Abstract
Sarcoids are benign and locally aggressive skin lesions that commonly affect horses and other equid species. Sarcoids are generally considered to be caused by bovine delta-papillomaviruses (δPVs) types 1 and 2 (BPV1 and BPV2, respectively). Moreover, while bovine δPV types 13 and 14 (BPV13 and BPV14, respectively) are also suspected to induce sarcoids, information regarding this possibility and the occurrence of multiple bovine δPV infections in sarcoids is scarce. This study aimed, for the first time, to assess BPV1, BPV2, BPV13, and BPV14 infections and co-infections in equine sarcoid samples of Austrian provenance, and to determine the intralesional DNA loads of the detected bovine δPV types using highly sensitive droplet digital polymerase chain reaction (ddPCR). BPV DNA was detected in 93 sarcoid samples. The analyses revealed that BPV1 was the predominant bovine δPV type in sarcoids from Austria, with 83/93 lesions testing BPV1-positive. Importantly, 66 tumors also contained BPV2 DNA. In six cases, a triple infection including BPV13 or BPV14 was noted, and one lesion showed a quadruple infection. This is the first ddPCR-based study to show multiple infections by all four bovine δPVs in equine sarcoids. Clinical data suggest that BPV1/2 co-infection may be associated with more severe and therapy-resistant disease. In-depth studies are required to investigate this possibility in greater detail.
Similar content being viewed by others
Introduction
Sarcoids are the most common tumors in equids, affecting up to 12% of horses worldwide1. Sarcoids are skin neoplasms that do not metastasize. Based on their clinical appearance, sarcoids are classified as mildly occult, verrucose, nodular, fibroblastic, mixed, or malevolent lesions2. Importantly, sarcoids are often resistant to therapy and tend to reoccur in more extensive and multiple forms following ineffective treatment. Consequently, sarcoid disease constitutes a serious welfare problem and negatively affects the horse industry3.
Sarcoids are generally believed to be caused by bovine papillomavirus (BPV) types 1 and 2 (BPV1 and BPV2, respectively), and are probably associated with accidental or iatrogenic trauma1,3. In addition, a growing body of evidence indicates that BPV type 13 (BPV13), and possibly type 14 (BPV14), can induce sarcoid lesions in equids4,5,6.
Papillomaviruses (PVs) are a family of small DNA viruses that cause benign skin lesions and malignant tumors in vertebrates7,8. Typically, PVs are strictly epitheliotropic and highly species-specific. The small genus of delta-papillomaviruses (δPVs) is an exception to this rule9. The members of this genus, including bovine δPVs 1, 2, 13, and 14 (BPV1, BPV2, BPV13, and BPV14, respectively) can also infect fibroblasts. This particular feature may explain their wide host range, which includes horses and other equid species3.
Sarcoid formation is primarily induced by BPV1 and/or BPV2. However, the prevalence and distribution of these two genotypes varies geographically. In Western United States of America (USA) and Oceania, sarcoids predominantly harbor BPV210,11. In Eastern USA and Europe, most sarcoids are induced by BPV1, and BPV2 is rarely detected3,10,12. In some cases, sarcoids contain both BPV types, yet al.most no information on bovine δPV co-infections in sarcoids has been reported so far13.
BPV13 was first identified and characterized in a cow cutaneous papilloma in Brazil. This virus is also involved in the pathogenesis of bladder tumors in European cattle14 as well as squamous and fibropapillomas in cattle rumen in Brazil15. Although BPV13 DNA was detected in sarcoids affecting horses in Brazil4,5, no BPB13 DNA has been detected in sarcoids affecting European horses so far16.
Little is known regarding the occurrence and geographical distribution of BPV14. Genomic BPV14 sequences have been detected in felid skin lesions in the USA and Oceania17,18,19, bovine papillomas and inflammatory skin conditions in Australia20, and urinary bladder tumors in Italy6. To date, no reports have described the detection of BPV14 in equine sarcoids. However, the almost identical genomic organizations of BPV1, BPV2, BPV13, and BPV14, and the high similarities among these PVs at the DNA and protein levels suggest that BPV14 can potentially infect and transform equine fibroblasts19.
