Abstract
PLXNA3 is a transmembrane protein essential for axon guidance and synapse formation, gaining attention in cancer research for its roles in cell migration, invasion, and signal transduction. However, its biological significance in breast cancer remains underexplored. Here, bulk RNA sequencing and survival analysis revealed that elevated PLXNA3 levels were correlated with increased malignancy and poorer survival in breast cancer. Single-cell RNA sequencing demonstrated that PLXNA3 was predominantly enriched in tumor cell clusters within the tumor immune microenvironment, rather than in immune cell clusters. In vitro experiments further demonstrated that knockdown and overexpression of PLXNA3 significantly altered the proliferative, invasive, and migratory capacities of breast cancer cells. Additionally, the overexpression of PLXNA3 was negatively correlated with immune activation status and served as a predictor of poor response to anti-PD1 immunotherapy. Our findings suggested that high expression of PLXNA3 is associated with poor prognosis in breast cancer and plays a crucial role in cancer immunity, making it a promising novel target for intervention in breast cancer.
Similar content being viewed by others
Introduction
Breast cancer is one of the most prevalent malignancies among women worldwide and remains a leading cause of cancer-related mortality1,2,3. Although substantial progress has been made in screening, diagnosis, and treatment of breast cancer over recent decades, challenges such as treatment resistance, metastatic recurrence, and unfavorable prognosis persist4,5. Thus, there is an urgent need to identify novel biomarkers for predicting clinical prognosis and treatment response.
Plexins are large transmembrane proteins distinguished by their complex architecture, consisting of ten extracellular domains, a transmembrane region, and a cytoplasmic domain6. The cytoplasmic domain harbors critical functional elements, such as a GTPase-activating protein (GAP) domain and a Rho GTPase-binding domain7. These domains are critical for plexin-mediated signal transduction, with GAP domain mutations leading to severe developmental dysfunctions8. Plexin receptors are classified into four groups: four type A, three type B, and individual type C and D plexins9. Among them, type A plexins (PLXNAs) uniquely interact with semaphorins and play essential roles in immune regulation, angiogenesis, and tumor progression10,11,12. Previous studies have shown that aberrant expression of certain PLXNAs, such as PLXNA1, is associated with increased tumor aggressiveness in various cancer types, including lung, prostate, and gastric cancers13,14,15.
Interestingly, not all PLXNAs exhibit oncogenic behavior. PLXNA3 has been reported to act as a tumor suppressor in endometrial and ovarian cancers, where its downregulation promotes tumor cell proliferation and invasion16,17. These contrasting roles underscore the context-dependent function of PLXNs in cancer biology.
Although initial studies have linked PLXNA3 to the progression of invasive ductal carcinoma, its expression profile and functional roles in breast cancer remain largely uncharacterized18. To bridge this gap, we evaluated the expression and function of PLXNA3 in breast cancer, particularly in tumor cell populations, through comprehensive bioinformatic analyses and cell experiments. We also investigated its clinical relevance and scrutinized its association with the tumor microenvironment (TME). These findings offer a fresh avenue for potential diagnosis and treatment of breast cancer, paving the way for future investigations and personalized therapies involving PLXNA3.
Results
The expression of PLXNA3 was upregulated in breast cancer tissues
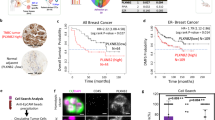
The UALCAN database showed that the expression of PLXNA3 in tumor tissues was significantly elevated compared to normal tissues, including bladder cancer, breast cancer, esophageal cancer, and others (Fig. 1A). Subsequently, we performed a comprehensive analysis of PLXNA3 expression in breast cancer. Subgroup analysis based on individual cancer stages, breast cancer subclasses, and lymph node metastatic status indicated that the mRNA levels of PLXNA3 in breast cancer patients were significantly higher than in healthy individuals (Fig. 1B). The positive expression rate of PLXNA3 gradually increased from Stage I and II BRCA to Stage III and IV BRCA (Fig. 1C). Among different breast cancer subtypes, the Luminal subtype showed a significantly higher proportion (Fig. 1D). Notably, the more severe the lymph node metastasis, the higher expression level of PLXNA3 in patients (Fig. 1E). Taken together, these findings suggest that the PLXNA3 expression is progressively upregulated during the malignant transformation of breast cancer.
