Abstract
Umbilical cord-derived human mesenchymal stem cells (UC-hMSCs) are multipotent stem cells with great potential for treating bone diseases. Although they can be easily isolated from umbilical cord tissue, their osteogenic differentiation is less efficient than differentiation of bone marrow-derived hMSCs (BM-hMSCs). Improving osteogenic differentiation of UC-hMSCs is essential for their clinical use. This study identified specific microRNAs (miRNAs) that inhibit osteogenic differentiation and explored their regulatory mechanisms to improve the osteogenic potential of UC-hMSCs. High-throughput miRNA expression analysis was performed to identify miRNAs involved in osteogenic differentiation. Quantitative real-time RT-PCR confirmed the expression levels of these miRNAs during osteogenic differentiation. The effects of specific anti-miRNAs on osteogenic differentiation were evaluated using alkaline phosphatase (ALP) activity, Alizarin Red S staining, and osteogenic gene expression assays. Analysis revealed significant differential expression of 806 miRNAs in high-osteogenic UC-hMSCs and 760 miRNAs in low-osteogenic UC-hMSCs. Four miRNAs—miR-21, miR-27b, miR-29a, and let-7b—were significantly down-regulated during osteogenic differentiation in high-osteogenic UC-hMSCs but remained elevated in low-osteogenic UC-hMSCs. Inhibition of these miRNAs using specific anti-miRs significantly increased osteogenic gene expression, ALP activity, and matrix mineralization. These effects could be partially mediated by modulation of the PI3K/Akt and Wnt/β-catenin signaling pathways, which led to the up-regulation of RUNX2 expression in UC-hMSCs. Our findings indicate that miR-21, miR-27b, miR-29a, and let-7b are important regulators of osteogenic differentiation in UC-hMSCs. Targeting these miRNAs could enhance osteogenic differentiation by modulating the PI3K/Akt and Wnt/β-catenin signaling pathways, leading to increased RUNX2 expression. These findings provide valuable insights into the role of specific miRNAs in regulating osteogenic differentiation of UC-hMSCs and highlight potential therapeutic strategies for bone regeneration through miRNA modulation.
Similar content being viewed by others
Introduction
People worldwide are living longer, with the global population older than 60 years expected to increase to 2.1 billion by 20501. Aging is a gradual process that results in a loss of tissue homeostasis, that leads to a progressive deterioration of tissue and organ functions2. The human skeleton undergoes significant changes over time, with bone loss beginning as early as the third decade of life, predisposing the elderly population to an increased risk of debilitating bone fractures3. Although osteoporosis can be treated with various medications, the efficacy of these treatments is limited and, in some circumstances, causes serious side effects, such as osteonecrosis and bone cancers4.
Mesenchymal stem cells (MSCs), the multipotent stem/progenitor cells found in various tissues of the adult body5are capable of differentiation into various functional cell types, including osteocytes that could be used to treat various bone defects6. Although the main source of human MSCs for most clinical applications is bone marrow (BM-hMSCs)5the derivation of BM-hMSCs requires an invasive procedure and their number and differentiation capacity gradually decrease with age7. Therefore, many attempts have been made to obtain hMSCs from other sources, such as various gestational tissues, including amnion, chorion, placenta, umbilical cord, and Wharton’s jelly8,9,10which are readily available in large quantities without the need for an invasive procedure. Among these gestational tissues, the umbilical cord, which is easy to handle and can be stored for later use, appears to be a promising option. Although umbilical cord-derived hMSCs (UC-hMSCs) exhibit typical hMSC characteristics, they generally have lower osteogenic differentiation capacity compared to BM-hMSCs and need longer time to differentiate into osteoblasts11. Therefore, finding a strategy to increase osteogenic differentiation of UC-hMSCs is crucial for their orthopaedic applications.
MicroRNA (miRNA), a short non-coding single-stranded RNAs, has recently emerged as important regulator of stem cell fate and behaviour12. These miRNAs modulate gene expression at the post-transcriptional level by binding to the 3’ untranslated regions (3’-UTR) of target mRNAs and inhibiting translation or promoting degradation of these mRNAs13. Although a previous study reported that several miRNAs play critical roles in the osteogenic differentiation of BM-hMSCs and human osteoblasts14, the roles of these miRNAs in osteogenic differentiation of gestational tissue-derived hMSCs, especially UC-hMSCs, have not yet been investigated. Therefore, this study aimed to discover miRNAs that play an important role in osteogenic differentiation of UC-hMSCs using differential expression analysis of miRNA and functional in vitro study. We believe that the obtained knowledge could be used to increase osteogenic differentiation of UC-hMSCs for future research and therapeutic use.
Materials and methods
Isolation and culture of UC-hMSCs
This study was approved by the Human Research Ethics Committee of Thammasat University No. 1 (Faculty of Medicine). Umbilical cords (length ~ 2–4 cm) obtained from pregnant women after normal delivery were thoroughly rinsed with 1XPBS at least 5 times before being cut into small pieces (approximately 1–2 mm3. The tissues were then washed twice with 1XPBS and digested with 0.5% trypsin-EDTA (Thermo Fisher Scientific, USA) for 3 h at 37 °C while shaking. After incubation, cells were washed twice with 1XPBS and cultured with a complete medium [DMEM + 10% fetal bovine serum (FBS, Thermo Fisher Scientific, USA)] in 25-cm2 tissue culture flasks. The cultures were maintained in a humidified tissue culture incubator at 37 °C with 5% CO2. The culture medium was refreshed every 3 days. Once the cells reached 80% confluence, they were subcultured using 0.25% trypsin-EDTA, replated at 1 × 104 cells/cm2 in a new flask, and incubated to expand further.
Immunophenotyping
UC-hMSCs (passages 3–6) were tested for the expression of typical hMSC surface markers following the recommendation of the International Society for Cell and Gene Therapy (ISCT)15. Cells were harvested with 0.25% trypsin-EDTA, washed, and resuspended in PBS. Then, 5 × 105 cells were incubated with fluorescein isothiocyanate (FITC) or phycoerythrin (PE) conjugated antibodies against human CD34, CD45, HLA-DR, CD73, CD90, and CD105 (Biolegend, USA) for 30 min at 4°C in the dark. After washing, cells were fixed with 1% paraformaldehyde in PBS. Flow cytometry (CytoFLEX, Beckman Coulter, USA) was used to acquire 2 × 104 labelled cells, and the data were analyzed using CytoExpert for DxFLEX version 2.0 (https://www.beckmancoulter.com/it/softwaredownloads, Beckman Coulter, USA) .
