0.
We also analyzed α-diversity using several indices, but only the Simpson_evenness index showed a significant difference across periods. This index, a Simpson index variant, measures α-diversity with a focus on species dominance and evenness. Kruskal–Wallis tests showed significant variable distribution differences among groups. Pairwise comparisons showed that 0 was significantly different to 60 (p value = 0.0032 and q-value = 0.0096, respectively). A reduced Simpson_evenness index indicated diversity loss or a shift in microbiota composition, resulting in lower uniformity at 60 (Fig. 4F).
In 60, a significant increase in Bariatricus and Butyricimonas genera and the Marinifilaceae family was observed when compared to 0. These taxa showed p- and adjusted p-values (p_adj) indicating statistical significance (p < 0.05), along with high Linear Discriminant Analysis (LDA) scores (log10), reinforcing their enrichment at this time point (Fig. 4G).
Discussion
In T2D patients (not well controlled with metformin), vildagliptin, in addition to improved glucose control, improved insulin resistance and modulated intestinal microbiota, accompanied by anti-inflammatory cytokine profiles and metabolomics. The integration of these data may have a role in neuroprotective effects of DPP-4i.
Improved insulin resistance, as demonstrated by increased HOMA-IR, was also accompanied by improvements in insulin resistance markers, such as BCAAs and/or their metabolites. BCAAs are clearly correlated with insulin resistance, and in some situations may predict T2D development37,38. We showed that vildagliptin reduced leucine and 2-oxoisocaproate (valine metabolite), suggesting that improved insulin resistance was not only related to glucose metabolism, but broadly, may also have affected amino acid metabolism.
In patients, the gut microbiota showed minimal changes after vildagliptin was introduced (60 days), though some important bacterial genera were increased. A reduced Simpson_e index at 60 days was likely due to increased Bariatricus and Butyricimonas genera and the Marinifilaceae family. This increased microbial subset suggested a distinctive microbial composition at 60 days, marked by higher genera levels. LDA scores indicated that taxa enriched at 60 days significantly contributed to the unique characteristics of this group. Such a microbial profile may be associated with beneficial functions, such as short-chain fatty acids (SCFA) production, particularly butyrate39, which exerts anti-inflammatory properties and has promotional effects on intestinal barrier integrity. Thus, microbiota changes by day 60 had positive implications for modulating inflammation and intestinal health.
Previous studies have also investigated the effects of DPP-4 inhibitors on gut microbiota modulation. For instance, Smits et al.26 reported no alterations in gut microbial diversity following treatment with liraglutide and sitagliptin, whereas in our study, vildagliptin treatment resulted in modest but significant changes, specifically increases in the genera Bariatricus and Butyricimonas, as well as the Marinifilaceae family. Furthermore, while Smits et al. observed no significant changes in choline or betaine levels, we found a significant increase in these neuroprotective metabolites following vildagliptin administration. Similarly, whereas Martínez-López et al.29 examined the effects of linagliptin combined with metformin, our study focused specifically on vildagliptin as add-on therapy.
Liao et al.23 investigated the effects of sitagliptin on gut microbiota and demonstrated that microbiota from sitagliptin-treated T2D patients, when transplanted into germ-free mice fed a high-fat diet (HFD), improved HFD-induced glucose intolerance in the recipient mice—suggesting that DPP-4 inhibitor-induced microbiota alterations may at least partially contribute to their hypoglycemic effects.
Recent data have shown that the intestinal microbiota express DPP-4, but human DPP-4i do not efficiently block this expression40,41,42. Although vildagliptin showed the lowest binding affinity to bacterial DPP-4 when compared with other human DPP-4i42, we cannot exclude the possibility that some of our data are secondary to partial bacterial DPP-4 blockage, in addition to efficient blockage by human DPP-4.
Growing evidence now indicates that gliptins exert neuroprotective effects in diabetes management; patients with T2D mellitus (T2DM) treated with DPP-4i showed improved functional outcomes and reduced mortality rates following acute ischemic stroke43,44. Additionally, elderly diabetic patients with Alzheimer’s disease receiving sitagliptin also demonstrated enhanced cognitive function45.