Digital polymerase chain reaction (dPCR) is a new-generation traditional quantitative PCR21. As such, droplet dPCR (ddPCR) provides more precise and reproducible detection of pathogen load in the clinical diagnosis of infectious diseases, including viral diseases22. ddPCR has been shown to be a very sensitive diagnostic tool for the detection and quantification of human PV (HPV) DNA23,24,25, BPV DNA in cattle26, as well as ovine (OaPV) and caprine PV (ChPV) DNA in small ruminants27,28. Given that ddPCR was previously shown to outperform qPCR in terms of sensitivity and yield significantly fewer false results23,26,27,28, we have used this method for the first time to assess BPV1, BPV2, BPV13, and BPV14 infections and co-infections in equine sarcoid samples of Austrian provenance, and to determine the intralesional DNA loads of the detected bovine δPV types.
Results
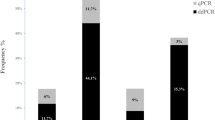
ddPCR detected and quantified BPV DNA in 93 of the 96 (~ 96.9%) equine sarcoid samples. BPV DNA was not detected in three (~ 3.1%) sarcoid samples. ddPCR detected BPV1, BPV2, BPV13, and BPV14 DNA in 83 (~ 89.3%), 66 (~ 71%), five (~ 5.4%), and 10 (~ 10.8%) samples, respectively (Table 1). Single infections were detected in 30 samples (~ 32.3%), and multiple infections were identified in 63 samples (~ 67.7%). No single infections caused by BPV13 or BPV14 were detected. BPV1 and BPV2 were responsible for 22 (~ 73.3%) and eight (~ 26.7%) single infections, respectively (Table 1).
Double infections were the most prevalent multiple infections and were detected in 56 patients (~ 89%). The BPV1/BPV2 genotype combination was observed in 49 samples (~ 87.5%). The remaining seven double infections (~ 12.5%) were characterized by the BPV1/13 and BPV1/14 genotype combinations in two and three samples, respectively, and the BPV2/13 and BPV2/14 genotype combinations in one sample each. ddPCR detected triple infections in six samples (~ 9.5%), five of which showed the BPV1/2/14 genotype combination, and one showed the BPV1/2/13 genotype combination. A quadruple infection was observed in only one sample (Table 2).
The Mann–Whitney test showed no significant differences in viral load between single and multiple infections. BPV1 showed the highest number of copies/µL. Seventy-one sarcoid samples showed a BPV1 DNA copy number varying from 259 to 20,585 copies/µL. Only 12 samples contained fewer than 259 BPV1 DNA copies/µL. BPV2 was detected and quantified at lower concentrations. Only isolated samples showed very high BPV2 DNA copy numbers (up to 19800 copies/µL). BPV13 and BPV14 DNA were quantified at low copy numbers, with the highest copy numbers for these genotypes being 7.7 and 8.2 copies/µL, respectively. BPV1 showed significant differences in viral load in comparison with the remaining BPV genotypes using the Friedman test (P < 0.05). Indeed, after adjusting for the Bonferroni multiple-comparison correction, the P value was ˂ 0.001. Finally, Fig. 1 shows the ddPCR rain plots for all delta-papillomaviruses. Supplemental Table S1 reports the raw data from all the 96 samples in which BPV infection was identified using ddPCR.
Equine sarcoid. QuantaSoft screenshots show the rain plots of the ddPCR results related to BPV genotypes. Positive plots are represented in blue, whereas negative droplets are in grey. BPV1–D09: positive samples; E09: is the positive control; H01: negative samples. BPV2–G04: positive samples; H03: is the positive control; H07: negative samples. BPV13–E07: positive samples; E09: is the positive control; B07: negative samples. BPV14–C03: positive samples; B03: is the positive control; C07: negative samples.
Discussion
Sarcoids are the most common tumor disease in horses and other equid species and have a major influence on the health and welfare of the affected animals. Although the causal association between BPV1/2 infection and sarcoid onset and progression is well established3, many aspects of this disease remain poorly understood. The unclear issues include (i) the prevalence of BPV13 and BPV14 in equine sarcoids, (ii) the incidence of co-infections by bovine δPVs in the lesions, and (iii) the possible pathobiological significance of bovine δPV co-infections in sarcoid disease. To address these gaps in knowledge, we tested tumor DNA isolates with confirmed PCR-compatible quality for the presence of bovine ðPV DNA by using ddPCR technology.