Trilineage differentiation assay
The potential for trilineage differentiation of UC-hMSCs (passages 3–6) was evaluated by inducing their differentiation into osteogenic, adipogenic and chondrogenic lineages using specific induction media. For osteogenic differentiation, UC-hMSCs were seeded at 5 × 103 cells/cm2 in a 35-mm2 dish (Costar, Corning, USA) and allowed to adhere overnight. After overnight incubation, the medium was replaced with osteogenic induction medium [complete medium supplemented with 100 nM dexamethasone (Sigma-Aldrich, USA), 10 mM β-glycerophosphate (Sigma-Aldrich, USA), and 50 µg/ml ascorbic acid (Sigma-Aldrich, USA)], and the cultures were maintained at 37 °C with 5% CO2. The medium was changed every 3 days. After 28 days, cells were fixed with 10% formalin solution, stained with 40 mM Alizarin Red S (Sigma-Aldrich, USA), and observed under an inverted microscope (Nikon Eclipse Ts2R, Japan). For adipogenic differentiation, UC-hMSCs were seeded at 5 × 103 cells/cm2 in a 35-mm2 dish and allowed to adhere overnight. The cells were then cultured in adipogenic induction medium [complete medium supplemented with 0.5 mM isobutylmethylxanthine (Sigma-Aldrich, USA), 1 µM dexamethasone, 10 µM insulin (Sigma-Aldrich, USA), 100 µM indomethacin (Sigma-Aldrich, USA), and 25 mM glucose (Ajax Finechem, Australia)] at 37 °C with 5% CO2, and the medium was changed every 3 days. After 28 days, cells were fixed with vapour of 37% formaldehyde, stained with 0.3% Oil Red O (Sigma-Aldrich, USA) in 60% isopropanol, and observed by inverted microscopy. For chondrogenic differentiation, UC-hMSCs were seeded at 3 × 106 cells/cm2 in a 96-well U bottom plate (Jet Biofil, China) and incubated overnight at 37 °C with 5% CO2. The following day, the medium was replaced with MSCgo™ Chondrogenic XF medium (Biological Industries, Israel) and the cells were cultured at 37 °C with 5% CO2, with medium changes every 3 days. After 14 days, the spheroidal masses were fixed with 10% formalin solution, stained with 1% Alcian Blue (HiMedia, India) in 0.1 N HCl and examined by inverted microscopy.
Screening of differentially expressed microRNAs
Sample preparation
High- and low-osteogenic UC-hMSCs were cultured with a complete medium in a 35-mm2 dish at a density of 5 × 103 cells/cm2 overnight and subjected to osteogenic differentiation for 28 days. At the end of the culture, alkaline phosphatase activity assay were performed and differentiated UC-hMSCs were stained with Alizarin Red S to identify their osteogenic differentiation capacity. The high- and low-osteogenic UC-hMSCs cultured in complete medium under the same conditions serve as controls. The UC-hMSCs were then classified into high- and low-osteogenic UC-hMSCs, based on the results of alkaline phosphatase (ALP) activity assay and Alizarin Red S staining.
RNA isolation
Total RNA was isolated from high- and low-osteogenic UC-hMSCs at the end of their osteogenic differentiation using Trizol® reagent (Invitrogen, USA). The amount and purity of isolated RNA were determined using NanoDrop™ 2000/2000c (Thermo Scientific, USA). One microgram of each RNA sample was sent to BGI, Hong Kong, for differential miRNA expression analysis. Briefly, RNA segments of different sizes were separated by polyacrylamide gel electrophoresis (PAGE), and 18–30 nucleotide RNA fragments were selected. Complementary DNA (cDNA) was then synthesized from purified and fragmented RNA samples following the BGI standard workflow and subsequently passed through DNA sequencing.
Bioinformatic analysis
The cDNAs were sequenced using the Illumina HiSeq 2000 instrument (Illumina Inc., USA). To adjust for multiple comparisons, the p-values for differential gene expression were corrected using the Bonferroni method. Differentially expressed genes (DEGs) were identified based on a false discovery rate (FDR) of ≤ 0.001, a Log2Ratio (fold change) of ≥ 1, and a p-value of ≤ 0.05. For even more precise DEG identification, stricter criteria were applied, with lower FDR values and greater fold changes. To predict target genes for the differentially expressed miRNAs, the tools RNAhybrid, miRanda, and TargetScan were used. Data were submitted and available in the NCBI database with accession number PRJNA1279367 at https://www.ncbi.nlm.nih.gov/bio-project/PRJNA1279367.
Functional and pathway enrichment analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of miRNAs were performed to determine their functions. GO enrichment analysis was performed using GOView.html (http://www.geneontology.org/), while KEGG pathway analysis was used to predict pathways associated with the selected miRNAs and KEGG (https://www.kegg.jp/kegg/kegg1.html). The term ‘osteogenic’ was entered in the KEGG pathway database search. Pathways related to osteogenesis were identified and the corresponding KO numbers were matched with NM numbers using Gene2KEGG. These NM numbers were further correlated with miRNAs through target filter data to identify relevant miRNAs involved in osteogenic pathways.
Determining the expression levels of selected MiRNAs during osteogenic differentiation of UC-hMSCs
To investigate the expression levels of selected miRNAs in high- and low-osteogenic UC-hMSCs during their osteogenic differentiation, 5 × 104 cells/cm2 UC-hMSCs (passages 3–5) were subjected to osteogenic differentiation for 28 days. The high- and low-osteogenic UC-hMSCs cultured in complete medium under the same conditions served as controls. On culture days 7, 14, 21, and 28, total RNA was isolated and the expression levels of hsa-miR-21, hsa-miR-27b, hsa-miR-29a, hsa-miR let-7b were determined by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). The mature miRNA sequences are shown in Table 1 (Table 1).
The effect of selected MiRNAs on osteogenic differentiation of UC-hMSCs
The role of selected miRNAs in osteogenic differentiation of UC-hMSCs was determined by a miRNA inhibition assay. UC-hMSCs (passages 3–5) were cultured overnight in complete medium at 5 × 103 cells/cm2 in a 6-well plate (Costar, Corning, USA). For anti-miR treatment, specific anti-miRs (miR-21 inhibitor, miR-27b inhibitor, miR-29a inhibitor and let-7b inhibitor) and lipofectamine® RNAiMAX reagent (Invitrogen, USA) were diluted with DMEM, mixed in a 1:1 ratio, and incubated at room temperature for 10 min. The mixtures containing 10 nM of each miRNA inhibitor were then added to an osteogenic differentiation medium and cells were subjected to osteogenic differentiation for an additional 28 days. UC-hMSCs transfected with 10 nM FAM-labeled miRNA negative control served as a negative control. On culture days 7, 14, 21, and 28 after transfection, the osteogenic differentiation capacity was determined using the ALP activity assay, Alizarin Red S staining, osteogenic gene expression, and Western blot analysis.
Alkaline phosphatase activity assay
ALP activity was measured on days 7, 14, 21, and 28 using the SensoLyte® pNPP ALP assay kit (AnaSpec, USA.), according to the manufacturer’s instructions. Briefly, cultured UC-hMSCs were washed twice with 1X PBS. Then, 100 µl of lysis buffer [0.1 M glycine (VWR Chemicals BDH®, USA), 1% Nonidet P-40 (USB Chemical, USA, 1mM MgCl2 (Sigma-Aldrich, USA), and 1mM ZnCl2 (EMSURE®, Germany), pH 9.6] was added. The samples were incubated on ice for 10 min. After centrifugation at 10,000 x g, the supernatant was collected. Next, p-Nitrophenyl Phosphate (pNPP) substrate solution was added, and the plate was incubated at room temperature for 60 min. ALP activity was measured using a microplate reader (BioTex, USA) at an absorbance of 405 nm. To normalize the results, total cellular protein was measured using a Bradford assay (Bio-Rad, USA), and ALP activity was calculated by comparing the optical density (O.D.) of the sample with a standard curve generated from 0 to 10 ng/ml of the ALP solution.