PLXNA3 was over-expressed in breast cancer tissues. (A) The expression level of PLXNA3 mRNA in various cancer tissues compared to normal tissue. (B) The protein expression level of PLXNA3 in breast cancer tissue and normal breast tissues. (C-E) The correlation between PLXNA3 mRNA expression and tumor stage, cancer subtypes, lymph node metastatic status in breast cancer. ****P < 0.0001; ns, no significance.
The expression of PLXNA3 was associated with poor prognosis in breast cancer patients
The increased expression of PLXNA3 in breast cancer prompted us to analyze its prognostic potential. We utilized the Kaplan-Meier plotter and divided the breast cancer patients in the TCGA datasets into low-risk and high-risk groups according to the median risk score. The results revealed that in patients with triple-negative breast cancer (TNBC), higher PLXNA3 expression was associated with shorter overall survival (OS) and distant metastasis-free survival (DMFS) (Fig. 2A, B). In luminal breast cancer patients, elevated PLXNA3 levels correlated with reduced recurrence-free survival (RFS), but no significant association was found with post-progression survival (PPS) (Fig. 2C, D). Furthermore, PLXNA3 expression didn’t correlate with prognosis in HER2-positive breast cancer patients (Fig. 2E, F). These results suggest that PLXNA3 may serve as a prognostic biomarker for breast cancer.
Single-cell level analysis of PLXNA3
TME plays a vital role in cancer progression19,20. To investigate the cellular distribution of PLXNA3 within the TME, we analyzed single-cell RNA sequencing data using the TISCH2 database. The heatmap illustrated the expression levels of the PLXNA3 gene across various breast cancer (BRCA) datasets and cell types (Fig. 3A). Notably, PLXNA3 exhibited elevated expression in malignant cells across multiple datasets, particularly in BRCA_GSE143423, BRCA_GSE148673, BRCA_GSE150660, BRCA_GSE161529, and BRCA_SRP114962 (Fig. 3B). Additionally, significant expression was also observed in monocytes/macrophages and T cells, including CD4 + conventional T cells and CD8 + T cells (Supplementary Fig. S1). These results indicate that PLXNA3 is predominantly enriched in tumor cells and may also be involved in modulating immune cell function within the breast cancer microenvironment.
PLXNA3 promotes the proliferation, invasion, and migration of breast cancer cells
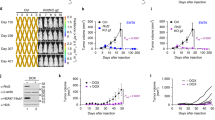
Based on these findings, we further validated the functional role of PLXNA3 in tumor cells by either silencing or overexpressing its expression in the triple-negative breast cancer cell lines MDA-MB-231 and MDA-MB-453 using small interfering RNA (siRNA) and plasmid transfection, respectively. Among the constructs tested, si-PLXNA3#3 and pcDNA-PLXNA3#3 demonstrated the highest transfection efficiency in both cell lines and were thus selected for subsequent experiments. (Supplementary Fig. S2). Cell Counting Kit-8 (CCK-8) assay and plate cloning revealed that PLXNA3 knockdown significantly inhibited the proliferation of both cell lines (Fig. 4A, B). Concurrently, the overexpression of PLXNA3 increased breast cancer cells’ replicative potential (Fig. 4C, D). In addition, scratch assays demonstrated a markedly reduced wound closure rate in the si3-PLXNA3 group compared to the control group (Fig. 5A). Transwell assays further indicated that PLXNA3 silencing substantially decreased the relative invasion rate of the cells (Fig. 5C). While, the overexpression of PLXNA3 has the opposite effect (Fig. 5B, D). Collectively, these findings suggest that PLXNA3 inhibition impairs the proliferation, migration, and invasion of breast cancer cells.
Effects of PLXNA3 modulation on the proliferation of breast cancer cells. (A) CCK-8 assay evaluating the impact of PLXNA3 knockdown (si-PLXNA3) on the proliferation of MDA-MB-231 cells. (B) Colony formation assay assessing the effect of si-PLXNA3 on the proliferative capacity of MDA-MB-231 and MDA-MB-453 cells. (C) CCK8 assay examining the impact of PLXNA3 overexpression (pcDNA-PLXNA3) on proliferation in MDA-MB-231 cells. (D) Colony formation assay evaluating the effect of pcDNA-PLXNA3 on proliferation in MDA-MB-231 and MDA-MB-453 cells. ∗ P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001. Data are the mean ± SEM of at least three independent experiments.
