Abstract
Accurate rectal tumor segmentation using magnetic resonance imaging (MRI) is paramount for effective treatment planning. It allows for volumetric and other quantitative tumor assessments, potentially aiding in prognostication and treatment response evaluation. Manual delineation of rectal tumors and surrounding structures is time-consuming and labor-intensive. Over the past few years, deep learning has shown strong results in automated tumor segmentation in MRI. Current studies on automated rectal tumor segmentation, however, focus solely on tumoral regions without considering the rectal anatomical entities and often lack a solid multicenter external validation. In this study, we improved rectal tumor segmentation by incorporating anomaly maps derived from anatomical inpainting. This inpainting was trained using a U-Net-based model and trained to reconstruct a healthy rectum and mesorectum from prostate T2-weighted images (T2WI). The rectal anomaly maps were generated from the difference between the original rectal and reconstructed pseudo-healthy slices. The derived anomaly maps were used in the downstream tumor segmentation tasks by fusing them as an additional input channel (AAnnUNet). Alternative methods for integrating rectal anatomical knowledge were evaluated as baselines, including Multi-Target nnUNet (MTnnUNet), which added rectum and mesorectum segmentation as auxiliary tasks, and Multi-Channel nnUNet (MCnnUNet), which utilized rectum and mesorectum masks as additional input channels. As part of this study, we benchmarked nine models for rectal tumor segmentation on a large multicenter (num = 705) dataset of preoperative T2WI and nnUNet outperformed the other eight models on the external test. The MTnnUNet demonstrated improvements in both fully-supervised and mixed-supervised settings where human-annoated tumor masks and AI-generated rectum and mesoretum masks were used compared to nnUNet, while the MCnnUNet showed benefits only in the setting where mixed-supervision were used. Importantly, anomaly maps were strongly associated with tumoral regions, and their integration within AAnnUNet led to the best tumor segmentation results across both settings. The effectiveness of AAnnUNet demonstrated the value of the anomaly maps, indicating a promising direction for improving rectal tumor segmentation and model robustness for multicenter data.
Similar content being viewed by others
Introduction
Magnetic Resonance Imaging (MRI) plays a pivotal role in staging rectal cancer and selecting treatment plans, providing valuable information regarding the extent of tumor infiltration within and beyond the bowel wall, and into critical anatomical structures, including perirectal vessels, the mesorectal fascia (MRF), peritoneum, and neighboring pelvic organs1,2. T2-weighted imaging (T2WI) forms the mainstay of the MRI protocol because of its superior soft tissue contrast to discern the different layers of the rectal wall, mesorectal fat, adjacent vessels, and MRF to allow for detailed local staging3,4.
Precisely segmenting the tumor is an important task in rectal cancer management. Tumor segmentations are utilized for several purposes including radiation treatment planning, volumetric analysis, and extraction of imaging biomarkers. Tumor delineation by radiologists is considered the current gold standard. Nevertheless, it is time-consuming and subject to substantial intra- and inter-observer variation5,6,7. Developing an accurate, generalizable, and robust rectal tumor segmentation model can help reduce this variability and assist in standardizing several steps of diagnostic and therapeutic rectal cancer management.
Deep learning in rectal tumor segmentation
Deep learning (DL) has seen a rapid uptake in several fields, achieving promising results in medical image analysis8. Convolutional Neural Network-based (CNN) based DL approaches excel in learning image representations from annotated data by using learnable feature extraction filters and sequential convolution, activation, and pooling operations. Among CNNs, the U-Net9 and its variants are the most popular architecture. Studies10,11,12 have explored the ability of 2D U-Net and other 2D CNNs variants. These CNNs achieved a Dice similarity coefficient score (DSC) ranging from 0.59 to 0.84 for tumor segmentation. Although 2D CNNs are less computationally expensive, 3D models can leverage richer context to improve predictions13.
Hamabe et al.14 implemented a 3D U-Net, achieving an averaged DSC of 0.73 (0.60–0.80) over 10-fold cross-validation. However, there was no external validation in their study. Besides CNN-based models, transformer-based architectures are also being applied in medical image analysis due to their ability to access long-range semantic information15. Li et al.16 proposed RTAU-Net, a novel 3D dual path fusion network containing a transformer encoder for extracting global contour information of the rectal tumor. RTAU-Net achieved an averaged DSC of 0.80 and 0.68 in data from two respective medical centers. However, RTAU-Net requires the manual removal of tumor-free slices, which hinders the fully automated implementation. Additionally, RTAU-Net was not compared against the state-of-the-art medical segmentation networks, such as nnUNet17, a self-configuring implementation of the U-Net architecture, or nnFormer18, which introduces 3D transformer blocks on top of nnUNet.
Deep learning in rectum and mesorectum segmentation
Besides rectal tumor delineation, some studies also demonstrated that CNNs can accurately delineate anatomical structures such as rectum and mesorectum or perirectal fat with DSC above 0.9014,19,20. Automated rectum and mesorectum delineation could potentially improve radiological evaluation. The prognosis for rectal cancer depends on how far the tumor has infiltrated the layers of the rectal wall and the mesorectum, and the successful attainment of negative circumferential resection margins (CRMs) through surgical intervention21. Additionally, Lee et al.22 demonstrated that a 2D model’s variance in tumor regions can be reduced by 90% by incorporating rectal segmentation on the model’s objective. Integrating rectal anatomical knowledge can provide a more comprehensive representation of the T2WI, allowing the model to learn richer and more nuanced patterns, leading to better performance on unseen data. However, the impact of adding rectal anatomical structures including mesorectum for rectal tumor segmentation has not been investigated in a multi-institutional setting. Also, incorporating rectal anatomical structure has so far been limited to adding additional segmentation tasks as presented in Lee et al.22.
Anatomy-aware inpainting for anomaly detection
Most automated medical image segmentation methods rely on supervised learning, which requires large volumes of reliably labeled data—often difficult to obtain in medical imaging. As a result, semi-supervised and unsupervised methods have gained increasing attention. Anomaly detection, which can operate in both settings, has been applied to tasks such as segmentation. Generative models—such as Generative Adversarial Networks (GANs)23, Autoencoders (AEs), and their variants including Variational Autoencoders (VAEs)24 and Vector Quantized VAEs (VQ-VAEs)25—have shown promise in this field26,27,28. These models are typically trained to reconstruct images from a distribution of normal tissue; when presented with anomalies (e.g., tumors), they often fail to reconstruct the affected regions, resulting in higher reconstruction errors.