As expected, consistent with data previously obtained in Austria16,29,30, the majority of sarcoid DNA isolates harbored BPV1. In 32.3% of the lesions, BPV1 represented the only detected PV type, whereas approximately 87.5% of tumor samples showing double infections contained the BPV1 and BPV2 genotype combination. The latter finding was surprising, since a single BPV2 infection was detected in only eight cases. Triple and quadruple infections were observed much less frequently and consistently showed BPV1 and BPV2 in combination with BPV13 and/or BPV14.
To the best of our knowledge, this is the first study to evaluate both the detection and virus load of bovine ðPVs associated with equine sarcoids using ddPCR technology. Notably, ddPCR has been already shown to be an accurate, sensitive, and specific molecular assay for detection and quantification of bovine ðPVs in healthy and diseased cattle26,31, sheep32, goats33, and healthy horses34 as well as of OaPV DNA in equine sarcoids35 and in commercial semen of stallions36.
Interestingly, some tumor DNA samples that had scored BPV1/2 PCR-negative in Vienna were subsequently tested using the ddPCR tool in Naples, and scored positive for BPV1 or showed a double BPV2/BPV14 co-infection. These findings agree with the concept that ddPCR outperforms other PCR-based approaches37, and indicate that ddPCR can detect otherwise undetectable bovine ðPVs25. Furthermore, ddPCR provided novel information about the presence of BPV13 and BPV14 DNA, co-infections, and type-specific viral DNA loads in the tested sarcoids, thereby providing a better understanding of geolocalization and territorial distribution of bovine ðPV genotypes.
To the best of our knowledge, this is the first study to provide evidence of BPV13 infection in sarcoid-affected horses from Austria and of BPV14 infections associated with neoplastic pathology in equids, thus confirming the findings of a recent study that included BPV13 and BPV14 in the equine biota34. However, the low BPV13 and BPV14 DNA loads indicated the possibility that their detection was the result of contamination events. Nevertheless, the detection rates (approximately 6.8% and 13.6%, respectively) were consistent with the previous findings and supported the suggestion that our findings were attributable to actual viral infections. Furthermore, the positive, negative, and no-template controls included in ddPCR yielded the expected results, indicating that the reaction was conducted accurately and corroborating our findings. Very little information is available on the epidemiology of BPV13 and BPV14 infections in horses, which could explain the poor understanding of the biological significance of cross-species transmission of these viruses. Therefore, more research is needed to define the role of BPV13 and BPV14 in the pathogenesis of equine sarcoids and to determine whether the clinical implications of sarcoids differ in relation to the causative BPVs.
In few BPV1/2-positive samples, ddPCR did not detect any BPV genotype. These samples were repeatedly subjected to virus PCR screening in Vienna; thus, the DNA was frozen and thawed several times prior to their transfer to Naples. Therefore, we assumed that the negative ddPCR results for these samples might be due to DNA degradation. Furthermore, we do not exclude the possibility that novel, unknown bovine δPVs and/or involvement of other bovine PV types that are not classified within δPV genus could have caused these neoplasms. It is conceivable that some equine sarcoids can be viral lesions characterized by a high genetic PV diversity, which may make current molecular tools enable to detect PV in some cases. Our suggestion appears to be corroborated by recent molecular studies based on PV amplification and sequencing38,39,40.
Our molecular findings also characterized multiple infections. Multiple PV infections have potential oncogenic risks that remain unclear, and their clinical significance in comparison with those of single infections remains controversial. Some studies have indicated that co-infection enhances the risk of PV-related disease and poorer clinical outcomes41,42; in contrast, other studies have shown that multiple infections have no synergistic or additive effects on the development of PV-related disease43,44,45,46.
Determining the epidemiology of single infection and co-infections of bovine ðPVs may be important with the aim of identifying territorial genotype distribution and establishing appropriate prevention strategies, such as designing new pharmacological and/or immune therapeutic protocols that could be tailored to each animal species.