Quantitative real-time reverse transcription polymerase chain reaction
UC-hMSCs treated with specific anti-miRs were examined for the expression of osteogenic genes, including runt-related transcription factor 2 (RUNX2 ), osterix (OSX), osteocalcin (OCN) and collagen 1A1 (COl IA1) on days 7, 14, 21, and 28 of their osteogenic differentiation using qRT-PCR. Briefly, treated hMSCs were washed with 1XPBS and then lysed with Trizol® reagent (Invitrogen, USA). Total RNA was reverse transcribed to cDNA using iScript™ Reverse Transcription Supermix for qRT-PCR (Bio-Rad, USA). qRT-PCR reactions were prepared using the Universal SYBR® Green Supermix iTaq™ (Bio-rad, USA) and amplified in the StepOne plus™ real-time PCR system (Applied Biosystems; ABI, USA) for 40 cycles (denaturation at 95 °C for 15 s, followed by annealing at 60 °C for 60 s). The primer sequences and product size are shown in Table 2 (Table 2). The mRNA expression levels of target genes were normalized to the mRNA expression level of the endogenous control gene (GAPDH), analyzed using the comparative threshold cycle value (ΔΔCT) method with StepOne™ software version 2.3 (https://www.thermofisher.com/id/en/home/technical-resources/software-downloads/StepOne-and-StepOnePlus-Real-Time-PCR-System.html, Applied Biosystems; ABI, USA.) and presented as the relative mRNA expression level.
Western blot analysis
The effect of miRNA inhibition on protein expression in the osteogenic pathway was analyzed by western blotting. UC-hMSCs were treated, then trypsinized and resuspended in 1X RIPA buffer (Cell Signaling Technology, USA) with a protease inhibitor cocktail (Cell Signaling Technology, USA) and 1 mM PMSF (Sigma-Aldrich, USA). After incubating on ice for 10 min, cells were sonicated and centrifuged at 12,000 g at 4 °C for 10 min. The supernatant was collected, and protein concentration was measured using a Bradford assay (Bio-Rad, USA) with BSA (Sigma-Aldrich, USA) as a standard. Next, 100 µg of total protein was mixed with 3X blue loading buffer (BioLabs, USA) and heated for 5 min. Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to nitrocellulose membranes (0.45 μm pore size, Hybond ECL; Amersham Pharmacia, USA) using a mini transblot electrophoretic transfer cell (Bio-Rad, USA) at 120 V for 1.5 h. Membranes were stained with 0.1% (w/v) Ponceau S in 5% acetic acid to confirm a complete protein transfer. The membranes were cut and blocked with 5% non-fat dry milk in Tris-buffered saline with 0.1% Tween® 20 detergent (TBST) for 1 h at room temperature, then incubated overnight at 4 °C with primary antibodies against PI3K, β-catenin and RUNX2 (Table 3) diluted in 3% BSA in TBST. After washing with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. Protein signals were detected using a Clarity Western ECL substrate (Bio-Rad, USA). Densitometric scanning and statistical analysis of immunoblots were performed using Odyssey LI-COR and Image Studio™ software (Biosciences, USA). Image studio version 6 (https://www.licorbio.com/image-studio).
Statistical analysis
All experiments were carried out on at least three different samples. Data were presented as mean ± standard error of the mean (SEM). Statistical analysis was performed using one-way analysis of variance (ANOVA). A p-value of less than 0.05 was considered statistically significant.
Results
Characterization of umbilical cord-derived human mesenchymal stem cells
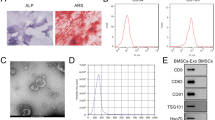
The umbilical cord-derived human mesenchymal stem cells (UC-hMSCs), isolated from umbilical cord tissue by enzymatic digestion, exhibited homogeneous fibroblast-like morphology (Fig. 1A). These cells expressed typical hMSC surface markers (CD73, CD90, and CD105) and lacked hematopoietic surface markers (CD34, CD45 and HLA-DR) (Fig. 1B). UC-hMSCs could also differentiate into osteoblasts, adipocytes, and chondrocytes, as determined by Alizarin Red S staining, Oil Red O staining, and Alcian Blue staining, respectively (Fig. 1C-E). These UC-hMSCs could be expanded up to 20 passages before showing a decrease in proliferative capacity.
The characteristics of umbilical cord-derived hMSCs (UC-hMSCs). (A) The spindle morphology of the UC-hMSCs cultured in DMEM + 10%FBS at passage 0 and passage 3 (B) Flow cytometry analysis showed positive expression of hMSC surface markers (CD73, CD90, CD105) and negative expression of hematopoietic surface markers (CD34, CD45, HLA-DR). (C) Osteogenic differentiation was confirmed by the presence of extracellular calcium deposits that marked positively with Alizarin Red S in UC-hMSCs cultured in osteogenic differentiation medium for 28 days. (D) After adipogenic induction for 28 days, lipid droplets in the cytoplasm of UC-hMSCs exhibited an orange-red color upon staining with Oil Red O. (E) The chondrogenic differentiation of UC-hMSCs was evidenced by positive Alcian blue staining. Data were obtained from 5 donors. Images a, c, and e were captured with a 10X magnification. Scale bar = 100 μm. Image d was captured with a 20X magnification. Scale bar = 100 μm.
High-osteogenic UC-hMSCs exhibited the same immunophenotypes, proliferative capacity and adipogenic differentiation capacity compared to low-osteogenic UC-hMSCs
UC-hMSCs were derived from six independent donors and expanded under identical conditions before subjected to osteogenic differentiation assays. The results showed that UC-hMSCs derived from three different donors exhibited significantly higher ALP activity (30.4 ± 2.7 vs. 15.8 ± 1.0 ng/mg protein; P < 0.05; Supplementary Fig. S1D) and matrix mineralization levels (0.52 ± 0.03 vs. 0.18 ± 0.02; P < 0.05; Supplementary Fig. S1E) after osteogenic induction compared to UC-hMSCs derived from other three donors. Based on their osteogenic differentiation capacity, the UC-hMSCs were then categorized into high-osteogenic UC-hMSCs (n = 3) and low-osteogenic UC-hMSCs (n = 3) (Supplementary Fig. S1).
To determine whether these two groups of UC-hMSCs exhibited other different characteristics apart from their osteogenic differentiation capacity, both groups were subjected to immunophenotyping, proliferation, and adipogenic differentiation assays. The results showed that both high- and low-osteogenic UC-hMSCs expressed similar percentages of all typical hMSC markers examined, including CD73 (99.24 ± 0.20% vs. 98.96 ± 0.61%), CD90 (97.86 ± 1.13% vs. 96.07 ± 1.19%) and CD105 (94.54 ± 1.72% vs. 96.25 ± 1.44%). Both groups of UC-hMSCs also expressed very low percentages of hematopoietic markers, CD34 (1.13 ± 0.80% vs. 2.88 ± 0.05%) and CD45 (0.39 ± 0.14% vs. 1.25 ± 0.30%) (Supplementary Fig. S1A). Similar to the immunophenotyping, both high- and low-osteogenic UC-hMSCs also exhibited the same level of adipogenic differentiation capacity, determined by Oil Red O after 28 days of induction (Supplementary Fig. S1C) and had similar proliferative capacity at passage 3 (Supplementary Fig. S1B).
Collectively, these results demonstrate that while high- and low-osteogenic UC-hMSCs are indistinguishable in terms of their immunophenotype, proliferation, and adipogenic differentiation capacity, they exhibit significant differences in their osteogenic differentiation capacity.