Gliptin neuroprotective effects are also substantiated in animal studies; vildagliptin and sitagliptin both mitigated hippocampal mitochondrial dysfunction and improved learning in adult rats on a high-fat diet9,10. Pretreatment with linagliptin also reduced brain infarction size following transient middle cerebral artery occlusion in T2DM mice11. Notably, gliptin-induced neuroprotection extended beyond diabetic models with dietary-induced abnormalities, as agents also showed protective effects in Alzheimer’s12,13 and Parkinson’s14 disease models. In non-diabetic animals, gliptins reduced neurodegeneration after acute focal cerebral ischemia11 and chronic cerebral hypoperfusion15. Recent research in a stereotactic cryo-lesion model showed that pre-surgical sitagliptin administration reduced lesion size and activated a neuroprotective cAMP response element-binding protein pathway in female C57BL/6J mice16.
Collectively, these findings suggest that gliptins may exert preventive neuroprotective effects in both diabetic and non-diabetic models. Moreover, chronic or prophylactic vildagliptin administration prevented neurodegeneration in animal models of Alzheimer’s disease13, Parkinson’s disease14, diabetic vascular dementia17, and insulin resistance induced by a high-fat diet9,10.
Our data showing increased circulating choline and betaine levels after vildagliptin administration suggested a possible mechanism that accounted, at least in part, for DPP-4i neuroprotective effects. These metabolites can be increased by the diet and endogenous sources, while betaine is produced by the intestinal microbiota46,47. Previous studies identified relationships between decreased choline intake and increased risks for cognitive decline and Alzheimer’s disease48,49,50, while a disease-associated reduction in serum choline in Alzheimer’s disease patients was identified when compared with controls48. Additionally, patients with cerebral amyloid angiopathy and cerebral white matter rarefaction had lower choline levels when compared with controls, emphasizing a putative choline “link” to white matter integrity. Patients with mild cognitive impairment showed reduced circulating choline levels, with acetylcholine levels unchanged, suggesting that initial Alzheimer’s disease development may be linked to reduced choline levels, but only in later disease stages is there a reduction in acetylcholine48. Recent data also suggested that betaine exerted beneficial effects on obesity, diabetes, cancer, and neurodegenerative diseases51. The neuroprotective role of betaine may be related to its anti-inflammatory effects, inhibiting nuclear factor-κB activity and NLRP3 inflammasome activation and counteracting oxidative stress. Additionally, betaine also alleviates endoplasmic reticulum stress and inhibits apoptosis52.
We also observed increased IL-10 and IL-4 levels accompanied by decreased MCP-1 and INF-α levels, which suggested an immunological profile with potentially neuroprotective effects53. IL-10 is an anti-inflammatory cytokine with neuroprotective effects against multiple sclerosis, traumatic brain injury, amyotrophic lateral sclerosis, Alzheimer´s disease, and Parkinson´s disease54, with effects partly mediated by attenuating neuroinflammatory responses55 and promoting axon remyelination56.
Reduced circulating MCP-1 levels induced by vildagliptin may also have contributed to its neuroprotection effects, as MCP-1 is a proinflammatory cytokine which breaks down the blood–brain barrier and recruits and activates glia cells and macrophages57. IL-4 is secreted by Th2 cells and has critical roles in modulating physiological functions in the central nervous system and mitigating neuroinflammatory processes58,59,60. IL-4 also modulates immune responses and neuroinflammation and was shown to induce neuroprotective and neurorepair effects in different central nervous system experimental disease models61. Such neuroprotection mechanisms are not completely understood, but may include an induced polarized microglial/macrophage phenotype and gene expression reflecting an M2 microglia phenotype62.
Although increased choline levels may exert neuroprotective effects, this increase, associated with increased dimethylamine levels, may also increase cardiovascular disease (CVD) risks. A recent meta-analysis showed that higher circulating choline levels were associated with a higher CVD risk and all-cause mortality63. Moreover, increased circulating dimethylamine, which is methylamine gut microbiota-dependent, may also have detrimental cardiovascular risk effects64. However, previous data also showed that circulating aspartate and serine levels had negative correlations with CVDs65,66. Thus, increased aspartate and serine levels induced by vildagliptin may exert beneficial effects on cardiovascular risk. Taken together, these data suggest that although DPP-4i improves glycemic control and insulin resistance and increases aspartate and serine levels, increased circulating choline and dimethylamine levels may prevent any beneficial cardiovascular effects, contributing to explain the neutral effect on MACE67,68,69.