The effect of PLXNA3 on the invasion and metastasis of breast cancer cells. (A-B) Cell scratch assay detecting migration ability in MDA-MB-231 cells. (C-D) Transwell test detecting migration ability in MDA-MB-231 cells. ∗ P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001. Data are the mean ± SEM of at least three independent experiments.
PLXNA3 expression was correlated with immune factors
To further explore the immunological relevance of PLXNA3, we assessed its correlation with tumor-infiltrating lymphocytes (TILs), immune stimulators, and human leukocyte antigens (HLAs) in breast cancer. As shown in figures, PLXNA3 was positively associated with several TIL subsets, including TemCD8, TcmCD4, Tfh, and NK (Fig. 6A and B). Additionally, it showed significant correlations with multiple immune stimulators, such as CD40, CD86, CXCL12, and TNFRSF17 (Fig. 6C, D), as well as HLA class I and II molecules, including B2M, HLA-DMA, HLA-DPA1, and HLA-DQB1 (Fig. 6E, F). These findings highlight that PLXNA3 is closely linked to key immune components within TME, indicating a potential role in modulating immune responses in breast cancer.
Correlation of PLXNA3 expression with immune factors. (A, C, E) The heatmap shows the correlation between PLXNA3 and tumor-infiltrating lymphocytes, immune stimulatory factors, and antigen presentation-related factors across different cancers. (B, D, F) Line graph shows the correlation of PLXNA3 with specific immune indicators in breast cancer.
Correlation between PLXNA3 expression and the efficacy of immunotherapy
Immune checkpoint inhibitors (ICIs) have emerged as a pivotal strategy in cancer immunotherapy21,22. Given the observed association between PLXNA3 expression and altered immune infiltration, we further examined its relationship with immunotherapy response. Analysis of clinical outcomes revealed that patients with high PLXNA3 expression exhibited significantly poorer responses to anti-PD-1 therapy, as reflected by reduced OS and progression-free survival (PFS) compared to those with low PLXNA3 levels (Fig. 7A, B). In contrast, no significant differences in OS or PFS were observed in patients receiving anti-CTLA-4 treatment based on PLXNA3 expression (Fig. 7C, D).
Discussion
In this study, we systematically investigated the expression profile, prognostic significance, biological function, and immunological relevance of PLXNA3 in breast cancer. Our results demonstrated that PLXNA3 is markedly upregulated in breast cancer tissues compared to normal counterparts and shows significant associations with advanced pathological features, including tumor stage, molecular subtype, and lymph node metastasis. Kaplan–Meier survival analyses further demonstrated that elevated PLXNA3 expression correlates with poorer clinical outcomes in specific subtypes, particularly triple-negative and luminal breast cancers, underscoring its potential as an independent prognostic biomarker.
To further verify its biological function in breast cancer, we performed in vitro experiments using human breast cancer cell lines MDA-MB-231 and MDA-MB-468. Both knockdown and overexpression assays confirmed that PLXNA3 promotes proliferation, migration, and invasion, supporting its tumor-promoting role in breast cancer cells.
The TME encompasses the growth environment of the tumor and increasing evidence has supported that immune cell infiltration is a critical factor in tumor progression and the effectiveness of immunotherapy23. Existing research has reported that plexins (PLXNs) can regulate immune responses by modulating immune cell activation, differentiation, and migration24. For example, PLXNA1 promotes dendritic cell–mediated T cell activation25 while PLXND1 inhibits dendritic cell migration upon ligand binding26. Notably, PLXNA3 expression is upregulated during the M2 polarization of monocytes, suggesting its involvement in immunosuppressive processes27. PLXNB1 has also been implicated in tumor-associated macrophage migration and angiogenesis28. However, the role of PLXNA3 in regulating immune cell infiltration in breast cancer remains unclear. Single-cell RNA-sequencing (scRNA-seq) showed that PLXNA3 is not only enriched in malignant epithelial cells but also expressed in T cells and monocyte/macrophage populations29,30. Importantly, PLXNA3 expression was negatively correlated with the infiltration of effector immune cells, such as CD8 + and CD4 + T cells, suggesting a potential role in shaping an immunologically “cold” tumor microenvironment. Previous studies have shown that Semaphorin 3 A, a ligand of PLXNA1, inhibits CD8 + T cell adhesion, motility, and immune synapse formation, thereby impairing anti-tumor immunity31. Whether PLXNA3 exerts similar immunosuppressive effects through direct modulation of T cell function remains to be determined. Moreover, PLXNA3 was inversely related to key immune stimulators (e.g., CD86, CXCL12) and antigen presentation molecules (e.g., B2M). β2-microglobulin (B2M), a core component of MHC-I complexes, is essential for proper antigen presentation to cytotoxic T cells. Dysregulation of B2M expression impairs MHC-I surface localization and has been widely implicated in immune escape across multiple tumor types32,33,34. Collectively, these findings suggest that PLXNA3 may contribute to immune evasion in breast cancer by reducing T cell infiltration and impairing antigen presentation, underscoring its potential as a target to overcome immune resistance.