Many microRNAs are differentially expressed in high-osteogenic compared to low-osteogenic UC-hMSCs
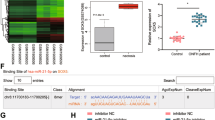
To identify microRNAs (miRNAs) that affect osteogenic differentiation of UC-hMSCs, we first identified miRNAs whose expression were significantly down-regulated upon osteogenic induction. After osteogenic induction, 806 miRNAs were down-regulated in high-osteogenic UC-hMSCs, while 760 miRNAs were down-regulated in low-osteogenic UC-hMSCs compared to the non-differentiated controls. Of these, 224 miRNAs were specifically down-regulated in high-osteogenic UC-hMSCs, 178 in low-osteogenic UC-hMSCs, and 582 in both groups (Fig. 2A, Supplementary Tab. S1-S3).
Hierarchical clustering of the data revealed significant changes in 31 miRNAs involved in the regulation of Wnt signaling pathway, TGF-β signaling pathway and RUNX2, that are known to play critical roles in the regulation of osteogenesis (Fig. 2B). These findings suggest that some of these miRNAs may play essential roles in osteogenic differentiation of UC-hMSCs.
Screening of differentially expressed miRNAs in UC-hMSCs. (A) The diagram showed the number of down-regulated miRNAs in high-osteogenic UC-hMSCs and low-osteogenic UC-hMSCs. (B) The heatmap diagram showed the results of the two-way hierarchical clustering of miRNAs. Each row represents one miRNA, and each column represents a sample. The color scale illustrates the relative expression level of a miRNA across all samples: green represents an expression level above the mean, and red represents an expression lower than the mean.
Gene ontology enrichment analysis
To better understand the functions of these miRNAs, we performed a gene ontology (GO) enrichment analysis to categorize their functions into three broad groups: biological process, cellular component, and molecular function.
The GO analysis for high-osteogenic UC-hMSCs versus control revealed 25 enriched GO terms in the “Biological Process” category, particularly in the cellular process (Fig. 3A), 15 enriched GO terms in the “Molecular Function” category, particularly in binding (Fig. 3B), and 18 enriched GO terms in the “Cellular Component” category, particularly in cells and cell parts (Fig. 3C). Similarly, the GO analysis for low-osteogenic UC-hMSCs versus control revealed 25 enriched GO terms in the “Biological Process” category, particularly in cellular process (Fig. 3A), 14 enriched GO terms in the “Molecular Function” category, particularly in binding (Fig. 3B), and 18 enriched GO terms in the “cellular component” category, particularly in cells and cell parts (Fig. 3C).
Interestingly, the number of genes in each functional category was lower for low-osteogenic UC-hMSCs compared to high-osteogenic UC-hMSCs. Comparison of high-osteogenic to low-osteogenic UC-hMSCs revealed similar enriched GO terms. Most of the genes in the biological process category were involved in cellular processes, while most genes in the cellular component category were associated with the cells and cell parts, and most genes in the molecular function category were involved in binding (Fig. 3A-C).
Pathway enrichment analysis of target genes
Genes typically interact with each other to perform certain biological functions. To further understand these functions, the pathway enrichment analysis of target genes for differentially expressed miRNAs was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Bar plots for KEGG pathway statistics were generated for each pairwise comparison: high-osteogenic UC-hMSCs versus control, low-osteogenic UC-hMSCs versus control, and high-osteogenic versus low-osteogenic UC-hMSCs (Fig. 4).
The KEGG pathway analysis across each pairwise comparison revealed six primary pathways: metabolism, human diseases, organismal systems, cellular processes, genetic information processing, and environmental information processing. Within these categories, multiple secondary KEGG pathway terms were identified. In particular, significant enrichment was observed in 12, 11, 10, 4, 4, and 3 pathways, respectively. These enriched pathways included signal transduction, cancer overview, and processes related to folding, sorting, and degradation, specifically within categories of environmental information processing, human diseases, and genetic information processing, compared to the whole genome background. These findings provide a deeper understanding of the biological functions and interactions of target genes associated with differentially expressed miRNAs during osteogenic differentiation of UC-hMSCs.
KEGG classification for high-osteogenic UC-hMSCs and control, low-osteogenic UC-hMSCs and control, and high-osteogenic UC-hMSCs and low-osteogenic UC-hMSCs (https://www.kegg.jp/kegg/kegg1.html). The X-axis indicates the number of genes. The Y-axis indicates the groups and subgroups of the KEGG pathway.
miR-21, miR-27b, miR-29a, and let-7b inhibit the osteogenic differentiation of low-osteogenic UC-hMSCs
To investigate the roles of miRNAs identified by the differential miRNA expression analysis in human osteogenesis, we first confirmed their expression levels during osteogenic differentiation of UC-hMSCs by qRT-PCR. We hypothesized that miRNAs expressed at higher levels in low-osteogenic UC-hMSCs compared to high-osteogenic UC-hMSCs might inhibit osteogenic differentiation of UC-hMSCs.
To prove this hypothesis, four miRNAs, including miR-21, miR-27b, miR-29a, and let-7b, with higher expression levels in low-osteogenic UC-hMSCs than in high-osteogenic UC-hMSCs, were selected for further study. Consistent with our hypothesis, the expression of these four miRNAs was significantly down-regulated during the osteogenic differentiation of high-osteogenic UC-hMSCs in a time-dependent manner (Fig. 5). In contrast, the expression levels of miR-29a and let-7b remained unchanged throughout osteogenic differentiation of low-osteogenic UC-hMSCs (Fig. 5). Although miR-21 and miR-27b levels were also down-regulated in low-osteogenic UC-hMSCs, they remained significantly higher than those in high-osteogenic UC-hMSCs under the same conditions (Fig. 5). These results suggest that elevated levels of these four miRNAs may inhibit the osteogenic differentiation of low-osteogenic UC-hMSCs.
Quantitative real-time RT-PCR showed the expression of miR-21, miR-27b, miR-29a, and let-7b during osteogenic differentiation of high- and low-osteogenic UC-hMSCs. UC-hMSCs cultured in DMEM + 10%FBS served as control. Data are presented as mean ± SEM (n = 3). *p < 0.05 compared to low-osteogenic UC-hMSCs cultured in OstDiff medium on day 7, #p < 0.05 compared to high-osteogenic UC-hMSCs cultured in OstDiff medium on day 7 and $p < 0.05 compared to low-osteogenic UC-hMSCs cultured in OstDiff medium on the same day.
Down-regulation of the miR-21, miR-27b, miR-29a, and let-7b expression increases the expression levels of osteogenic genes
To further examine the role of selected miRNAs in osteogenic differentiation of UC-hMSCs, we first reduced the expression of miR-21, miR-27b, miR-29a, and let-7b in UC-hMSCs using specific anti-miRs (anti-miR-21, anti-miR-27b, anti-miR-29a, and anti-let-7b). Anti-miR treatment significantly decreased the expression levels of each miRNA in UC-hMSCs throughout osteogenic differentiation (Fig. 6).
Quantitative real-time RT-PCR analysis showed the relative expression levels of miR-21 (A), miR-27b (B), miR-29a (C), and let-7b (D) in UC-hMSCs during osteogenic differentiation after transfection with anti-miR-21, anti-miR-27b, anti-miR-29a, and anti-let-7b on days 3, 7, 14, 21, and 28. Data are presented as mean ± SEM. *p < 0.05: compared to UC-hMSCs cultured in osteogenic differentiation medium with negative control.
To investigate whether reducing the expression of miR-21, miR-27b, miR-29a, and let-7b affects the expression of osteogenic genes (RUNX2, OSX, OCN, and COL1) during osteogenic differentiation of UC-hMSCs, we treated cells with specific anti-miRs targeting each miRNA. The results revealed that inhibition of miR-21, miR-27b, miR-29a, and let-7b significantly increased the expression of all osteogenic genes in a time-dependent manner compared to the untreated control (Fig. 7).