While our study did not include functional assays such as fecal microbiota transplantation, our findings reveal a correlation between vildagliptin treatment and an immunometabolic profile consistent with neuroprotection. This correlation, supported by existing literature on the individual roles of the metabolites and cytokines we observed, may contribute to the growing body of evidence regarding the neuroprotective potential of DPP-4 inhibitors.
The present study has certain limitations that should be acknowledged. First, we did not assess the isolated effects of vildagliptin on microbiota modulation and the immunometabolic profile, as all patients were also receiving metformin, which is known to significantly influence microbiota composition70,71. Additionally, we cannot exclude the possibility that improvements in glycemic control themselves contributed to changes in gut microbiota and intestinal metabolites. Previous studies have demonstrated that glycemic control, independent of the therapeutic agent used, can modulate the gut microbiota72,73. However, the patterns of modulation reported in prior studies differ from those observed with vildagliptin in our study, suggesting that the effects we describe may be specifically attributable to vildagliptin.
Moreover, the lack of a placebo control group (due to ethical constraints) is another limitation of our study. Nonetheless, our findings provide valuable insights into the combined effects of vildagliptin on metabolic control, microbiota modulation, and inflammatory profiles, which collectively may contribute to neuroprotection in a real-world clinical setting for T2D patients inadequately controlled with metformin alone.
Therefore, despite the absence of a placebo control group, our findings advance the understanding of the multifaceted actions of vildagliptin—particularly its unique effects on the metabolome and gut microbiota within this specific patient cohort—which were not fully elucidated in previous studies. We believe that these novel correlations offer important insights into the potential mechanisms underlying the neuroprotective effects of DPP-4 inhibitors.
In summary, we showed that vildagliptin improved insulin resistance, induced an intestinal microbial profile associated with beneficial functions (SCFA production), and an anti-inflammatory cytokine profile. A slightly modulated microbiota was also accompanied by increased metabolite levels associated with possible roles in neuroprotection. Combined, these data may contribute to explain some of the neuroprotective effects associated with DPP-4i (Fig. 5).
Vildagliptin improved insulin resistance and promoted an intestinal microbial profile linked to beneficial functions, including SCFA production and an anti-inflammatory cytokine profile. Additionally, the slight modulation of the microbiota was accompanied by increased metabolite levels associated with neuroprotection. Together, these findings provide a potential explanation for the neuroprotective effects observed with DPP-4 inhibitors.
Data availability
All the data produced or examined during this study are presented within this article. The 16S rRNA sequence dataset has been deposited in the BioProject repository (PRJNA1219131 https://dataview.ncbi.nlm.nih.gov/object/PRJNA1219131?reviewer=q28vjp4m2q43pc7mkpbenfq2fk). Licensing rights Figure Title: Fig. 5 Created in BioRender. Saad, M. (2025) https://BioRender.com/m07k176.
References
Carr, R. D. & Solomon, A. Inhibitors of dipeptidyl peptidase-4 as therapeutic agents for individuals with type 2 diabetes: A 25-year journey. Diabet. Med. 37(8), 1230–1233. https://doi.org/10.1111/dme.14325 (2020).
Mulvihill, E. E. & Drucker, D. J. Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr. Rev. 35(6), 992–1019. https://doi.org/10.1210/er.2014-1035 (2014).
Kushwaha, R. N., Haq, W. & Katti, S. B. Sixteen-years of clinically relevant dipeptidyl peptidase-IV (DPP-IV) inhibitors for treatment of type-2 diabetes: A perspective. Curr. Med. Chem. 21(35), 4013–4045. https://doi.org/10.2174/0929867321666140915143309 (2014).
Waget, A. et al. Physiological and pharmacological mechanisms through which the DPP-4 inhibitor sitagliptin regulates glycemia in mice. Endocrinology 152(8), 3018–3029. https://doi.org/10.1210/en.2011-0286 (2011).
Mulvihill, E. E. et al. Cellular sites and mechanisms linking reduction of dipeptidyl peptidase-4 activity to control of incretin hormone action and glucose homeostasis. Cell Metab. 25(1), 152–165. https://doi.org/10.1016/j.cmet.2016.10.007 (2017).
He, Y. L. Clinical pharmacokinetics and pharmacodynamics of vildagliptin. Clin. Pharmacokinet 51(3), 147–162. https://doi.org/10.2165/11598080-000000000-00000 (2012).