When trained on full MRI slices, these models are required to reconstruct the entire image—including both relevant and irrelevant anatomy—which can impair accurate modeling of healthy structures and lead to errors in both normal and abnormal regions. Consequently, anomaly maps—pixel-wise error maps designed to localize abnormalities—may incorrectly flag normal anatomical variations or imaging artifacts as anomalies, reducing specificity.
Unlike image-to-image reconstruction, inpainting focuses on restoring missing or occluded image regions using surrounding context, typically with dataset-independent masks. Nguyen et al.29 applied inpainting for brain tumor segmentation in T1-weighted MRI by identifying regions with the highest reconstruction loss. Incorporating anatomical priors allows inpainting to target high-risk areas, minimizing background influence. Yeganeh et al.30 introduced an anatomy-aware masking strategy to improve organ shape learning, while Woo et al.31 proposed a UNet-based model for bone lesion detection in knee MRI via inpainting, showing that identified anomalies can support downstream segmentation.
In anatomical inpainting based anomaly detection, the region of interest (ROI) is masked—typically covering relevant structures—and the model is trained to reconstruct these regions assuming normal anatomy. The difference between the original and reconstructed images is then used to localize anomalies.
Our contributions
In this study, we present several key contributions aimed at advancing rectal tumor segmentation in T2WI. We developed and evaluated a rectal tumor segmentation model that incorporates anomaly maps generated through anatomical inpainting, achieving improved segmentation performance. These anomaly maps were derived using a novel end-to-end inpainting model trained exclusively on prostate T2-weighted imaging (T2WI)32 and applied to rectal T2WI, challenging conventional domain-specific practices and demonstrating the potential of transfer learning. The generated maps can also support various clinical downstream tasks.
Unlike for medical challenges with public datasets like clinically significant prostate lesion segmentation (PICAI)32 or brain tumor segmentation33, there is no large multicenter publicly available MRI dataset for rectal cancer studies. This makes it difficult to benchmark different models. An extensive external validation study with multicenter data is highly desirable to compare different deep learning segmentation approaches. We benchmarked nine state-of-the-art 3D deep learning models for rectal tumor segmentation using a large multicenter dataset, offering comprehensive comparative insights. A 3D deep learning model was specifically developed to segment rectal anatomical structures, including the rectum and mesorectum (fatty tissue surrounding the rectum). Additionally, we explored multiple strategies for integrating rectal anatomical information into tumor segmentation, including the use of anomaly maps, auxiliary segmentation tasks, and prior knowledge, demonstrating their potential to both improving segmentation accuracy and clinical utility. These contributions collectively address critical challenges in rectal MRI analysis, advancing the field toward more robust and clinically applicable solutions.
Results
Dataset and patient characteristics
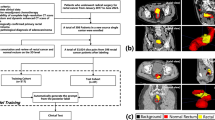
As a part of a previous institutional review board approved multicenter study project34,35,36,37,38,39, the clinical and imaging data from 1426 patients with biopsy-proven rectal cancer were retrospectively collected from ten medical centers between 2011 and 2018. The original study was conducted in accordance with the Declaration of Helsinki and informed consent was waived due to the retrospective nature of the study. For the current study, the baseline staging T2WI of 705 patients (from 9 centers) was selected from this previous dataset. Patients were excluded if they had one of the following properties: non-diagnostic image quality issues, multiple tumors in the field of view, abscesses surrounding the tumor, unavailability of pre-treatment T2WI, or the unavailability of axial images. Table 1 shows the patient characteristics of all the rectal cancer patients including age, gender, clinical T and N staging, tumor location, and EMVI status. To train the anatomical inpainting model for reconstructing the healthy rectum and mesorectum, 300 samples with healthy rectum and mesorectum were randomly selected from the PICAI dataset.
Rectal anomaly detection
According to Table 2, the inpainting model demonstrated superior performance and less variance in reconstructing the rectum and mesorectum in prostate T2WI, with an averaged SSIM (aSSIM) of 86.72 and an averaged PSNR (aPSNR) of 25.87, compared to in-house rectal T2WI, which had an aSSIM of 83.38 and an aPSNR of 23.87. These results aligned with expectations. Prostate T2WI was in-distribution and represented healthy samples of the rectum and mesorectum. Conversely, rectal T2WI with tumor regions were out-of-distribution data and exhibited greater discrepancies from their corresponding pseudo-healthy counterparts. Importantly, we observed that aSSIM, 39.32, and aPSNR, 17.50 of tumoral regions are noticeably lower than the tumor-free regions, indicating effective anomaly detection. The anomaly map can be generated from the differences between original and pseudo-healthy images (Fig. 1).
The visualization of anatomical inpainting. The columns from left to right are original T2WI slices, masked slices (grey: rectum; black: mesorectum), inpainted slices, anomaly map, which is the difference between original and reconstructed slices, and tumor masks. The first two rows are prostate T2WI (without tumor), and the last two rows are from rectal T2WI (with tumor). Colorbar: Shows pixel-wise reconstruction error; higher values indicate greater differences and potential anomalies.
The comparison of nine DL models for rectal tumor segmentation
In the internal dataset (Training Cohort 1, 5-fold cross-validation), MedFormer delivered the best overall performance, with an average DSC (aDSC) of 66.3 and a median 95% HD (mHD) of 6.39 mm. U-mamba achieved the highest median DSC (mDSC), while ResUNet excelled in the average 95% HD (aHD), see Table S1. However, in the external testing data (num = 666, 9 centers), as is displayed in Table 3, S2, and Fig. 2a, nnUNet achieved the best results, with aDSC 62.8 and aHD 17.28 mm, significantly better than others. We observed that nnUNet consistently outperformed other models in the external test when the training cases was increased to 132 from a single center, see Tables S3, S4. Additionally, transformer-based networks including UNetR, SwinUNet, and nnFormer or the SSMs-inspired architecture like U-Mamba underperformed with respect to CNN-based architectures in external test. We also compared the size of trainable parameters and Floating Point Operations (FLOPs) for each model (Fig. 2b). In summary, nnUNet demonstrated the highest DSC despite having relatively few model parameters and a low number of FLOPs. From Fig. 3, nnUNet showed better prediction performance in identifying rectal tumors in the external test. nnUNet exhibited a significantly reduced false positive voxel classification in these displaying cases, especially in the last row of Fig. 3, which was a tumor-free slice.