Quantitative real-time RT-PCR analysis demonstrated the relative expression levels of RUNX2 (A), Osterix (OSX) (B), Osteocalcin (OCN) (C), and Collagen 1 (COL1) in UC-hMSCs after transfection with anti-miR-21, anti-miR-27b, anti-miR-29a, and anti-let-7b on days 7, 14, 21, and 28. Data are presented as mean ± SEM. *p < 0.05 compared to UC-hMSCs cultured in osteogenic differentiation medium with negative control.
Down-regulation of miR-21, miR-27b, miR-29a, and let-7b expression increases alkaline phosphatase activity and matrix mineralization
To further investigate the roles of miR-21, miR-27b, miR-29a, and let-7b in the osteogenic differentiation of UC-hMSCs, we assessed whether reducing the expression of these miRNAs affects ALP activity and matrix mineralization. ALP activity was measured using an ALP activity assay, and matrix mineralization was evaluated by Alizarin Red S staining.
Consistent with their effects on osteogenic gene expression, reduction of miR-21, miR-27b, miR-29a, and let-7b by specific anti-miRs significantly increased ALP activity and matrix mineralization levels in a time-dependent manner compared to the untreated control (Figs. 8 and 9). Also similar to the effects on osteogenic gene expression, there was no significant difference in the enhancement of ALP activity and matrix mineralization between the four miRNAs examined (Figs. 8 and 9).
Alkaline phosphatase activity during osteogenic differentiation of UC-hMSCs was assessed after transfection with anti-miR-21, anti-miR-27b, anti-miR-29a, and anti-let-7b on days 7, 14, 21, and 28. Data are presented as mean ± SEM. *p < 0.05 compared to MSCs cultured in osteogenic differentiation medium with negative control.
Alizarin Red S staining was performed on UC-hMSCs transfected with anti-miRNAs on days 3, 7, 14, 21, and 28 to assess osteogenic differentiation. The control group was cultured in DMEM supplemented with 10% FBS. Red stain indicates positive osteogenic differentiation (A). Quantitative analysis of Alizarin Red S staining in UC-hMSCs after anti-miRNA transfection on days 3, 7, 14, 21, and 28 is shown in (B). Data are presented as mean ± SEM. *p < 0.05 compared to UC-hMSCs cultured in osteogenic differentiation medium with negative control.
Down-regulation of miR-21, miR-27b, miR-29a, and let-7b expression increases osteogenic differentiation of UC-hMSCs through PI3K and Wnt/β-catenin signaling pathways
To determine whether the enhanced osteogenic differentiation of UC-hMSCs after reduction of miR-21, miR-27b, miR-29a, and let-7b expression involves the PI3K and Wnt/β-catenin signaling pathways—key regulators of osteogenic differentiation—we examined the expression levels of PI3K and β-catenin proteins in anti-miR-treated UC-hMSCs. The results showed that inhibition of miR-21, miR-27b, miR-29a, and let-7b using specific anti-miRs significantly increased the levels of PI3K and β-catenin proteins during early osteogenic differentiation, compared to untreated control groups. Moreover, reduction of these miRNAs also elevated RUNX2 protein levels, a critical transcription factor for osteogenesis, in a time-dependent manner compared to controls (Fig. 10A, Supplementary Fig. S2).
Quantitative analysis of protein levels from Western blotting showed a significant increase in the expression of PI3K (Fig. 10B), β-catenin (Fig. 10C), and RUNX2 (Fig. 10D) in the anti-miR-treated groups relative to controls at all time points. Similar to their effects on osteogenic gene expression, ALP activity, and matrix mineralization, there were no significant differences in the expression of PI3K, β-catenin, and RUNX2 protein between individual anti-miRs.
Western blot analysis revealed the expression levels of PI3K, β-catenin, and RUNX2 in UC-hMSCs transfected with anti-miR-21, anti-miR-27b, anti-miR-29a, and anti-let-7b (A). The number specify the band intensity. The blotted membranes were cropped before hybridization with primary antibodies. The original blots of Fig. 10A are shown in Supplementary Fig. S2. Quantitative analysis of the expression of PI3K, β-catenin, and RUNX2 in UC-hMSCs after transfection with anti-miRNAs is shown in (B). Data are presented as mean ± SEM. * p < 0.05: compared to MSCs cultured in osteogenic differentiation medium with negative control.
Discussion
hMSCs, a type of multipotent non-hematopoietic stem/progenitor cell, have the potential to differentiate into various types of mesodermal cells, including osteoblasts, chondrocytes, and adipocytes16. Although BM-hMSCs are commonly used for most research and therapeutic applications, their extraction requires an invasive procedure, and the quantity is limited, especially in aging donors17. Therefore, this study isolated UC-hMSCs from the umbilical cord, which can be easily collected in large amounts by a non-invasive procedure. Consistent with the criteria of the ISCT for defining hMSCs, the UC-hMSCs collected in this study exhibited a fibroblast-like morphology, expressed typical hMSC surface markers, and were capable of differentiation into adipocytes, chondrocytes, and osteoblasts.
Despite several benefits, UC-hMSCs have lower osteogenic differentiation capacity than BM-hMSCs, which limits their use in orthopedic applications17. In the present study, UC-hMSCs were isolated from six independent donors using an identical harvesting protocol and expanded under the same culture conditions. Phenotypic and functional characterization showed that while high- and low-osteogenic UC-hMSCs are indistinguishable in terms of their immunophenotype, proliferation, and adipogenic differentiation capacity, they exhibit significant differences in their osteogenic differentiation capacity. Our subsequent analysis shows that the difference in osteogenic differentiation between these UC-hMSCs is caused, at least in part, by differences in the miRNA expression profile that could lead to a different response to the osteogenic inducing signal under our condition. Although several miRNAs have recently been shown to play a critical role in osteogenic differentiation of BM-hMSCs and pre-osteoblastic cell lines18,19 their role in osteogenic differentiation of other hMSC sources, particularly UC-hMSCs, has not yet been determined. The differential expression of miRNAs in UC-hMSCs with varying osteogenic potential underscores their essential role in the regulation of osteogenic differentiation. We identified miRNAs with significantly lower expression levels in differentiated high-osteogenic UC-hMSCs compared to low-osteogenic UC-hMSCs. We observed significant down-regulation of numerous miRNAs during osteogenic differentiation, with distinct profiles emerging between high-osteogenic and low-osteogenic groups. This finding is consistent with recent research indicating that specific miRNAs can function as key regulators in MSC fate decisions, particularly in osteogenic differentiation20.
The substantial down-regulation of 806 miRNAs in high-osteogenic UC-hMSCs compared to their undifferentiated controls and 760 miRNAs in low-osteogenic UC-hMSCs highlights the complexity of miRNA regulation during differentiation. In particular, the identification of 224 miRNAs specifically down-regulated in high-osteogenic UC-hMSCs suggests that these miRNAs may serve as negative regulators of osteogenic differentiation, while the 178 miRNAs unique to low-osteogenic UC-hMSCs may play a role in inhibiting osteogenesis. This selective down-regulation aligns with previous findings that miRNAs can modulate osteogenic potential by targeting critical signaling pathways21.