In this context, it is worthwhile remembering that bovine ðPV infection impairs immune response of the host that plays an important role in the pathogenesis of equine sarcoids. Bovine ðPVs develop mechanisms via their E5 oncoprotein that lead to the shutdown of the host immune system, which allows these viruses to escape immune response and facilitates persistent viral infection47,48.
Methods
Ethics statement
This study was conducted in accordance with relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
The animal collection and handling and tissue sampling procedures were performed in accordance with the ethics guidelines of the Veterinary University Vienna and Austrian Law. Permission to collect material was obtained by oral informed consent from the animals’ owners who were previously informed and in agreement with the purpose and methods used.
Sample collection
The 96 samples included in this study were obtained from 95 equids residing in Austria. They consisted of tissue aliquots, superficial swabs, or scrapings from lesions with diagnosis of equine sarcoids. The samples were collected between 2018 and 2024 during therapeutic surgical excision at the Centre of Equine Health and Research (CEHR) of the University of Veterinary Medicine in Vienna, Austria, or sent to the University’s Research Group Oncology (RGO) by referral vets for diagnostic purposes. The horse and sample specifications are listed in Supplemental Table S2.
Standard PCR screening
DNA was extracted from all tissue samples using a DNeasy Blood & Tissue kit in accordance with the manufacturer’s instructions (Qiagen, Hilden, Germany). Subsequently, the DNA isolates were tested using equine beta-actin PCR (EBA PCR), as described previously49, to confirm their PCR compatibility. In short, primers 5′EBA and 3′EBA (5′-TCACCCACACTGTGCCCATCTACG-3′ and 5′-CGTCRTACTCCTGCTTGCTGATCC-3′; made by Eurofins, Vienna, Austria) were selected from the sequence corresponding to GenBank accession no. AF035774 and used for amplification of a 614-bp sequence using Phusion™ Hot Start II DNA Polymerase (Thermo Fisher Scientific, Vienna, Austria) according to the manufacturer’s instructions. The cycling program was performed in a Bioer LifeECO cycler (Biozym, Hessisch-Oldendorf, Germany) and consisted of 98 °C for 2 min followed by 40 amplification cycles (98 °C for 15 s, 69 °C for 30 s, 72 °C for 30 s) and a final elongation step at 72 °C for 5 min.
In the next step, all EBA-positive DNA isolates (100%) were screened for the presence of BPV1/2 E5 DNA using an established PCR protocol with primers 5′BPV1/2-E5 and 3′BPV1/2-E5 (5′-CACTACCTCCTGGAATGAACATTTCC-3′; 5′-CTACCTTWGGTATCACATCTGGTGG-3′; Eurofins) selected from homologous regions up- and downstream of the E5 ORF of BPV1 and BPV2 (Genbank IDs X02346 and M20219.1). This system does not allow for distinction between BPV1 and BPV250. Confirmed BPV1-positive sarcoid DNA, non-infected equine skin DNA, and sterile water were included in each reaction as positive, negative, and no-template controls, respectively. The reactions were performed as described above. EBA and E5 PCR products (16 µL) were separated by gel electrophoresis on 1.5% Tris-acetic acid-ethylenediaminetetraacetic acid (TAE) gels and then visualized by ethidium bromide staining. In the final step, 50-µL aliquots of all EBA-positive DNA isolates were consecutively sent in two batches (batch #1: fall 2023, batch #2: spring 2024) to the Università degli Studi di Napoli Federico II for ddPCR-based analyses.
Droplet digital PCR
The samples obtained from Vienna were photometrically analyzed using a NanoVue Plus system (GE Healthcare, Boston, MA, USA) to determine their respective DNA concentrations. Then, aliquots were adjusted to concentrations of 100 ng/7 µL. ddPCR was performed as described previously51 using the QX100 ddPCR System according to the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA, USA). Each reaction was performed in a final volume of 22 µL, which consisted of 11 µL of ddPCR 2x Supermix for Probes (Bio-Rad Laboratories, Hercules, CA, USA), 0.9 µM of each primer, 0.25 µM of probe, and 7 µL of sample DNA (100 ng).