The visualization of the segmentation performance of all nine DL models using T2WI. Each row is a different patient from a different center (external set). The columns from left to right are original T2WI, GT: ground truth, tumor prediction masks from UNet, ResUNet, UNetR, SwinUNetR, Atten-UNet, MedFormer, nnFormer, U-Mamba, and nnUNet.
Rectum and mesorectum segmentation
nnUNet demonstrated superior performance for rectum segmentation with an aDSC of 0.87 and mHD 10.15 mm, better than the mesorectum segmentation with an aDSC of 0.81, aHD 10.57 mm in the external cohort containing 141 samples see Table S5, Fig. S1. This disparity in performance between the rectum and mesorectum is attributed to the rectum’s more regular shape than that of the mesorectum. This is consistent with the findings by Hamabe et al.14. Overall, the segmentation tasks for rectal anatomical structures were significantly easier than for tumors.
MTnnUNet, MCnnUNet and AAnnUNet (Fully-Supervision)
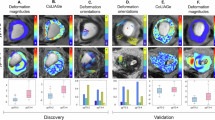
From Table 4, S6 and Fig. 4a, MTnnUNet and AAnnUNet significantly outperformed both nnUNet and MCnnUNet. Even though MCnnUNet showed the best results in the internal validation (Table S7), it exhibited the lowest aDSC and aHD in the external test. This may be due to MCnnUNet’s reliance on accurate anatomical inputs. It was trained with ground truth anatomical masks but used AI-generated masks (rectum and mesorectum) during inference, introducing inconsistencies that likely contributed to the performance drop on the external test. Unlike MCnnUNet, MTnnUNet uses anatomical knowledge only during training. Although AAnnUNet—fusing anomaly maps that highlight tumoral regions—also relied on the quality of rectum and mesorectum masks, it slightly outperformed MTnnUNet in terms of aDSC. Unlike MCnnUNet, which directly incorporated anatomical masks as input channels, AAnnUNet utilized anomaly maps derived from healthy distributions. We also introduced a union ensemble combining MTnnUNet and AAnnUNet, which improved aDSC by 3%. However, it did not reduce HD. Figure 5 shows that AAnnUNet effectively segmented both small and large tumors, demonstrating the benefit of anomaly fusion. Furthermore, the Grad-CAM saliency map in Fig. S2 indicates that AAnnUNet more effectively captures tumoral features. However, its performance declined when anomaly maps were suboptimal, as seen in the last row. In some cases, all models failed to detect the tumor, though high anomaly map intensities—illustrated in Fig. 6—still indicated potential abnormalities.
The visualization of the segmentation performance of nnUNet, MTnnUNet, MCnnUNet, AAnnUNet, and Ensemble using T2WI, supervised setting. Each row is a different sample from the external test set. The columns from left to right are original T2WI, ground truth, tumor prediction masks from nnUNet, MTnnUNet, MCnnUNet, AAnnUNet, and Ensemble. Colorbar: Shows pixel-wise reconstruction error; higher values indicate greater differences and potential anomalies.
MTnnUNet, MCnnUNet and AAnnUNet (Mixed-Supervision)
Mixed-supervision, using manually annotated tumors and AI-generated rectum and mesorectum masks, was applied during training on cohort 2, which included 141 cases from a single center for nnUNet, MTnnUNet, MCnnUNet, and AAnnUNet. In the internal validation set, MTnnUNet achieved the best performance, while AAnnUNet performed best on the external test. Unlike the fully supervised setting, MCnnUNet showed improved aDSC and aHD on the external test. This may be attributed to the consistent use of AI-generated rectum and mesorectum masks during both training and inference, leading to more stable performance across datasets (Table 5, Tables S8, S9, Fig. 4b).
In both fully- and mixed-supervision, MTnnUNet outperformed the baseline nnUNet. MCnnUNet showed improvement only in mixed-supervision. Notably, incorporating anomaly maps enhanced tumor localization accuracy across both settings. As illustrated in the first row of Fig. S3, while nnUNet misclassified part of the bladder as tumor due to similar intensities, MTnnUNet, MCnnUNet, and AAnnUNet—fusing anatomical masks or anomaly maps—correctly identified the tumor regions.
Methods
Ground truth segmentation
The ground truth segmentation masks were annotated by a GIT radiologist (M.A.A) with 6–7 years of radiological experience in interpreting rectal MRI. Masks for the rectum and mesorectum were annotated using 180 randomly selected rectal cases and 100 prostate T2W images. Specifically, the entire rectum was annotated, including its lumen from the anorectal junction to the recto-sigmoid junction, following the definition of the sigmoid take-off as the upper anatomical landmark of the rectum. The mesorectal fat, enveloped by the mesorectal fascia, was identified as the high T2 signal area surrounding the rectum on all sides, thinner anteriorly than postero-laterally40,41. Moving caudally towards the lower rectum, the thickness of the mesorectal fat enveloping the rectal wall decreases due to the gradual tapering of the mesorectum41. Tumor segmentation was annotated for all the cases (n = 705), where tumors were labeled as abnormal mural growth within the rectal lumen and extending outside into the mesorectum41,42. See Table S10 for annotation details.