Tian, S. et al. Comparison on cognitive outcomes of antidiabetic agents for type 2 diabetes: A systematic review and network meta-analysis. Diabetes Metab. Res. Rev. 39(7), e3673. https://doi.org/10.1002/dmrr.3673 (2023).
Zhou, J. B. et al. Impact of antidiabetic agents on dementia risk: A Bayesian network meta-analysis. Metabolism 109, 154265. https://doi.org/10.1016/j.metabol.2020.154265 (2020).
Pipatpiboon, N. et al. DPP4-inhibitor improves neuronal insulin receptor function, brain mitochondrial function and cognitive function in rats with insulin resistance induced by high-fat diet consumption. Eur. J. Neurosci. 37(5), 839–849. https://doi.org/10.1111/ejn.12088 (2013).
Pintana, H. et al. DPP-4 inhibitors improve cognition and brain mitochondrial function of insulin-resistant rats. J. Endocrinol. 218(1), 1–11. https://doi.org/10.1530/JOE-12-0521 (2013).
Darsalia, V. et al. The DPP-4 inhibitor linagliptin counteracts stroke in the normal and diabetic mouse brain: A comparison with glimepiride. Diabetes 62(4), 1289–1296. https://doi.org/10.2337/db12-0988 (2013).
Kosaraju, J. et al. Saxagliptin: A dipeptidyl peptidase-4 inhibitor ameliorates streptozotocin induced Alzheimer’s disease. Neuropharmacology 72, 291–300. https://doi.org/10.1016/j.neuropharm.2013.04.008 (2013).
Kosaraju, J. et al. Vildagliptin: An anti-diabetes agent ameliorates cognitive deficits and pathology observed in streptozotocin-induced Alzheimer’s disease. J. Pharm. Pharmacol. 65(12), 1773–1784. https://doi.org/10.1111/jphp.12148 (2013).
Abdelsalam, R. M. & Safar, M. M. Neuroprotective effects of vildagliptin in rat rotenone Parkinson’s disease model: Role of RAGE-NFκB and Nrf2-antioxidant signaling pathways. J. Neurochem. 133(5), 700–707. https://doi.org/10.1111/jnc.13087 (2015).
Tsai, T. H. et al. Sitagliptin attenuated brain damage and cognitive impairment in mice with chronic cerebral hypo-perfusion through suppressing oxidative stress and inflammatory reaction. J. Hypertens. 33(5), 1001–1013. https://doi.org/10.1097/HJH.0000000000000529 (2015).
DellaValle, B. et al. Oral administration of sitagliptin activates CREB and is neuroprotective in murine model of brain trauma. Front. Pharmacol. 7, 450. https://doi.org/10.3389/fphar.2016.00450 (2016).
Jain, S. & Sharma, B. Neuroprotective effect of selective DPP-4 inhibitor in experimental vascular dementia. Physiol. Behav. 152(Pt A), 182–193. https://doi.org/10.1016/j.physbeh.2015.09.007 (2015).
Reimer, R. A. et al. Combining sitagliptin/metformin with a functional fiber delays diabetes progression in Zucker rats. J. Endocrinol. 220(3), 361–373. https://doi.org/10.1530/JOE-13-0484 (2014).
Yan, X. et al. Microflora disturbance during progression of glucose intolerance and effect of sitagliptin: An animal study. J. Diabetes Res. 2016, 2093171. https://doi.org/10.1155/2016/2093171 (2016).
Wang, L. et al. Structural modulation of the gut microbiota and the relationship with body weight: Compared evaluation of liraglutide and saxagliptin treatment. Sci. Rep. 6, 33251. https://doi.org/10.1038/srep33251 (2016).
Zhang, Q. et al. Vildagliptin increases butyrate-producing bacteria in the gut of diabetic rats. PLoS ONE 12(10), e0184735. https://doi.org/10.1371/journal.pone.0184735 (2017).
Olivares, M. et al. The DPP-4 inhibitor vildagliptin impacts the gut microbiota and prevents disruption of intestinal homeostasis induced by a Western diet in mice. Diabetologia 61(8), 1838–1848. https://doi.org/10.1007/s00125-018-4647-6 (2018).
Liao, X. et al. Alteration of gut microbiota induced by DPP-4i treatment improves glucose homeostasis. EBioMedicine 44, 665–674. https://doi.org/10.1016/j.ebiom.2019.03.057 (2019).