The hierarchical clustering analysis that revealed significant changes in 31 miRNAs associated with Wnt and TGF-β signaling pathways further supports the notion that these pathways are central to osteogenic differentiation. Wnt signaling, in particular, has been shown to promote osteogenic differentiation by stabilizing β-catenin, which in turn regulates RUNX2 expression22. This correlation emphasizes the importance of miRNAs in modulating these signaling pathways during osteogenic differentiation, as they may influence the expression of key transcription factors involved in the process.
The Gene Ontology (GO) enrichment analysis revealed a rich landscape of biological processes and functions associated with these differentially expressed miRNAs. The identification of numerous enriched terms, particularly in cellular processes and binding, indicates that the regulatory networks that govern osteogenic differentiation are extensive and complex. Recent studies have highlighted the importance of miRNAs in coordinating these networks, suggesting that they not only fine-tune gene expression but also participate in broader regulatory circuits that influence cellular behavior23.
Additionally, the pathway enrichment analysis using the KEGG database identified significant pathways associated with the target genes of differentially expressed miRNAs. The enriched pathways related to metabolism, signal transduction, and human diseases underscore the multifaceted roles that these miRNAs play in cellular function and development. Recent work has shown that miRNAs involved in these pathways can influence various aspects of MSC biology, including proliferation, survival, and differentiation24,25.
These findings reveal a complex interplay of miRNAs in the regulation of osteogenic differentiation in UC-hMSCs. The different miRNA expression patterns and the identification of key involved pathways provide insights into the molecular mechanisms underlying MSC behavior. This understanding may pave the way for the development of targeted therapies aimed at enhancing bone regeneration and repair by manipulating miRNA networks in stem cells. This study further explains the mechanisms by which specific miRNAs regulate osteogenic differentiation and how these insights can be translated into therapeutic strategies for bone-related diseases.
In this study, four selected miRNAs, miR-21, miR-27b, miR-29a and let-7b, whose expressions are either significantly down-regulated in both high- and low-osteogenic UC-hMSCs after their osteogenic differentiation or specifically down-regulated in high-osteogenic UC-hMSCs after their osteogenic differentiation, and are predicted to target RUNX2, a critical osteogenic regulator, are chosen for subsequent functional study to demonstrate their effects on osteogenic differentiation of UC-hMSCs. The subsequent qRT-PCR results confirmed that the expression of these four miRNAs was greatly down-regulated during osteogenic differentiation of high-osteogenic UC-hMSCs in a time-dependent manner but not in low-osteogenic UC-hMSCs. This pattern of expression suggests that these miRNAs may function as negative regulators of osteogenic differentiation, which is reinforced by a previous study which highlighted that elevated levels of specific miRNAs can inhibit the expression of osteogenic markers, thereby affecting the differentiation capacity of MSCs20. The significant down-regulation of the selected miRNAs during osteogenic differentiation of high-osteogenic UC-hMSCs underscores the critical role of miRNA expression dynamics in stem cell differentiation. Specifically, the reduction in these miRNAs suggests an active repression mechanism, allowing for the upregulation of osteogenic genes such as RUNX2, OSX, OCN, and COL1. Conversely, the relative stability of miR-29a and let-7b in low-osteogenic UC-hMSCs suggests that these miRNAs may inhibit osteogenic processes when overexpressed.
Consistent with this, our functional assay using specific anti-miRs also shows that reduction of miR-21, miR-27b, miR-29a, and let-7b significantly increases osteogenic gene expression, ALP activity, and matrix mineralization of UC-hMSCs throughout their osteogenic differentiation. These results are consistent with previous studies showing that miR-21 inhibition significantly increased osteogenesis of human periodontal ligament stem cells26 and down-regulation of miR-27b increased ALP activity and matrix mineralization in maxillary sinus membrane stem cells by up-regulating the expression of the osteogenic transcription factor, OSX27. The significant increase in ALP activity and matrix mineralization after the inhibition of these miRNAs provides further evidence of their inhibitory role in osteogenic differentiation. ALP is a well-known marker of early osteogenic differentiation and its up-regulation is often correlated with increased mineralization, which is essential for bone formation28. The consistent enhancement in these functional assays reinforces the notion that targeted manipulation of specific miRNAs can effectively promote osteogenic differentiation in UC-hMSCs, aligning with findings from recent studies that emphasize the therapeutic potential of miRNA modulation in regenerative medicine29.
Our results also suggest that downregulation of miR-21, miR-27b, miR-29a, and let-7b enhances osteogenic differentiation of UC-hMSCs, probably mediated through the activation of PI3K and Wnt/β-catenin signaling pathways. These pathways have been shown to play critical roles in promoting osteogenesis, particularly by regulating key transcription factors and signaling cascades necessary for bone formation30,31. PI3K signaling is widely recognized for its role in cell survival, proliferation, and differentiation, and recent studies emphasize its involvement in osteogenic differentiation. Activation of the PI3K/Akt pathway enhances osteoblast differentiation by increasing the expression of osteogenic genes, such as RUNX2 and OSX, and promoting mineralization32. Our observation that anti-miR treatment increases PI3K expression aligns with these findings and further supports the hypothesis that PI3K signaling is crucial for the osteogenic potential of MSCs.
The Wnt/β-catenin signaling pathway is also crucial for bone formation. Activation of β-catenin promotes osteogenic differentiation by regulating the transcription of genes associated with bone development, including RUNX222. The suppression of miR-21, miR-27b, miR-29a, and let-7b resulted in elevated levels of β-catenin, which aligns with previous studies indicating that Wnt/β-catenin signaling fosters osteogenesis by stabilizing β-catenin and enhancing the expression of osteogenic markers33.
The reduction in miR-21, miR-27b, miR-29a, and let-7b levels also increased the expression of RUNX2, a critical transcription factor for osteogenic differentiation, in UC-hMSCs. These results are consistent with previous studies showing that miR-29a regulates the expression of RUNX2 in human osteoblasts and subchondral hMSCs by modulating the Wnt/β-catenin and PI3K/Akt signaling pathways in these cells34,35 while miR-27b regulates the differentiation of BM-hMSCs into hypertrophic chondrocytes by modulating core-binding factor subunit beta (CBFB) and RUNX2 expression36. RUNX2 is a master transcription factor essential for the commitment of MSCs to the osteogenic lineage, and its expression is vital for the subsequent stages of bone formation37. The time-dependent elevation of RUNX2 in response to anti-miR treatment further underscores the potential of modulating miRNA levels to influence osteogenic outcomes in UC-hMSCs.
The lack of significant differences in the enhancement of protein expression levels of PI3K, β-catenin, and RUNX2 between individual anti-miRs suggests that these miRNAs may operate within overlapping regulatory networks. Previous research has indicated that many miRNAs target common pathways and genes involved in osteogenesis, leading to similar outcomes regardless of the specific miRNA being inhibited38. This observation is consistent with previous studies indicating that miRNAs often converge on common regulatory networks involved in MSC osteogenesis39. Furthermore, the consistent results across ALP activity, matrix mineralization, and the expression of key signaling proteins indicated that targeting these miRNAs not only promotes osteogenic gene expression but also translates into functional outcomes critical for bone formation25.
These results suggest that the reduction of miR-21, miR-27b, miR-29a, and let-7b enhances osteogenic differentiation in UC-hMSCs by modulating PI3K and Wnt/β-catenin signaling. This highlights the potential of miRNA inhibition as a strategy to improve bone regeneration and repair using MSCs.