Immune checkpoint blockade is an irreplaceable approach in cancer immunotherapy, and its growing importance in breast cancer treatment has brought new hope to patients35. Our study showed that patients with higher levels of PLXNA3 displayed poorer clinical outcomes from immunotherapy. This suggests that targeting PLXNA3 with inhibitors, or using them in combination with immune checkpoint inhibitors, could potentially enhance the anti-cancer effects in breast cancer patients.
In summary, this study elucidates the prognostic and biological roles of PLXNA3 in breast cancer. It was found that silencing PLXNA3 markedly suppresses the proliferation, migration, and invasion of breast cancer cells, whereas its overexpression exerts the opposite effect. Moreover, the significant correlations between PLXNA3 expression, immune cell infiltration, and immune-related factors suggest that PLXNA3 could serve as a novel therapeutic target for overcoming breast cancer resistance.
Nevertheless, this study has several limitations. Firstly, functional validation was confined to in vitro assays using TNBC cell lines, thereby excluding HER2-positive and Luminal subtypes and constraining the broader applicability of our findings. Additionally, we did not conduct in vivo experiments, such as testing PLXNA3 inhibitors or combined with PD-1 inhibitors in animal models, to further assess therapeutic potential. Additionally, while a significant correlation between PLXNA3 expression and immune cell infiltration was observed, the precise molecular mechanisms remain elusive and merit detailed exploration through pathway analyses and co-culture experiments. In conclusion, PLXNA3 has great potential as a prognostic marker and therapeutic target for breast cancer.
Materials and methods
PLXNA3 expression level analysis
The expression levels of PLXNA3 in various tumor tissues and their corresponding adjacent normal tissues were analyzed using UALCAN (http://ualcan.path.uab.edu/)36. Additionally, we compared PLXNA3 expression across different stages, subtypes, and lymph node metastatic statuses in breast cancer patients.
Survival analysis
Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p) is a publicly accessible online database that integrates gene expression data and survival information from cancer patients, including breast cancer37. This database was utilized to investigate the association between PLXNA3 expression and survival rates in breast cancer patients as well as their responses to immunotherapy, and 95% confidence interval hazard ratios and log-rank p values were calculated.
Cell culture and transfection
The humanized triple-negative breast cancer cell lines MDA-MB-231 and MDA-MB-453 (obtained from the American Type Culture Collection) were cultured in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) at 37 °C in a 5% CO2 atmosphere. The culture medium was additionally fortified with streptomycin (100 µg/mL) and penicillin (100 IU/mL) to inhibit bacterial growth. Small interfering RNAs (siRNAs) targeting PLXNA3 (si-PLXNA3) and a non-targeting control siRNA (si-NC) were synthesized by GenePharma (Shanghai, China). Hongxin Biological Technology Co., Ltd. (Jiangsu, China) offered the control plasmid vector pcDNA, as well as the PLXNA3 overexpression plasmid pcDNA-PLXNA3. Nearly, 1 × 106 cells received the seeding process inside 6-well plates and the 24 h culturing process. Following the manufacturer’s guidelines, cells were transiently transfected with siRNA or overexpression plasmids using Lipofectamine 2000 (Invitrogen, USA) in serum-free medium for 48 h.