Ryan, P. M. et al. Metformin and dipeptidyl peptidase-4 inhibitor differentially modulate the intestinal microbiota and plasma metabolome of metabolically dysfunctional mice. Can. J. Diabetes 44(2), 146–155. https://doi.org/10.1016/j.jcjd.2019.05.008 (2020).
Zhang, M. et al. Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in Zucker diabetic fatty rats. BMJ Open Diabetes Res. Care 7(1), e000717. https://doi.org/10.1136/bmjdrc-2019-000717 (2019).
Smits, M. M. et al. Liraglutide and sitagliptin have no effect on intestinal microbiota composition: A 12-week randomized placebo-controlled trial in adults with type 2 diabetes. Diabetes Metab. 47(5), 101223. https://doi.org/10.1016/j.diabet.2021.101223 (2021).
Silva-Veiga, F. M. et al. Peroxisome proliferator-activated receptor-alpha activation and dipeptidyl peptidase-4 inhibition target dysbiosis to treat fatty liver in obese mice. World J. Gastroenterol. 28(17), 1814–1829. https://doi.org/10.3748/wjg.v28.i17.1814 (2022).
Zhang, X. et al. Metagenomic analysis reveals crosstalk between gut microbiota and glucose-lowering drugs targeting the gastrointestinal tract in Chinese patients with type 2 diabetes: A 6 month, two-arm randomised trial. Diabetologia 65(10), 1613–1626. https://doi.org/10.1007/s00125-022-05768-5 (2022).
Martínez-López, Y. E. et al. Effect of metformin and metformin/linagliptin on gut microbiota in patients with prediabetes. Sci. Rep. 14(1), 9678. https://doi.org/10.1038/s41598-024-60081-y (2024).
Tang, Y. et al. Effects of metformin, saxagliptin and repaglinide on gut microbiota in high-fat diet/streptozocin-induced type 2 diabetic mice. BMJ Open Diabetes Res. Care 12(3), e003837. https://doi.org/10.1136/bmjdrc-2023-003837 (2024).
Assalin, H. B. et al. Proton pump inhibitor pantoprazole modulates intestinal microbiota and induces TLR4 signaling and fibrosis in mouse liver. Int. J. Mol. Sci. 23(22), 13766. https://doi.org/10.3390/ijms232213766 (2022).
Tobar, N. et al. Metformin acts in the gut and induces gut-liver crosstalk. Proc. Natl. Acad. Sci. U.S.A. 120(4), e2211933120. https://doi.org/10.1073/pnas.2211933120 (2023).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6(8), 1621–1624. https://doi.org/10.1038/ismej.2012.8 (2012).
McDonald, D. et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 42(5), 715–718. https://doi.org/10.1038/s41587-023-01845-1 (2024).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13(7), 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
Kuczynski, J. et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. 27, 1–5. https://doi.org/10.1002/9780471729259.mc01e05s27 (2012).
Wang, T. J. et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 17(4), 448–453. https://doi.org/10.1038/nm.2307 (2011).
Langenberg, C. & Savage, D. B. An amino acid profile to predict diabetes?. Nat. Med. 17(4), 418–420. https://doi.org/10.1038/nm0411-418 (2011).
Yuille, S. et al. Human gut bacteria as potent class I histone deacetylase inhibitors in vitro through production of butyric acid and valeric acid. PLoS ONE 13(7), e0201073. https://doi.org/10.1371/journal.pone.0201073 (2018).
Clausen, D. J., Kanitra, J. & Bendix, S. Hybrid staged approach to subclavian artery aneurysm repair with aberrant dominant left vertebral artery. J. Surg. Case Rep. 2023(7), rjad405. https://doi.org/10.1093/jscr/rjad405 (2023).
Yasuda, T., Harada, N. & Inagaki, N. Effects of dipeptidyl peptidase-4 inhibition in vivo: Dipeptidyl peptidase-4 inhibitor/gut microbiome crosstalk suggests novel therapeutic options for diabetes management. J. Diabetes Investig. 15(6), 704–706. https://doi.org/10.1111/jdi.14157 (2024).
Carpio, L. E. et al. Comparative binding study of gliptins to bacterial DPP4-like enzymes for the treatment of type 2 diabetes mellitus (T2DM). Int. J. Mol. Sci. 25(11), 5744. https://doi.org/10.3390/ijms25115744 (2024).