Rectal anomaly detection
The overall pipeline, inspired by43,44, for detecting rectal anomalies is shown in Fig. 7a. A single inpainting model was trained to generate both the rectum and mesorectum. Prostate T2WI with masked healthy rectum and mesorectum were used to train the model for their reconstruction. The inpainting model was adapted from Han et al.43, an end-to-end MRI sequence generation framework. The framework consists of two stages. In the first stage, only the reconstruction loss is optimized for the encoder and generator. In the second stage, both adversarial loss and cycle-consistent loss are incorporated in addition to the reconstruction loss, with the optimization applied to the encoder, generator, and discriminator. The training was based on 2D slices. The inpainting model contains an encoder \({\varvec{E}}\) and a decoder \({\varvec{G}}\). A masked (rectum and mesorectum) 2D T2WI slice X, can be compressed by \({\varvec{E}}\) into a latent space \(z={\varvec{E}}\left( X \right)\) and \({\varvec{G}}\) can reconstruct the original slice from the latent representation z.The skip connections were added to recover fine-grained details. To enforce similarity between the generated slices and the actual slices, a supervised reconstruction loss is used:
where X is the original slice, and \(X^{\prime}={\varvec{G}}\left( {{\varvec{E}}\left( X \right)} \right)\) is the restored image. \({\left\| \cdot \right\|_1}\) is the \({L_1}\) loss and \({L_p}\) is the perceptual loss from pre-trained VGG19, which involves comparing high-level features (not just pixel values) from both the generated and reference images45. Instead of measuring raw pixel differences, it evaluates how similar the images are in terms of their content and style, based on features extracted from different layers of the VGG19 network. \({{\text{\varvec{\uplambda}}}_r}\) and \({{\text{\varvec{\uplambda}}}_p}\) are weight factors, for which the respective values 10 and 0.01 were chosen empirically.
(a) The anatomical inpainting workflow. The inpainting model containing an encoder E and a decoder G was trained using prostate T2WI with a healthy rectum and mesorectum. The trained model was inferred across the rectal dataset to generate a reconstructed pseudo healthy rectum and mesorectum. The difference between the reconstructed and the original image is the anomaly map. Ground Truth tumor is in red. Colorbar: Shows pixel-wise reconstruction error; higher values indicate greater differences and potential anomalies. (b) The workflow figure connecting anatomy nnUNet, inpainting and AAnnUNet/MCnnUNet during inference.
For the second stage of the training, the adversarial loss and cycle-consistent loss46 were added on top of the reconstruction loss to ensure that the inpainted images were both realistic and consistent with the original images. The adversarial loss helps to ensure that the completed regions look realistic and blend seamlessly with the surrounding areas and the cycle-consistent loss focuses on preserving the original structure by ensuring the inpainted image can be accurately reconstructed back to the original.
where \(X^{\prime\prime}=~~\) \({\varvec{G}}\left( {{\varvec{E}}\left( {X^{\prime}} \right)} \right)\) and \({\left\| \cdot \right\|_2}\) is the \({L_2}\) loss and \({\varvec{D}}\) is the discriminator. The anomaly maps are then defined as the absolute differences between the reconstructed slice and the original slice,
Let I be an image with intensity values. The normalization scheme before training involved the following steps:
\(l=Percentil{e_{0.5}}\left( I \right)\)
\(h=Percentil{e_{99.5}}\left( I \right)\)
\({I_{norm}}=\frac{{max\left( {I,l} \right) - l}}{{h~ - ~l}}\)
First Compute the 0.5th percentile (l) and the 99.5th percentile (h) of the intensity values in the image and normalize the intensity values in the image using the computed percentiles. The rectum was masked with a value of 0, while the mesorectum was masked with a value of 0.5.
To train the inpainting model, 100 prostate T2WI with manually segmented healthy rectum and mesorectum masks were split into 80 for training and 20 for internal validation. The model was externally tested on all slices of 200 prostate T2WIs and 705 in-house rectal T2WIs. The inference also requires rectum and mesorectum masks. However, only 180 rectal samples have radiologist-annotated rectum and mesorectum masks. To overcome this, the anatomy nnUNet (see Sect. 3.3) was used to generate the required masks (Fig. 7b).
Rectum and mesorectum segmentation
A 3D nnUNet model, referred to as anatomy nnUNet, was specifically trained to segment the rectum and mesorectum using Training Cohort 1 with five-fold cross-validation, as illustrated in Fig. 8a. The model was then externally evaluated on the rest 141 cases from different centers. The anatomy nnUNet was then used to infer all the rectum and mesorectum masks. The predicted rectum and mesorectum masks were defined as AI-generated rectum and mesorectum masks.
(a) Model definition. A: nnUNet for tumor segmentation only; B: MTnnUNet: multi-target nnUNet, segmenting tumor, rectum, and mesorectum simultaneously, C: MCnnUNet: multi-channel nnUNet with rectum and mesorectum masks, and T2WI as input, D: AAnnUNet: Anomaly-Aware nnUNet, with anomaly maps as additional input E: Anatomy nnUNet, to segment rectum and mesorectum. F: MTnnUNet but trained with AI-generated rectum and mesorectum, G: MCnnUNet but using AI-generated rectum and mesorectum, H: AAnnUNet but using AI-generated rectum and mesorectum (b) The training scheme for different models. 5 F-CV: 5-fold cross-validation. (b) Fully-Supervised: Training with entirely manual annotations. Mixed-Supervised: Training with manually annotated tumors and AI-generated rectum and mesorectum.
MTnnUNet, MCnnUNet and AAnnUNet (Fully-Supervision)
We incorporated anomaly maps from the inpainting model into rectal tumor segmentation by adding them as an additional input to nnUNet, referred to as Anomaly-Aware nnUNet (AAnnUNet). This approach was compared with two alternative strategies for integrating anatomical knowledge: Multi-Target nnUNet (MTnnUNet), which added rectum and mesorectum segmentation as auxiliary tasks, and Multi-Channel nnUNet (MCnnUNet), which used rectum and mesorectum masks as additional input channels (Fig. 8a). Same as the benchmark settings, MTnnUNet, MCnnUNet, and AAnnUNet were trained on training cohort 1 using 5-fold cross-validation and externally tested on 666 samples from 9 centers, as illustrated in Fig. 8b. For the inference of MCnnUNet and AAnnUNet, AI-generated rectum and mesorectum were used. We also included ensemble results, which is a union of MTnnUNet and AAnnUNet. Results from both 5-fold internal validation and external testing are presented.
MTnnUNet, MCnnUNet and AAnnUNet (Mixed-Supervision)
Using AI-generated pseudo-anatomical structures, MTnnUNet, MCnnUNet, and AAnnUNet were trained on Training Cohort 2 (141 cases) with 5-fold cross-validation (see Fig. 8). Instead of ground truth rectum and mesorectum masks, AI-generated annotations were used, combining them with manually labeled tumors. We also included ensemble results from the union of MTnnUNet and AAnnUNet. Results from both internal validation and external testing (564 cases from eight centers) were presented.