RNA extraction and qRT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Germany) following the provided protocol. The RNA purity was determined by measuring absorbance at 260/280 nm. Reverse transcription of RNA into cDNA was carried out using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). Transcript quantification was performed using specific primers from GeneRay (Shanghai, China) and qPCR was conducted on 96-well or 384-well plates (Applied Biosystems, USA) with SYBR Premix Ex Taq II (TaKaRa). The primer sequences for PLXNA3 were as follows: GAPDH-F: GGTGAAGGTCGGTGTGAACG, GAPDH-R: CTCGCTCCTGGAAGATGGTG; PLXNA3-F: TGGTGTAAGTACCGCCACAC, PLXNA3-R: GAGGTAGGTTCTTAGCCCGC-3. Relative expression levels were normalized against the internal control and calculated using the 2 − ΔΔCt method.
Colony formation assay
Inoculating 3,000 breast cancer cells onto six-well plates, they were then cultivated for 14 days. Crystal violet (Beyotime, China) was used to dye the cell colonies after they had been fixed by 4% polyacetal soaking for 10 min. Images and counts of the cell colonies were taken. All experiments were performed in triplicate to ensure reproducibility.
CCK-8 assay
MDA-MB-231 and MDA-MB-453 breast cancer cell lines were harvested post-transfection and incubated at 37 °C. Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8, Biosharp, China) according to the manufacturer’s instructions. Cells were seeded into 96-well plates and maintained at 37 °C in a humidified atmosphere containing 5% CO₂. At 0, 24, 48 and 72 h post-transfection, CCK-8 reagent was added to each well and incubated for 2 h. Absorbance was measured at 450 nm using a microplate reader. Each experiment was independently repeated three times to confirm reproducibility.
Wound healing assay
MDA-MB-231 cells were transfected and subsequently seeded into 6-well plates. Once the cells reached confluence, a sterile 20 µL pipette tip was used to create a scratch in the cell monolayer. The wells were rinsed with PBS to remove detached cells and debris, and fresh serum-free medium was added. Cell migration was observed at 0, 24, and 48 h, with images captured using an inverted microscope. The experiment was performed in triplicate, and the migration rate was quantified accordingly. All experiments were performed in triplicate to ensure reproducibility.
Transwell test
Cell invasion assays were performed using 24-well Transwell chambers (Corning, USA). A total of 50,000 cells from the PLXNA3-transfected group and control cells, suspended in 200 µL of medium, were seeded in the upper chambers coated with Matrigel. The lower chambers were filled with 600 µL of medium containing 10% FBS. After 24 h of incubation at 37 °C, non-invasive cells on the upper surface of the membrane were carefully removed. The invaded cells on the lower membrane surface were fixed with 4% paraformaldehyde and stained with crystal violet for 10 min. Photographs were captured, and the number of invaded cells were counted using ImageJ software. All experiments were conducted in triplicate to ensure the reliability and reproducibility of the results.
Immune-related analysis
The Tumor Immune Single-Cell Hub (TISCH) serves as a comprehensive single-cell RNA-sequencing (scRNA-seq) resource, specifically targeting the tumor microenvironment (TME). It offers intricate annotations of cell types at the single-cell level, facilitating the investigation of TME in various cancers, including breast cancer. TISIDB (http://cis.hku.hk/TISIDB/index.php) is a web-based platform that integrates diverse data types to study interactions between tumors and the immune system. In our analysis, we explore the Spearman correlations between PLXNA3 expression and tumor-infiltrating lymphocytes, immune stimulators, and human leukocyte antigens (HLAs) across a range of human cancers.
Statistical analysis
All analyses were conducted using GraphPad Prism (v9.0.0). Data from experimental replicates are expressed as mean ± standard deviation. Student’s t-test or one-way analysis of variance (ANOVA) were used for statistical analysis of all cell line experiments. Gene expression correlations were assessed using Spearman’s correlation. -values denoted by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Change history
07 January 2026
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-30560-x
References
Nicholson, W. K. et al. Screening for breast cancer: US preventive services task force recommendation statement. JAMA. 331, 1918–1930. https://doi.org/10.1001/jama.2024.5534 (2024).
Delman, K. A. Introducing the virtual tumor board series in CA: A cancer journal for clinicians. CA Cancer J. Clin. 70, 77. https://doi.org/10.3322/caac.21598 (2020).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Garcia-Martinez, L., Zhang, Y., Nakata, Y., Chan, H. L. & Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 12, 1786. https://doi.org/10.1038/s41467-021-22024-3 (2021).