Chen, D. Y. et al. Sitagliptin after ischemic stroke in type 2 diabetic patients: A nationwide cohort study. Medicine (Baltimore) 94(28), e1128. https://doi.org/10.1097/MD.0000000000001128 (2015).
Tziomalos, K. et al. Prior treatment with dipeptidyl peptidase 4 inhibitors is associated with better functional outcome and lower in-hospital mortality in patients with type 2 diabetes mellitus admitted with acute ischaemic stroke. Diabetes Vasc. Dis. Res. 12(6), 463–466. https://doi.org/10.1177/1479164115597867 (2015).
Isik, A. T. et al. The effects of sitagliptin, a DPP-4 inhibitor, on cognitive functions in elderly diabetic patients with or without Alzheimer’s disease. Diabetes Res. Clin. Pract. 123, 192–198. https://doi.org/10.1016/j.diabres.2016.12.010 (2017).
Schratz, W. W. Pulse oximetry: A review with emphasis on applications in dentistry. Anesth. Prog. 34(3), 100–101 (1987).
Haikonen, R. et al. Diet- and microbiota-related metabolite, 5-aminovaleric acid betaine (5-AVAB), in health and disease. Trends Endocrinol. Metab. 33(7), 463–480. https://doi.org/10.1016/j.tem.2022.04.004 (2022).
Judd, J. M. et al. Inflammation and the pathological progression of Alzheimer’s disease are associated with low circulating choline levels. Acta Neuropathol. 146(4), 565–583. https://doi.org/10.1007/s00401-023-02616-7 (2023).
Yuan, J. et al. Is dietary choline intake related to dementia and Alzheimer’s disease risks? Results from the Framingham Heart Study. Am. J. Clin. Nutr. 116(5), 1201–1207. https://doi.org/10.1093/ajcn/nqac193 (2022).
Liu, L. et al. Choline intake correlates with cognitive performance among elder adults in the United States. Behav. Neurol. 2021, 2962245. https://doi.org/10.1155/2021/2962245 (2021).
Zhang, A. et al. Causal association between plasma metabolites and neurodegenerative diseases. Prog. Neuropsychopharmacol. Biol. Psychiatry 134, 111067. https://doi.org/10.1016/j.pnpbp.2024.111067 (2024).
Bhatt, S. P. et al. Dupilumab for COPD with type 2 inflammation indicated by eosinophil counts. N. Engl. J. Med. 389(3), 205–214. https://doi.org/10.1056/NEJMoa2303951 (2023).
Hung, Y. W., Wang, Y. & Lee, S. L. DPP-4 inhibitor reduces striatal microglial deramification after sensorimotor cortex injury induced by external force impact. FASEB J. 34(5), 6950–6964. https://doi.org/10.1096/fj.201902818R (2020).
Porro, C., Cianciulli, A. & Panaro, M. A. The regulatory role of IL-10 in neurodegenerative diseases. Biomolecules 10(7), 1017. https://doi.org/10.3390/biom10071017 (2020).
Ewen, T. et al. Neuroprotective effect of atorvastatin involves suppression of TNF-α and upregulation of IL-10 in a rat model of intracerebral hemorrhage. Cell Biochem. Biophys. 66(2), 337–346. https://doi.org/10.1007/s12013-012-9453-z (2013).
Atkins, S. et al. Interleukin-10 reduces scarring and enhances regeneration at a site of sciatic nerve repair. J. Peripher. Nerv. Syst. 12(4), 269–276. https://doi.org/10.1111/j.1529-8027.2007.00148.x (2007).
Manley, N. C. et al. Characterization of monocyte chemoattractant protein-1 expression following a kainate model of status epilepticus. Brain Res. 1182, 138–143. https://doi.org/10.1016/j.brainres.2007.08.092 (2007).
Zhang, J. et al. IL4-driven microglia modulate stress resilience through BDNF-dependent neurogenesis. Sci. Adv. 7(12), eabb9888. https://doi.org/10.1126/sciadv.abb9888 (2021).
Hasan, M. et al. Increased levels of brain serotonin correlated with MMP-9 activity and IL-4 levels resulted in severe experimental autoimmune encephalomyelitis (EAE) in obese mice. Neuroscience 319, 168–182. https://doi.org/10.1016/j.neuroscience.2016.01.045 (2016).