The comparison of nine DL models for rectal tumor segmentation
We established a baseline for rectal tumor segmentation by evaluating the performance of nine 3D deep learning models (see Fig. 9), including UNet47, ResUNet48, UNetR49, SwinUNetR50, AttentionUNet (Atten-UNet)51, MedFormer52, nnFormer18, U-Mamba (bot)53 and nnUNet17.
The rectal tumor segmentation performances of nine 3D DL models were compared. All the models were trained with training cohort 1 including 39 patients from Center 9 only and externally tested with 666 samples from 9 centers. Manual annotated tumor segmentation by the expert was used for both training and external test.
-
1.
UNet47, extends the previous u-net architecture from Ronneberger et al.9 by replacing all 2D operations with their 3D counterparts.
-
2.
ResUNet48, is a modified version of UNet. It replaces double convolution layers of UNet with residual blocks from ResNet54, incorporating shortcut connections for faster convergence. This adaptation works in both 2D and 3D settings, enhancing performance in capturing complex patterns.
-
3.
UNETR49, adopts a ViT-inspired encoder and employs a CNN decoder for 3D image segmentation. The images are initially divided into patches, which are linearly transformed into token embeddings. These tokens undergo processing through a self-attention block, akin to ViT. To manage the quadratic complexity of self-attention, the patch size is set to be relatively large (16) to prevent overly long sequence lengths.
-
4.
SwinUNETR50, reformulates the segmentation task as a sequence-to-sequence prediction using a Swin Transformer as the encoder. The encoder is then connected to a Fully Convolutional Neural Network (FCNN)-based decoder through skip connections.
-
5.
Attention UNet51, introduces an attention-gating module to UNet to enhance its ability to suppress irrelevant regions and highlight salient features crucial for a given task.
-
6.
nnFormer18, is a 3D transformer for volumetric medical image segmentation nnFormer combines interleaved convolution and self-attention operations. It introduces a special self-attention mechanism to understand both local and global aspects of the image volume. To improve efficiency, it also uses skip attention instead of the usual concatenation or summation operations.
-
7.
MedFormer52, is a transformer-based designed to handle scalable 3D medical image segmentation, including three crucial components: a beneficial inductive bias, hierarchical modeling using linear-complexity attention, and multi-scale feature fusion that combines spatial and semantic information globally. MedFormer can learn from both small and large scale data without pre-training.
-
8.
U-Mamba53, is inspired by the State Space Sequence Models (SSMs)55, which are known for their ability to handle long sequences. The model is designed specifically for biomedical image segmentation with the hybrid CNN-SSM block that integrates the local feature extraction power of convolutional layers with the abilities of SSMs to capture the long-range dependency.
-
9.
nnUNet17, is a self-configuring framework for medical image segmentation. It utilizes UNet as its architecture but offers a specialized preprocessing, training technique, and hyper-parameter configuration. nnUNet achieves state-of-the-art performance on several medical image segmentation challenges with a relatively simple architectural design.
-
10.
Anatomy nnUNet, is the nnUNet trained to segment rectal-related anatomical structures including the rectum and mesorectum.
-
11.
MTnnUNet, is the nnUNet trained to segment rectum, mesorectum, and rectal tumors.
-
12.
MCnnUNet, is the nnUNet trained to segment rectal tumors with rectum and mesorectum masks as additional input channels.
-
13.
AAnnUNet, is the nnUNet trained to segment rectal tumors with anomaly maps \(\:M\) derived from anatomical inpainting as an additional input channel.
All the models underwent training using training cohort 1 (39 cases) with a 5-fold cross-validation. Subsequently, all models were externally tested on the remaining 666 samples from nine centers. Training cohort 1 is relatively small. To assess the effect of training size, we repeated the experiments using 132 cases from the same center to evaluate whether increasing the number of cases would influence the performance of the nine benchmarked models. Results from both 5-fold internal validation and external test were presented.
Implementation details
For the training of the inpainting models, the input patch size was (384, 384), with a batch size of 1. We used the AdamW optimizer to train the network (both stages) with \(\:{\upbeta\:}\) 0.9 and 0.95, an initial learning rate of 0.0001, a weight decay factor of 0.05, and following a polynomial decay.
For tumor segmentation, the imaging preprocessing was adopted from nnUNet, which included ZScoreNormalization for standardizing intensities, uniform resampling of all images, and a cropping process. All the segmentation models were trained with randomly initialized weights without transfer learning. The batch size was set to 2 and the models were trained for 1000 epochs with the SGD optimizer. The loss function is the sum of the cross-entropy and Dice loss. During inference, predictions were obtained by averaging the outputs of each model resulting from the 5-fold cross-validation procedure. All the models were implemented in PyTorch (Torch version 2.1.2), and the training was conducted on an NVIDIA RTX A6000 GPU.
Evaluation metrics and statistical analysis
Statistical analysis was conducted in Python (version 3.9) with the SciPy package (version 1.13.1). To assess reconstruction performance, Structural Similarity Index (SSIM) and Peak Signal-to-Noise Ratio (PSNR) were calculated between the reconstructed and original images. To measure the segmentation performance, the Dice Similarity Coefficient score (DSC) and 95% Hausdorff Distance (HD) were utilized on both cross-validation and external tests. To provide a comprehensive comparison of the nine benchmarked models, we report the number of trainable parameters and inference-time floating point operations (FLOPs) for each. The characteristic differences of cohorts were compared by the Kruskal-Wallis test. The Mann–Whitney U-test was used to compare the difference of indicators among different methods. The model performance differences were calculated using the paired sample t-test. All statistical analyses were two-sided and p-values below 0.05 were regarded as statistically significant. 95% confidence intervals were generated using the bootstrap method with 10,000 replications.