Conclusions
In conclusion, our study demonstrates that specific miRNAs—miR-21, miR-27b, miR-29a, and let-7b—play pivotal roles in regulating the osteogenic differentiation of UC-hMSCs through the PI3K and Wnt/β-catenin signaling pathways. The differential expression analysis reveals that these miRNAs are significantly down-regulated during osteogenic differentiation in high-osteogenic UC-hMSCs, suggesting that they act as negative regulators of this process. Functional experiments, including the application of anti-miRs, confirmed that reducing the expression of these miRNAs enhances the expression of key osteogenic genes (RUNX2, OSX, OCN, and COL1), and increases ALP activity and matrix mineralization. Collectively, these findings provide valuable insights into the molecular mechanisms underlying osteogenic differentiation in UC-hMSCs and suggest that targeted inhibition of specific miRNAs could represent a promising strategy to improve bone regeneration.
Data availability
The datasets generated and analysed during the current study are uploaded and available in the NCBI database with accession number PRJNA1279367 at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1279367.
Abbreviations
- 3’-UTR:
-
3’-Untranslated Region
- ALP:
-
Alkaline Phosphatase
- ANOVA:
-
Analysis of Variance
- anti-miR:
-
Anti-microRNA
- BM-hMSCs:
-
Bone Marrow-derived Human Mesenchymal Stem Cells
- BMP:
-
Bone Morphogenetic Protein
- BSA:
-
Bovine Serum Albumin
- CBFB:
-
Core-Binding Factor Subunit Beta
- CD:
-
Cluster of Differentiation
- cDNA:
-
Complementary DNA
- Col I:
-
Collagen Type I
- DEGs:
-
Differentially Expressed Genes
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- ECL:
-
Enhanced Chemiluminescence
- EDTA:
-
Ethylenediaminetetraacetic Acid
- FDR:
-
False Discovery Rate
- FITC:
-
Fluorescein Isothiocyanate
- GAPDH:
-
Glyceraldehyde 3-Phosphate Dehydrogenase
- GO:
-
Gene Ontology
- HDAC4:
-
Histone Deacetylase 4
- HRP:
-
Horseradish Peroxidase
- ISCT:
-
International Society for Cell and Gene Therapy
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KO:
-
KEGG Orthology
- miRNA:
-
MicroRNA
- MSCs:
-
Mesenchymal Stem Cells
- NM:
-
Nucleotide
- O.D.:
-
Optical Density
- Ocn:
-
Osteocalcin
- Osx:
-
Osterix
- PBS:
-
Phosphate-Buffered Saline
- PCR:
-
Polymerase Chain Reaction
- PE:
-
Phycoerythrin
- PI3K:
-
Phosphoinositide 3-Kinase
- PMSF:
-
Phenylmethylsulfonyl Fluoride
- pNPP:
-
p-Nitrophenyl Phosphate
- qRT-PCR:
-
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
- RIPA:
-
Radioimmunoprecipitation Assay
- RNAiMAX:
-
RNA Iinterference MAX
- RUNX2:
-
Runt-related Transcription Factor 2
- SDS-PAGE:
-
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
- SEM:
-
Standard Error of the Mean
- TBST:
-
Tris-Buffered Saline with Tween® 20 Detergent
- TGF-β:
-
Transforming Growth Factor Beta
- TPM:
-
Transcripts Per Kilobase Million
- UC-hMSCs:
-
Umbilical Cord-derived Human Mesenchymal Stem Cells
- ΔΔCT:
-
Comparative Threshold cycle
References
United Nations Department of Economic and Social, Affairs, P. D. & World Population, A. : Challenges and opportunities of population ageing in the least developed countries. UN DESA/ (2023). POP/2023/TR/NO.5 (2023).
Guo, J. et al. Aging and aging-related diseases: from molecular mechanisms to interventions and treatments. Signal. Transduct. Target. Ther. 7, 391. https://doi.org/10.1038/s41392-022-01251-0 (2022).
Infante, A. & Rodriguez, C. I. Osteogenesis and aging: lessons from mesenchymal stem cells. Stem Cell. Res. Ther. 9, 244. https://doi.org/10.1186/s13287-018-0995-x (2018).
LeBoff, M. S. et al. The clinician’s guide to prevention and treatment of osteoporosis. Osteoporos. Int. 33, 2049–2102. https://doi.org/10.1007/s00198-021-05900-y (2022).
Hoang, D. M. et al. Stem cell-based therapy for human diseases. Signal. Transduct. Target. Ther. 7, 272. https://doi.org/10.1038/s41392-022-01134-4 (2022).
Malige, A., Gates, C. & Cook, J. L. Mesenchymal stem cells in orthopaedics: A systematic review of applications to practice. J. Orthop. 58, 1–9. https://doi.org/10.1016/j.jor.2024.06.026 (2024).
Handke, M. et al. Bone marrow from periacetabular osteotomies as a novel source for human mesenchymal stromal cells. Stem Cell. Res. Ther. 14, 315. https://doi.org/10.1186/s13287-023-03552-9 (2023).
Manochantr, S., Marupanthorn, K., Tantrawatpan, C. & Kheolamai, P. The expression of neurogenic markers after neuronal induction of chorion-derived mesenchymal stromal cells. Neurol. Res. 37, 545–552. https://doi.org/10.1179/1743132815Y.0000000019 (2015).
Meesuk, L., Tantrawatpan, C., Kheolamai, P. & Manochantr, S. The immunosuppressive capacity of human mesenchymal stromal cells derived from Amnion and bone marrow. Biochem. Biophys. Rep. 8, 34–40. https://doi.org/10.1016/j.bbrep.2016.07.019 (2016).
Marupanthorn, K., Tantrawatpan, C., Kheolamai, P., Tantikanlayaporn, D. & Manochantr, S. MicroRNA treatment modulates osteogenic differentiation potential of mesenchymal stem cells derived from human chorion and placenta. Sci. Rep. 11, 7670. https://doi.org/10.1038/s41598-021-87298-5 (2021).
Marupanthorn, K., Tantrawatpan, C., Kheolamai, P., Tantikanlayaporn, D. & Manochantr, S. Bone morphogenetic protein-2 enhances the osteogenic differentiation capacity of mesenchymal stromal cells derived from human bone marrow and umbilical cord. Int. J. Mol. Med. 39, 654–662. https://doi.org/10.3892/ijmm.2017.2872 (2017).
Estrada-Meza, C. et al. Recent insights into the MicroRNA and long non-coding RNA-mediated regulation of stem cell populations. 3 Biotech. 12, 270. https://doi.org/10.1007/s13205-022-03343-8 (2022).
Ranganathan, K. & Sivasankar, V. MicroRNAs - Biology and clinical applications. J. Oral Maxillofac. Pathol. 18, 229–234. https://doi.org/10.4103/0973-029X.140762 (2014).
Bottani, M., Banfi, G. & Lombardi, G. Perspectives on MiRNAs as epigenetic markers in osteoporosis and bone fracture risk: A step forward in personalized diagnosis. Front. Genet. 10, 1044. https://doi.org/10.3389/fgene.2019.01044 (2019).
Dominici, M. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 8, 315–317. https://doi.org/10.1080/14653240600855905 (2006).
Zhang, Y., Khan, D., Delling, J. & Tobiasch, E. Mechanisms underlying the osteo- and adipo-differentiation of human mesenchymal stem cells. ScientificWorldJournal 2012 (793823). https://doi.org/10.1100/2012/793823 (2012).
Manochantr, S. et al. Immunosuppressive properties of mesenchymal stromal cells derived from amnion, placenta, wharton’s jelly and umbilical cord. Intern. Med. J. 43, 430–439. https://doi.org/10.1111/imj.12044 (2013).