The ddPCR primers and probes for amplification of 67- to 98-bp sequences were selected within the E5 coding and non-coding regions according to the following sequences deposited in the GenBank: MH197482.1 (BPV1), M20219.1 (BPV2), JQ798171.1 (BPV13), and KR868228.1 (BPV14)26 (Table 3).
The positive controls included BPV-1 DNA from a zebra sarcoid (gifted by Dr. Altamura, University of Naples, Italy), BPV-2 clone DNA (gifted by Dr. A. Venuti, IRCSS Regina Elena, National Cancer Institute, Rome, Italy), and BPV-13 and BPV-14 DNA from bovine bladder tumors from our laboratories6,14. A BPV-negative sample and non-template control were included in each run.
Each sample was analyzed in triplicate to ensure accuracy. Samples were considered BPV-positive if at least three droplets containing BPV amplicons were present, consistent with the criterion for PV infections in human and veterinary medicine25,26,51. Furthermore, samples with < 20 positive droplets were re-analyzed to ensure that these low copy number samples were not caused by cross-contamination. Sterile water served as a no-template control. Data acquisition and analyses were performed using the CFX Maestro software (Bio-Rad Laboratories).
Statistical analyses
The Friedman test was used to compare the median viral load of all bovine ðPV genotypes. Furthermore, to determine the significance relative to the number of copies of BPV DNA detected, the Friedman test was used after adjusting for Bonferroni multiple-comparison correction of the median. The Mann–Whitney U test was used to evaluate the viral load of single and multiple BPV infections. In all comparisons, statistical significance was defined by a P value < 0.05. Statistical analyses were performed using RStudio® software (version 4.2; The RStudio, Boston, MA, USA).
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
References
Nasir, L. & Campo, M. S. Bovine papillomaviruses: their role in the aetiology of cutaneous tumour of Bovids and equids. Vet. Dermatol. 19, 243–254. https://doi.org/10.1111/j.1365-3164.2008.00683.x (2008).
Knottenbelt, D. C. The equine sarcoid -Why are there so many treatment options. Vet. Clin. North. Am. Equine Pract. 35, 243–262. https://doi.org/10.1016/j.cveq.2019.03.006 (2019).
Chambers, G. et al. Association of bovine papillomavirus with the equine sarcoid. J. Gen. Virol. 84, 1055–1062. https://doi.org/10.1099/vir.0.18947-0 (2003).
Lunardi, M. et al. Genetic characterization of a novel bovine papillomavirus member of the deltapapillomavirus genus. Vet. Microbiol. 162, 207–213. https://doi.org/10.1016/j.vetmic.2012.08.030 (2013).
Lunardi, M. et al. Bovine papillomavirus type 13 DNA in equine sarcoids. J. Clin. Microbiol. 51, 2167–2171. https://doi.org/10.1128/jcm.00371-13 (2013).
Roperto, S., Munday, J. S., Corrado, F., Goria, M. & Roperto, F. Detection of bovine papillomavirus type 14 DNA sequences in urinary bladder tumors in cattle. Vet. Microbiol. 190, 1–4. https://doi.org/10.1016/j.vetmic.2016.04.007 (2016).
zur Hausen, H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92, 690–698. https://doi.org/10.1093/jnci/92.9.690 (2000).
Rector, A. & Van Ranst, M. Animal papillomaviruses. Virology 445, 213–223. https://doi.org/10.1016/j.virol.2013.05.007 (2013).
de Villiers, E. M., Fauquet, C., Broker, T. R. & Bernard, H. U. Zur hausen, H. Classification of papillomaviruses. Virology 324, 17–27. https://doi.org/10.1016/j.virol.2004.03.033 (2004).
Carr, E. A., Theon, A. P., Madewell, B. R., Griffey, S. M. & Hitchcock, M. E. Bovine papillomavirus DNA in neoplastic and nonneoplastic tissues obtained from horses with and without sarcoids in the Western united States. Am. J. Vet. Res. 62, 741–744. https://doi.org/10.2460/ajvr.2001.62.741 (2001).
Wobeser, B. K. et al. Epidemiology of equine sarcoids in horses in Western Canada. Can. Vet. J. 51, 1103–1108 (2010).