Discussion and conclusion
In this study, we successfully developed a rectal anomaly detection model by training the anatomical inpainting model using prostate T2WI with healthy rectum and mesorectum. The model was then applied to the rectal T2WI to generate pseudo-healthy rectal structures and the anomaly map was the differences between the pseudo-healthy and original slices. The derived anomaly maps were used in the downstream tumor segmentation tasks and outperformed the baseline nnUNet, MTnnUNet, which jointly predicts tumor, rectum, and mesorectum, and MCnnUNet including rectum and mesorectum as additional input channels, in both the fully-supervised and mixed-supervised settings. To automatically generate the rectum and mesorectum masks, we trained an nnUNet to effectively delineate the normal rectum (without tumoral involvement) and mesorectum.
As part of this research, we benchmarked nine DL models, including CNN-based, transformer-based, and Mamba-based architectures, on a large multicenter dataset. nnUNet achieved the best results in the external test set, despite having a relatively lower model complexity, indicating that increased model complexity does not guarantee improved results. By fusing anatomical knowledge into the tumor segmentation model, MTnnUNet, MCnnUNet, and AAnnUNet demonstrated improved performance compared to nnUNet. Research in medical image analysis with AI bears many promises to improve patients’ health. However, Varoquaux et al.56 have pointed out that in academia, even though the goal is to solve scientific problems, the emphasis on publication quantity is influenced by Goodhart’s law57, which can compromise scientific quality. Researchers, in pursuit of novelty, may introduce unnecessary complexity in methods, contributing to technical debt without substantial improvement in predictions. Isensee et al.58 has conducted an extensive benchmarking of current segmentation methods across different dataset and their results revealed a concerning trend that most models introduced in recent years fail to outperform the nnUNet. This is consistent with the findings of this study. Recently published models with higher complexity did not exhibit higher tumor segmentation performance or better generalization ability. Although ViT and SSMs have demonstrated promising results in the natural image classification15, in rectal tumor contouring, transformer-based networks or SSMs-inspired networks do not show better results than CNN-based architectures. The reasons could be, that the transformer relies heavily on large-scale training and remains inferior to CNNs when training data is scarce. Unlike natural images, medical datasets are smaller, typically in the hundreds or thousands59. Secondly, the quadratic complexity of self-attention poses challenges when dealing with long token sequences50, especially for 3D T2WI.
Beyond architectural optimization, a profound comprehension of medical imaging is integral to advancing tumor segmentation. The rectum and mesorectum provide crucial anatomical context for the localization of tumors. Several studies60,61 have proved that mesorectal fat and tumor neighboring regions contain important prognostic information in rectal cancer patients. Therefore, there is a substantial demand for a rectum and mesorectum segmentation model that is accurate and robust. While previous studies14,19,20,22 have proposed segmentation networks for these structures, an extensive multi-center external test is highly desirable, and the majority employed 2D models. In this study, we externally tested the anatomical 3D nnUNet and observed successful rectal structure contouring.
Currently, most rectal tumor segmentation studies rely on retrospective datasets, which do not provide a healthy representation of the rectum. Abdominal imaging, particularly prostate MRI, shares overlapping anatomical structures with rectal MRI, and most prostate MRIs display healthy rectal structures. As a cross-domain application, we utilized a public prostate dataset to train the inpainting model, allowing it to learn the distribution of a healthy rectum and mesorectum. This design ensures that, during inference, any deviation from the learned healthy patterns—such as tumor tissue—results in a higher reconstruction error, which is reflected in the anomaly maps. Compared to simpler fusion strategies like MCnnUNet and MTnnUNet, AAnnUNet demonstrated a stronger anatomical understanding of the rectum and mesorectum, leading to improved tumor localization. MCnnUNet uses rectum and mesorectum masks to guide the model toward relevant anatomy, improving spatial specificity but depending heavily on mask accuracy. MTnnUNet promotes anatomical context learning via multi-task prediction, which can benefit complex cases but may underperform on small tumors. AAnnUNet incorporates anomaly maps to detect tumors—including those missed by other models—and is particularly valuable when manual annotations are limited. These maps also offer diagnostic value to radiologists and hold potential for downstream clinical tasks such as lymph node assessment, treatment response prediction, and staging stratification. However, they may introduce false positives due to reconstruction errors or anatomical variability.
There are some limitations of this study. First, all the T2WI was annotated by one radiologist. Multiple readers would add extra value to our research. Second, this study exclusively included T2W-MRI, with scans from 2011 to 2018. Some of them exhibited relatively low resolution and larger slice thicknesses, which would not meet the high-resolution criteria established by current protocol recommendations, see Table S11. Third, other MRI sequences like DWI and ADC can potentially improve the segmentation performance. Dou et al.62 have demonstrated by combining T2WI, ADC, and DWI, their model showed the best tumor segmentation performance. Fourth, although we have a relatively large and heterogeneous cohort, the data was solely from the Netherlands. Fifth, concerning rectal anatomical structures, the segmentation process focused solely on the rectum and mesorectum. In future studies, other anatomical structures such as the lumen could contribute to the performance. Lastly, the training and testing were performed exclusively on pre-treatment T2WI. The rectal environment in pre-treatment MRI is visually and pathologically distinct from post-treatment MRI. Incorporating post-treatment T2WI can enhance tumor characterization and improve downstream analytical workflow.
We proposed an anatomical inpainting model trained on prostate MRI to generate pseudo-healthy rectal images. The resulting anomaly maps, highlighting differences from original images, strongly aligned with tumor regions. Integrated into a segmentation model (AAnnUNet), they improved accuracy over other anatomy-informed models (MTnnUNet and MCnnUNet). These results show that anomaly maps enhance segmentation and have potential for broader rectal cancer monitoring. Code and annotated prostate masks are publicly available. (https://github.com/Liiiii2101/Anatomy-aware-nnUNet-for-Rectal-Tumor-Segmentation).
Data availability
Code and the annotations of prostate T2WI used in this study can be found: https://github.com/Liiiii2101/Anatomy-aware-nnUNet-for-Rectal-Tumor-Segmentation.
References
Beets-Tan, R. G. et al. Magnetic resonance imaging for clinical management of rectal cancer: updated recommendations from the 2016 European society of Gastrointestinal and abdominal radiology (ESGAR) consensus meeting. Eur. Radiol. 28, 1465–1475 (2018).
MERCURY Study Group. Extramural depth of tumor invasion at thin-section MR in patients with rectal cancer: results of the MERCURY study. Radiology 243, 132–139 (2007).