Huang, C., Geng, J. & Jiang, S. MicroRNAs in regulation of osteogenic differentiation of mesenchymal stem cells. Cell. Tissue Res. 368, 229–238. https://doi.org/10.1007/s00441-016-2462-2 (2017).
Gao, J. et al. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J. Cell. Biochem. 112, 1844–1856. https://doi.org/10.1002/jcb.23106 (2011).
Wang, J., Liu, S., Li, J., Zhao, S. & Yi, Z. Roles for MiRNAs in osteogenic differentiation of bone marrow mesenchymal stem cells. Stem Cell. Res. Ther. 10, 197. https://doi.org/10.1186/s13287-019-1309-7 (2019).
Ghafouri-Fard, S. et al. Contribution of MiRNAs and LncRNAs in osteogenesis and related disorders. Biomed. Pharmacother. 142, 111942. https://doi.org/10.1016/j.biopha.2021.111942 (2021).
Hu, L., Chen, W., Qian, A. & Li, Y. P. Wnt/beta-catenin signaling components and mechanisms in bone formation, homeostasis, and disease. Bone Res. 12, 39. https://doi.org/10.1038/s41413-024-00342-8 (2024).
Cora, D., Re, A., Caselle, M. & Bussolino, F. MicroRNA-mediated regulatory circuits: outlook and perspectives. Phys. Biol. 14, 045001. https://doi.org/10.1088/1478-3975/aa6f21 (2017).
Guo, R. et al. Hsa-miR-27b-5p suppresses the osteogenic and odontogenic differentiation of stem cells from human exfoliated deciduous teeth via targeting BMPR1A: an ex vivo study. Int. Endod J. 56, 1284–1300. https://doi.org/10.1111/iej.13959 (2023).
Iaquinta, M. R. et al. The role of MicroRNAs in the osteogenic and chondrogenic differentiation of mesenchymal stem cells and bone pathologies. Theranostics 11, 6573–6591. https://doi.org/10.7150/thno.55664 (2021).
Wei, F. et al. MicroRNA-21 regulates osteogenic differentiation of periodontal ligament stem cells by targeting Smad5. Sci. Rep. 7, 16608. https://doi.org/10.1038/s41598-017-16720-8 (2017).
Peng, W., Zhu, S., Li, X., Weng, J. & Chen, S. miR-27b-3p suppressed osteogenic differentiation of maxillary sinus membrane stem cells by targeting Sp7. Implant Dent. 26, 492–499. https://doi.org/10.1097/ID.0000000000000637 (2017).
Golub, E. & Boesze-Battaglia, K. The role of alkaline phosphatase in mineralization. Curr. Opin. Orthop. 18, 444–448. https://doi.org/10.1097/BCO.0b013e3282630851 (2007).
Iranmanesh, P. et al. MicroRNAs-mediated regulation of the differentiation of dental pulp-derived mesenchymal stem cells: a systematic review and bioinformatic analysis. Stem Cell. Res. Ther. 14, 76. https://doi.org/10.1186/s13287-023-03289-5 (2023).
Wang, H. et al. Exploring the effects of naringin on oxidative stress-impaired osteogenic differentiation via the Wnt/beta-catenin and PI3K/Akt pathways. Sci. Rep. 14, 14047. https://doi.org/10.1038/s41598-024-64952-2 (2024).
Wu, M., Wu, S., Chen, W. & Li, Y. P. The roles and regulatory mechanisms of TGF-beta and BMP signaling in bone and cartilage development, homeostasis and disease. Cell Res. 34, 101–123. https://doi.org/10.1038/s41422-023-00918-9 (2024).
Gao, S. et al. PI3K-Akt signaling regulates BMP2-induced osteogenic differentiation of mesenchymal stem cells (MSCs): A transcriptomic landscape analysis. Stem Cell. Res. 66, 103010. https://doi.org/10.1016/j.scr.2022.103010 (2023).
Liu, J. et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal. Transduct. Target. Ther. 7 https://doi.org/10.1038/s41392-021-00762-6 (2022).
Lian, W. S. et al. Subchondral mesenchymal stem cells from Osteoarthritic knees display high osteogenic differentiation capacity through microRNA-29a regulation of HDAC4. J. Mol. Med. (Berl). 95, 1327–1340. https://doi.org/10.1007/s00109-017-1583-8 (2017).
Kapinas, K., Kessler, C., Ricks, T., Gronowicz, G. & Delany, A. M. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 285, 25221–25231. https://doi.org/10.1074/jbc.M110.116137 (2010).
Lv, S. et al. MicroRNA-27b targets CBFB to inhibit differentiation of human bone marrow mesenchymal stem cells into hypertrophic chondrocytes. Stem Cell. Res. Ther. 11, 392. https://doi.org/10.1186/s13287-020-01909-y (2020).
Gomathi, K., Akshaya, N., Srinaath, N., Moorthi, A. & Selvamurugan, N. Regulation of Runx2 by post-translational modifications in osteoblast differentiation. Life Sci. 245, 117389. https://doi.org/10.1016/j.lfs.2020.117389 (2020).
Doghish, A. S. et al. A spotlight on the interplay of signaling pathways and the role of MiRNAs in osteosarcoma pathogenesis and therapeutic resistance. Pathol. Res. Pract. 245, 154442. https://doi.org/10.1016/j.prp.2023.154442 (2023).
Balachander, G. M., Nilawar, S., Meka, S. R. K., Ghosh, L. D. & Chatterjee, K. Unravelling MicroRNA regulation and miRNA-mRNA regulatory networks in osteogenesis driven by 3D nanotopographical cues. Biomater. Sci. 12, 978–989. https://doi.org/10.1039/d3bm01597a (2024).
Acknowledgements
The authors would like to acknowledge Michael Jan Everts, from the Clinical Research Center, Faculty of Medicine, Thammasat University, for English editorial assistance. The authors declare that they have not used AI-generated work in this manuscript. The authors thank the staff of the delivery room, Thammasat University Hospital, for facilitating the specimen collection, and all volunteers for kindly donating tissues for this study.
Funding
This work was supported by the Thailand Science Research and Innovation Fundamental Fund fiscal year 2024, Thammasat University (Grant No.TUFF 52/2567), the Center of Excellence in Stem Cell Research and Innovation, Thammasat University and Thailand Graduate Institute of Science and Technology, National Science and Technology Development (Contract No. SCA-CO-2560-4471-TH). The funding body played no role in the design of the study and the collection, analysis, and interpretation of the data, nor in the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
LM conducted the experiments and drafted the manuscript. PK contributed to the data interpretation and revised the manuscript. CT contributed to the data interpretation. JS contributed to the data interpretation. SM contributed to conceptualization, study design, resources, supervision, data analysis and interpretation, draft, and completed manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experimental procedures were conducted in accordance with the Declaration of Helsinki and the Belmont report. This study was approved by the Human Research Ethics Committee of Thammasat University No. 1 (Faculty of Medicine) [Approval title: The improvement of proliferation and osteogenic differentiation of human mesenchymal stem cells.] [Approval number: 171/2018] (Date of approval: August 23, 2018). All samples were obtained from donors with their written informed consent.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Meesuk, L., Kheolamai, P., Tantrawatpan, C. et al. Improvement of osteogenic differentiation in umbilical cord-derived human mesenchymal stem cells through specific MiRNA inhibition. Sci Rep 15, 28485 (2025). https://doi.org/10.1038/s41598-025-13093-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13093-1