Hainisch, E. K. et al. Bovine papillomavirus type 1 and 2 virion-infected primary fibroblasts constitute a near-natural equine sarcoid model. Viruses 14, 2658. https://doi.org/10.3390/v14122658 (2022).
Gysens, L., Vanmechelen, B., Haspeslagh, M., Maes, P. & Martens, A. New approach for genomic characterisation of equine sarcoid-derived BPV-1/-2 using nanopore-based sequencing. Virol. J. 19, 8. https://doi.org/10.1186/s12985-021-01735-5 (2022).
Roperto, S. et al. Bovine papillomavirus type 13 expression in the urothelial bladder tumours of cattle. Transbound. Emerg. Dis. 63, 628–634. https://doi.org/10.1111/tbed.12322 (2016).
Gasparotto, G. et al. Characterization of bovine papillomavirus types detected in cattle rumen tissues from Amazon region, Brazil. Animals 14, 2262. https://doi.org/10.3390/ani14152262 (2024).
Jindra, C., Kamjunke, A. K., Jones, S. & Brandt, S. Screening for bovine papillomavirus type 13 (BPV13) in a European population of sarcoid-bearing equids. Equine Vet. J. 54, 662–669. https://doi.org/10.1111/evj.13501 (2021).
Munday, J. S., Knight, C. G. & Howe, L. The same papillomavirus is present in feline sarcoids from North America and new Zealand but not in any non-sarcoid feline samples. J. Vet. Diagn. Invest. 22, 97–100. https://doi.org/10.1177/104063871002200119 (2010).
Orbell, G. M., Young, S. & Munday, J. S. Cutaneous sarcoids in captive African lions associated with feline sarcoid-associated papillomavirus infection. Vet. Pathol. 48, 1176–1179. https://doi.org/10.1177/0300985810391111 (2011).
Munday, J. S. et al. Genomic characterisation of the feline sarcoid-associated papillomavirus and proposed classification as Bos Taurus papillomavirus type 14. Vet. Microbiol. 177, 289–295. https://doi.org/10.1016/j.vetmic.2015.03.019 (2015).
Munday, J. S. & Knight, C. G. Amplification of feline sarcoid-associated papillomavirus DNA sequences from bovine skin. Vet. Dermatol. 21, 341–344. https://doi.org/10.1111/j.1365-3164.2010.00872.x (2010).
Kojabad, A. A. et al. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 93, 4182–4197. https://doi.org/10.1002/jmv.26846 (2021).
Li, H. et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep. 38, BSR20181170. https://doi.org/10.1042/BSR20181170 (2018).
Biron, V. L. et al. Detection of human papillomavirus type16 in oropharyngeal squamous cell carcinoma using droplet digital polymerase chain reaction. Cancer 122, 1544–1551. https://doi.org/10.1002/cncr.29976 (2016).
Isaac, A. et al. Ultrasensitive detection of oncogenic human papillomavirus in oropharyngeal tissue swabs. J. Otolaryngol. Head Neck Surg. 46, 5. https://doi.org/10.1186/s40463-016-0177-8 (2017).
Lillsunde Larsson, G. & Helenius, G. Digital droplet PCR (ddPCR) for the detection and quantification of HPV 16, 18, 33 and 45 – a short report. Cell. Oncol. 40, 521–527. https://doi.org/10.1007/s13402-017-0331-y (2017).
De Falco, F., Corrado, F., Cutarelli, A., Leonardi, L. & Roperto, S. Digital droplet for detection and quantification of Circulating bovine deltapapillomavirus. Transbound. Emerg. Dis. 68, 1345–1352. https://doi.org/10.1111/tbed.13795 (2021).
De Falco, F. et al. Molecular epidemiology of ovine papillomavirus infection in Southern Italy. Front. Vet. Sci. 8, 7903922. https://doi.org/10.3389/fvets.2021.790392 (2021).
Cutarelli, A. et al. Prevalence and genotype distribution of caprine papillomavirus in peripheral blood of healthy goats in farms from three European countries. Front. Vet. Sci. 10, 1213150. https://doi.org/10.3389/fvets.2023.1213150 (2023).