Horvat, N., Rocha, C. T., Clemente Oliveira, C., Petkovska, B., Gollub, M. J. MRI of rectal cancer: tumor staging, imaging techniques, and management. Radiographics 39, 367–387 (2019).
Suzuki, C. et al. The importance of rectal cancer MRI protocols on iInterpretation accuracy. World J. Surg. Onc. 6, 89 (2008).
Hearn, N. et al. Manual and semi-automated delineation of locally advanced rectal cancer subvolumes with diffusion-weighted MRI. Br. J. Radiol. 93, 20200543 (2020).
Irving, B. et al. Pieces-of-parts for supervoxel segmentation with global context: application to DCE-MRI tumour delineation. Med. Image. Anal. 32, 69–83 (2016).
Trebeschi, S. et al. Deep learning for Fully-Automated localization and segmentation of rectal cancer on multiparametric MR. Sci. Rep. 7, 5301 (2017).
Razzak, M. I., Naz, S. & Zaib, A. Deep learning for medical image processing: overview, challenges and the future. In Classification in BioApps: Automation of Decision Making (eds. Dey, N., Ashour, A. S. & Borra, S.) 323–350 (Springer International Publishing, 2018). https://doi.org/10.1007/978-3-319-65981-7_12.
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (eds. Navab, N. et al.) vol. 9351 234–241 (Springer International Publishing, 2015).
Jian, J. et al. Fully convolutional networks (FCNs)-based segmentation method for colorectal tumors on T2-weighted magnetic resonance images. Australas Phys. Eng. Sci. Med. 41, 393–401 (2018).
Knuth, F. et al. MRI-based automatic segmentation of rectal cancer using 2D U-Net on two independent cohorts. Acta Oncol. 61, 255–263 (2022).
Wang, J. et al. Technical note: a deep learning-based autosegmentation of rectal tumors in MR images. Med. Phys. 45, 2560–2564 (2018).
Mlynarski, P., Delingette, H., Criminisi, A. & Ayache, N. 3D convolutional neural networks for tumor segmentation using long-range 2D context. Comput. Med. Imaging Graph. 73, 60–72 (2019).
Hamabe, A. et al. Artificial intelligence–based technology for semi-automated segmentation of rectal cancer using high-resolution MRI. PLoS One. 17, e0269931 (2022).
Xiao, H., Li, L., Liu, Q., Zhu, X. & Zhang, Q. Transformers in medical image segmentation: a review. Biomed. Signal Process. Control. 84, 104791 (2023).
Li, D. et al. RTAU-Net: a novel 3D rectal tumor segmentation model based on dual path fusion and attentional guidance. Comput. Methods Programs Biomed. 242, 107842 (2023).
Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods. 18, 203–211 (2021).
Zhou, H. Y. et al. NnFormer: volumetric medical image segmentation via a 3D transformer. IEEE Trans. Image Process. 32, 4036–4045 (2023).
DeSilvio, T. et al. Region-specific deep learning models for accurate segmentation of rectal structures on post-chemoradiation T2w MRI: a multi-institutional, multi-reader study. Front. Med. 10, 1422 (2023).
Kim, J. et al. Rectal cancer: toward fully automatic discrimination of T2 and T3 rectal cancers using deep convolutional neural network. Int. J. Imaging Syst. Technol. 29, 247–259 (2019).
Nagtegaal, I. et al. Morphological changes in tumour type after radiotherapy are accompanied by changes in gene expression profile but not in clinical behaviour. J. Pathol. 204, 183–192 (2004).
Lee, J., Oh, J. E., Kim, M. J., Hur, B. Y. & Sohn, D. K. Reducing the model variance of a rectal cancer segmentation network. IEEE Access. 7, 182725–182733 (2019).
Goodfellow, I. et al. Generative adversarial Nets. In Advances in Neural Information Processing Systems vol. 27 (Curran Associates, Inc., 2014).
Kingma, D. P. & Welling, M. Auto-encoding variational bayes (2022). http://arxiv.org/abs/1312.6114.
van den Oord, A. & Vinyals, O. & kavukcuoglu, koray. Neural discrete representation learning. In Advances in Neural Information Processing Systems vol. 30 (Curran Associates, Inc., 2017).
Baur, C., Denner, S., Wiestler, B., Navab, N. & Albarqouni, S. Autoencoders for unsupervised anomaly segmentation in brain MR images: a comparative study. Med. Image Anal. 69, 101952 (2021).
Chen, X., You, S., Tezcan, K. C. & Konukoglu, E. Unsupervised lesion detection via image restoration with a normative prior (2020). http://arxiv.org/abs/2005.00031.
Pinaya, W. H. L. et al. Unsupervised brain imaging 3D anomaly detection and segmentation with Transformers. Med. Image Anal. 79, 102475 (2022).
Nguyen, B., Feldman, A., Bethapudi, S., Jennings, A. & Willcocks, C. G. Unsupervised region-based anomaly detection In brain MRI With adversarial image inpainting. In IEEE 18th International Symposium on Biomedical Imaging (ISBI) 1127–1131 (2021). (2021). https://doi.org/10.1109/ISBI48211.2021.9434115.
Yeganeh, Y., Farshad, A. & Navab, N. Shape-aware masking for inpainting in medical imaging (2022). http://arxiv.org/abs/2207.05787.
Woo, B. et al. Automated anomaly-aware 3D segmentation of bones and cartilages in knee MR images from the osteoarthritis initiative. Med. Image Anal. 93, 103089 (2024).
Saha, A. et al. Artificial intelligence and radiologists at prostate cancer detection in MRI — The PI-CAI challenge (2023).
Menze, B. H. et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging. 34, 1993–2024 (2015).
Bogveradze, N. et al. Evolutions in rectal cancer MRI staging and risk stratification in the Netherlands. Abdom. Radiol. 47, 38–47 (2022).
Schurink, N. W. et al. Sources of variation in multicenter rectal MRI data and their effect on radiomics feature reproducibility. Eur. Radiol. 32, 1506–1516 (2022).
Schurink, N. W. et al. Development and multicenter validation of a multiparametric imaging model to predict treatment response in rectal cancer. Eur. Radiol. 2023, 1–10 (2023).