Jing, Y. et al. Diagnostic value of 5 MiRNAs combined detection for breast cancer. Front. Genet. 15, 1482927. https://doi.org/10.3389/fgene.2024.1482927 (2024).
Oleari, R. et al. PLXNA1 and PLXNA3 cooperate to pattern the nasal axons that guide gonadotropin-releasing hormone neurons. Development. 146. https://doi.org/10.1242/dev.176461 (2019).
Kong, Y. et al. Structural basis for Plexin activation and regulation. Neuron. 91, 548–560. https://doi.org/10.1016/j.neuron.2016.06.018 (2016).
Worzfeld, T. et al. Genetic dissection of Plexin signaling in vivo. Proc. Natl. Acad. Sci. U S A. 111, 2194–2199. https://doi.org/10.1073/pnas.1308418111 (2014).
Guttmann-Raviv, N. et al. The neuropilins and their role in tumorigenesis and tumor progression. Cancer Lett. 231, 1–11. https://doi.org/10.1016/j.canlet.2004.12.047 (2006).
Neufeld, G. & Kessler, O. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat. Rev. Cancer. 8, 632–645. https://doi.org/10.1038/nrc2404 (2008).
Neufeld, G. et al. The semaphorins and their receptors as modulators of tumor progression. Drug Resist. Updat. 29, 1–12. https://doi.org/10.1016/j.drup.2016.08.001 (2016).
Franzolin, G. & Tamagnone, L. Semaphorin signaling in Cancer-Associated inflammation. Int. J. Mol. Sci. 20, 377. https://doi.org/10.3390/ijms20020377 (2019).
Yamada, D., Watanabe, S., Kawahara, K. & Maeda, T. Plexin A1 signaling confers malignant phenotypes in lung cancer cells. Biochem. Biophys. Res. Commun. 480, 75–80. https://doi.org/10.1016/j.bbrc.2016.10.006 (2016).
Ren, S. et al. Whole-genome and transcriptome sequencing of prostate cancer identify new genetic alterations driving disease progression. Eur. Urol. 73, 322–339. https://doi.org/10.1016/j.eururo.2017.08.027 (2018).
Zhao, X. Y., Chen, L., Li, Y. H. & Xu, Q. PlexinA1 expression in gastric carcinoma and its relationship with tumor angiogenesis and proliferation. World J. Gastroenterol. 13, 6558–6561. https://doi.org/10.3748/wjg.v13.i48.6558 (2007).
Nguyen, H. et al. Progesterone and 1,25-dihydroxyvitamin D₃ inhibit endometrial cancer cell growth by upregulating semaphorin 3B and semaphorin 3F. Mol. Cancer Res. 9, 1479–1492. https://doi.org/10.1158/1541-7786.Mcr-11-0213 (2011).
Syed, V. et al. Profiling estrogen-regulated gene expression changes in normal and malignant human ovarian surface epithelial cells. Oncogene. 24, 8128–8143. https://doi.org/10.1038/sj.onc.1208959 (2005).
Staton, C. A. et al. Expression of class 3 semaphorins and their receptors in human breast neoplasia. Histopathology. 59, 274–282. https://doi.org/10.1111/j.1365-2559.2011.03922.x (2011).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. https://doi.org/10.1038/nm.3394 (2013).
de Visser, K. E. & Joyce, J. A. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 41, 374–403. https://doi.org/10.1016/j.ccell.2023.02.016 (2023).
Dall’Olio, F. G. et al. Tumour burden and efficacy of immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 19, 75–90. https://doi.org/10.1038/s41571-021-00564-3 (2022).
Wang, S. J., Dougan, S. K. & Dougan, M. Immune mechanisms of toxicity from checkpoint inhibitors. Trends Cancer. 9, 543–553. https://doi.org/10.1016/j.trecan.2023.04.002 (2023).
Sadeghi Rad, H. et al. Understanding the tumor microenvironment for effective immunotherapy. Med. Res. Rev. 41, 1474–1498. https://doi.org/10.1002/med.21765 (2021).
Worzfeld, T. & Offermanns, S. Semaphorins and plexins as therapeutic targets. Nat. Rev. Drug Discov. 13, 603–621. https://doi.org/10.1038/nrd4337 (2014).
Kumanogoh, A. & Kikutani, H. Immunological functions of the neuropilins and plexins as receptors for semaphorins. Nat. Rev. Immunol. 13, 802–814. https://doi.org/10.1038/nri3545 (2013).