Wang, J. et al. Interleukin-4 modulates neuroinflammation by inducing phenotypic transformation of microglia following subarachnoid hemorrhage. Inflammation 47(1), 390–403. https://doi.org/10.1007/s10753-023-01917-z (2024).
Gärtner, Y. et al. Interleukin-4 as a therapeutic target. Pharmacol. Ther. 242, 108348. https://doi.org/10.1016/j.pharmthera.2023.108348 (2023).
Zhao, X. et al. Neuronal interleukin-4 as a modulator of microglial pathways and ischemic brain damage. J. Neurosci. 35(32), 11281–11291. https://doi.org/10.1523/JNEUROSCI.1685-15.2015 (2015).
Yang, Q. et al. Association of choline and betaine with the risk of cardiovascular disease and all-cause mortality: Meta-analysis. Eur. J. Clin. Investig. 53(10), e14041. https://doi.org/10.1111/eci.14041 (2023).
Hsu, C. N. et al. Association of trimethylamine, trimethylamine N-oxide, and dimethylamine with cardiovascular risk in children with chronic kidney disease. J. Clin. Med. 9(2), 336. https://doi.org/10.3390/jcm9020336 (2020).
Hu, S. et al. Causal relationships of circulating amino acids with cardiovascular disease: A trans-ancestry Mendelian randomization analysis. J. Transl. Med. 21(1), 699. https://doi.org/10.1186/s12967-023-04580-y (2023).
Prechtl, L. et al. Circulating amino acid signature features urea cycle alterations associated with coronary artery disease. Sci. Rep. 14(1), 25848. https://doi.org/10.1038/s41598-024-76835-7 (2024).
Green, J. B. et al. Effect of sitagliptin on cardiovascular outcomes in type 2 diabetes. N. Engl. J. Med. 373(3), 232–242. https://doi.org/10.1056/NEJMoa1501352 (2015).
Scirica, B. M. et al. Saxagliptin and cardiovascular outcomes in patients with type 2 diabetes mellitus. N. Engl. J. Med. 369(14), 1317–1326. https://doi.org/10.1056/NEJMoa1307684 (2013).
Scirica, B. M., Braunwald, E. & Bhatt, D. L. Saxagliptin, alogliptin, and cardiovascular outcomes. N. Engl. J. Med. 370(5), 483–484. https://doi.org/10.1056/NEJMc1313880 (2014).
Bauer, P. V. et al. Metformin alters upper small intestinal microbiota that impact a glucose-SGLT1-sensing glucoregulatory pathway. Cell Metab. 27(1), 101–17.e5. https://doi.org/10.1016/j.cmet.2017.09.019 (2018).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528(7581), 262–266. https://doi.org/10.1038/nature15766 (2015).
Anhê, F. F., Barra, N. G. & Schertzer, J. D. Glucose alters the symbiotic relationships between gut microbiota and host physiology. Am. J. Physiol. Endocrinol. Metab. 318(2), E111–E116. https://doi.org/10.1152/ajpendo.00485.2019 (2020).
Napoli, T. F. et al. Unveiling contrasts in microbiota response: A1c control improves dysbiosis in low-A1c T2DM, but fails in high-A1c cases-a key to metabolic memory?. BMJ Open Diabetes Res. Care 12(3), e003964. https://doi.org/10.1136/bmjdrc-2023-003964 (2024).
Acknowledgements
This study was supported by funding from INCT Obesidade e Diabetes CNPq (465693/2014-8) and FAPESP (2014/50907-5).
Author information
Authors and Affiliations
Contributions
JCCM, M.J.A.S. Methodology: JCCM, M.J.A.S. Investigation: A.S., D.G., H.B.A., D.O.M., E.S.O. Data analysis: A.S., D.G., H.B.A., D.O.M., E.S.O. Metabolomic analysis: M.R.A., M.L.S., S.A.R. Writing—original draft: JCCM, A.S., M.J.A.S. Writing—review and editing: JCCM, A.S., M.J.A.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
de Macedo, J.C.C., Guadagnini, D., Assalin, H.B. et al. Vildagliptin modulates the microbiota and induces an immunometabolic profile compatible with neuroprotection in type 2 diabetes. Sci Rep 15, 27932 (2025). https://doi.org/10.1038/s41598-025-12990-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12990-9