Cai, L. et al. An automated deep learning pipeline for EMVI classification and response prediction of rectal cancer using baseline MRI: a multi-centre study. Npj Precis Onc. 8, 1–11 (2024).
El Khababi, N. et al. Predicting response to chemoradiotherapy in rectal cancer via visual morphologic assessment and staging on baseline MRI: a multicenter and multireader study. Abdom. Radiol. 48, 3039–3049 (2023).
ESGAR. Book of Abstracts. Insights into Imaging 11, 64 (2020).
FACG, M. A. M. MD, FACR. Dynamic Radiology of the Abdomen: Normal and Pathologic Anatomy (Springer Science & Business Media, 2006).
Lambregts, D. M. J., Maas, M. & Beets-Tan, R. G. H. MRI of the Rectum. In MRI of the Gastrointestinal Tract (ed. Stoker, J.) 205–227 (Springer, 2010). https://doi.org/10.1007/978-3-540-85532-3_13.
Bogveradze, N. et al. MRI anatomy of the rectum: key concepts important for rectal cancer staging and treatment planning. Insights into Imaging. 14, 13 (2023).
Han, L. et al. Synthesis-based imaging-differentiation representation learning for multi-sequence 3D/4D MRI. Med. Image Anal. 92, 103044 (2024).
Han, L. et al. An explainable deep framework: towards Task-Specific fusion for Multi-to-One MRI synthesis. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2023 (ed. Greenspan, H.) 45–55 (Springer Nature Switzerland, C2023). https://doi.org/10.1007/978-3-031-43999-5_5.
Johnson, J., Alahi, A. & Fei-Fei, L. Perceptual losses for Real-Time style transfer and Super-Resolution. In Computer Vision – ECCV 2016 (eds. Leibe, B. et al.) 694–711 (Springer International Publishing, 2016). https://doi.org/10.1007/978-3-319-46475-6_43.
Zhu, J. Y., Park, T., Isola, P. & EfrosA. A. Unpaired Image-to-Image translation using Cycle-Consistent adversarial networks. In 2017 IEEE Int. Conf. Comput. Vis. (ICCV) 2242-2251 (2017). https://doi.org/10.1109/ICCV.2017.244.
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016 (eds. Ourselin, S. et al.) 424–432 (Springer International Publishing, 2016). https://doi.org/10.1007/978-3-319-46723-8_49.
Diakogiannis, F. I., Waldner, F., Caccetta, P. & Wu, C. ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogrammetry Remote Sens. 162, 94–114 (2020).
Hatamizadeh, A. et al. UNETR: Transformers for 3D medical image segmentation 574–584 (2022).
Hatamizadeh, A. et al. Swin UNETR: Swin Transformers for semantic segmentation of brain tumors in MRI images. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (eds. Crimi, A. & Bakas, S.) 272–284 (Springer International Publishing, 2022). doi:https://doi.org/10.1007/978-3-031-08999-2_22.
Oktay, O. et al. Attention U-Net: learning where to look for the pancreas (2018). https://doi.org/10.48550/arXiv.1804.03999.
Gao, Y. et al. A Data-scalable transformer for medical image segmentation: architecture, model efficiency, and benchmark (2023). https://doi.org/10.48550/arXiv.2203.00131.
Ma, J., Li, F. & Wang, B. U-Mamba: enhancing long-range dependency for biomedical image segmentation (2024). https://doi.org/10.48550/arXiv.2401.04722.
He, K., Zhang, X., Ren, S. & Sun, J. Identity mappings in deep residual networks. In Computer Vision – ECCV 2016 (eds. Leibe, B. et al.) 630–645 (Springer International Publishing, 2016). https://doi.org/10.1007/978-3-319-46493-0_38.
Gu, A. et al. Combining recurrent, convolutional, and continuous-time models with linear state space layers. In Advances in Neural Information Processing Systems, vol. 34 572–585 (2021).
Varoquaux, G. & Cheplygina, V. Machine learning for medical imaging: methodological failures and recommendations for the future. Npj Digit. Med. 5, 1–8 (2022).
Teney, D. et al. On the value of out-of-distribution testing: an example of Goodhart’ s Law. In Advances in Neural Information Processing Systems, vol. 33 407–417 (2020).
Isensee, F. et al. nnU-Net Revisited: A Call for Rigorous Validation in 3D Medical Image Segmentation (2024). http://arxiv.org/abs/2404.09556.
Willemink, M. J. et al. Preparing medical imaging data for machine learning. Radiology 295, 4–15 (2020).
Jayaprakasam, V. S. et al. MRI radiomics features of mesorectal fat can predict response to neoadjuvant chemoradiation therapy and tumor recurrence in patients with locally advanced rectal cancer. Eur. Radiol. 2022, 1–10 (2022).
Defeudis, A. et al. MRI-based radiomics to predict response in locally advanced rectal cancer: comparison of manual and automatic segmentation on external validation in a multicentre study. Eur. Radiol. Exp.. 6, 19 (2022).
Dou, M. et al. Segmentation of rectal tumor from multi-parametric MRI images using an attention-based fusion network. Med. Biol. Eng. Comput. 61, 2379–2389 (2023).
Acknowledgements
This study has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement No 857894. Research at the Netherlands Cancer Institute is supported by institutional grants of the Dutch Cancer Society and of the Dutch Ministry of Health, Welfare and Sport. The study was supported by the Research High Performance Computing (RHPC) facility of the Netherlands Cancer Institute.
Author information
Authors and Affiliations
Contributions
A short paragraph detailing individual contributions is provided for this research paper. L.C. developed the original idea and contributed to the study’s research design. D.M.J.L., J.v.G., and R.G.H.B. were responsible for data collection, while M.A.A. annotated the data. J.B., J.T., and S.B. provided supervision, guidance, and constructive feedback. L.H. and E.P. contributed to the writing process, with L.H. also providing input on the inpainting part of this study. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cai, L., Abdelatty, M.A., Han, L. et al. Improving rectal tumor segmentation with anomaly fusion derived from anatomical inpainting: a multicenter study. Sci Rep 16, 1545 (2026). https://doi.org/10.1038/s41598-025-14265-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-14265-9