Walzer, T., Galibert, L., Comeau, M. R. & De Smedt, T. Plexin C1 engagement on mouse dendritic cells by viral semaphorin A39R induces actin cytoskeleton rearrangement and inhibits integrin-mediated adhesion and chemokine-induced migration. J. Immunol. 174, 51–59. https://doi.org/10.4049/jimmunol.174.1.51 (2005).
Ji, J. D., Park-Min, K. H. & Ivashkiv, L. B. Expression and function of semaphorin 3A and its receptors in human monocyte-derived macrophages. Hum. Immunol. 70, 211–217. https://doi.org/10.1016/j.humimm.2009.01.026 (2009).
Ch’ng, E. S. & Kumanogoh, A. Roles of Sema4D and Plexin-B1 in tumor progression. Mol. Cancer. 9, 251. https://doi.org/10.1186/1476-4598-9-251 (2010).
Picelli, S. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol. 14, 637–650. https://doi.org/10.1080/15476286.2016.1201618 (2017).
Potter, S. S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 14, 479–492. https://doi.org/10.1038/s41581-018-0021-7 (2018).
Barnkob, M. B. et al. Semmaphorin 3 A causes immune suppression by inducing cytoskeletal paralysis in tumour-specific CD8 + T cells. Nat. Commun. 15, 3173. https://doi.org/10.1038/s41467-024-47424-z (2024).
Li, D. et al. β2-Microglobulin maintains glioblastoma stem cells and induces M2-like polarization of Tumor-Associated macrophages. Cancer Res. 82, 3321–3334. https://doi.org/10.1158/0008-5472.Can-22-0507 (2022).
Fu, X. et al. Med23 deficiency reprograms the tumor microenvironment to promote lung tumorigenesis. Br. J. Cancer. 130, 716–727. https://doi.org/10.1038/s41416-023-02556-9 (2024).
Wu, X. et al. Targeting MHC-I molecules for cancer: function, mechanism, and therapeutic prospects. Mol. Cancer. 22, 194. https://doi.org/10.1186/s12943-023-01899-4 (2023).
Hargadon, K. M., Johnson, C. E. & Williams, C. J. Immune checkpoint Blockade therapy for cancer: an overview of FDA-approved immune checkpoint inhibitors. Int. Immunopharmacol. 62, 29–39. https://doi.org/10.1016/j.intimp.2018.06.001 (2018).
Huang, X. et al. Association of DBNDD1 with prognostic and immune biomarkers in invasive breast cancer. Discov Oncol. 16, 218. https://doi.org/10.1007/s12672-025-01990-w (2025).
Luo, Z. et al. Cytokine-induced apoptosis inhibitor 1: a comprehensive analysis of potential diagnostic, prognosis, and immune biomarkers in invasive breast cancer. Transl Cancer Res. 12, 1765–1786. https://doi.org/10.21037/tcr-23-34 (2023).
Funding
The study was supported in part by the National Natural Science Foundation of China (81771953 W.Z.), the Natural Science Foundation of Jiangsu Province (BK20180108 W.Z., BK20230017 W.Z.), a project funded by Jiangsu province college students’ practice innovation training programs (202210312026Z W.Z.) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (W.Z.).
Author information
Authors and Affiliations
Contributions
Design of the experiments: WS, YFX, WBZ and SW; Acquisition of data: WS, GSS, YFX, YQ and XWS; Analysis and interpretation of data: WS, SW, QQ and WYZ; Validation: WS, YFX, QQ and WYZ; Writing-Original Draft: WS, QQ, YFX and WYZ; Funding Acquisition: WBZ; Writing-Review & Editing: WS, YFX, QQ and WBZ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained errors in Figure 5 where the image representing the ‘vector group at 0h’ was inadvertently duplicated from the ‘pcDNA-PLXNA3 group at 0h’ in panel (b), and the layout for each group incorrectly contained two images, resulting in panel (d) displaying the 'pcDNA-PLXNA3 group' with two images instead of one.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, W., Xu, Y., Qi, Q. et al. Comprehensive analysis of the PLXNA3 gene on prognosis and immune characteristics in breast cancer. Sci Rep 15, 28356 (2025). https://doi.org/10.1038/s41598-025-13843-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13843-1