Brandt, S. et al. BPV-1 infection is not confined to the dermis but also involves the epidermis of equine sarcoids. Vet. Microbiol. 150, 35–40. https://doi.org/10.1016/j.vetmic.2010.12.021 (2011).
Hainisch, E. K. & Brandt, S. Equine Sarcoids. In: Robinson’s Current Therapy in Equine Medicine, (eds. Robinson, N.E. & Sprayberry, K.A.) Vol.1Saunders Elsevier. St Louis, MO, USA,. (2015).
De Falco, F., Cutarelli, A., Fedele, M. L., Catoi, C. & Roperto, S. Molecular findings and virological assessment of bladder papillomavirus infection in cattle. Vet. Q. 44, 1–7. https://doi.org/10.1080/01652176.2024.2387072 (2024).
Roperto, S., Cutarelli, A., Corrado, F., De Falco, F. & Buonavoglia, C. Detection and quantification of bovine papillomavirus DNA by digital droplet PCR in sheep blood. Sci. Rep. 11, 10292. https://doi.org/10.1038/s41598-021-89782-4 (2021).
Cutarelli, A., De Falco, F., Uleri, V., Buonavoglia, C. & Roperto, S. The diagnostic value of the droplet digital PCR for the detection of bovine deltapapillomavirus in goats by liquid biopsy. Transbound. Emerg. Dis. 68, 3624–3630. https://doi.org/10.1111/tbed.13971 (2021).
Cutarelli, A. et al. Ultrasensitive detection and quantification of bovine deltapapillomavirus in the semen of healthy horses. Sci. Rep. 15, 769. https://doi.org/10.1038/s41598-024-81682-7 (2025).
De Falco, F., Cutarelli, A., Pellicanò, R., Brandt, S. & Roperto, S. Molecular detection and quantification of ovine papillomavirus DNA in equine sarcoid. Transbound. Emerg. Dis. 2024 (6453158). https://doi.org/10.1155/2024/6453158 (2024).
Cutarelli, A. et al. Molecular detection of transcriptionally active ovine papillomaviruses in commercial equine semen. Front. Vet. Sci. 11, 1427370. https://doi.org/10.3389/fvets.2024.1427370 (2024).
Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610. https://doi.org/10.1021/ac202028g (2011).
Daudt, C. et al. How many papillomavirus species can go undetected in papilloma lesions. Sci. Rep. 6, 36480. https://doi.org/10.1038/srep36480 (2017).
Sauthier, J. T. The genetic diversity of papillomavirome in bovine teat papilloma lesions. Anim. Microbiome. 3, 51. https://doi.org/10.1186/s42523-021-00114-3 (2021).
dos Souza, A. Characterization of papillomatous lesions and genetic diversity of bovine papillomavirus from the Amazon region. Viruses 17, 719. https://doi.org/10.3390/v17050719 (2025).
Chaturvedi, A. K. et al. Human papillomavirus infection with multiple types: pattern of coinfection and risk of cervical disease. J. Infect. Dis. 203, 910–920. https://doi.org/10.1093/infdis/jiq139 (2011).
Akinjyi, I. et al. HPV infection patterns and viral load distribution: implication on cervical cancer prevention in Western Kenia. Eur. J. Cancer Prev. 34, 329–336. https://doi.org/10.1097/CEJ.0000000000000920 (2025).
Guo, W. et al. Epidemiological study of human papillomavirus infection in 105,679 women in wuhan, China. BMC Infect. Dis. 24, 1111. https://doi.org/10.1186/s12879-024-10011-0 (2024).
Herrero, R. et al. Population-based study of human papillomavirus infection and cervical neoplasia in rural Costa Rica. J. Natl. Cancer Inst. 9, 464–474. https://doi.org/10.1093/jnci.92.6.464 (2020).
Luo, Q. et al. Epidemiologic characteristics of high-risk HPV and the correlation between multiple infections and cervical lesions. BMC Infect. Dis. 23, 667. https://doi.org/10.1186/s12879-023-08634-w (2023).
Capparelli, R. et al. Mannose-binding lectin haplotypes influence Brucella abortus infection in the water Buffalo (Bubalus bubalis). Immunogenetics 60, 157–165. https://doi.org/10.1007/s00251-008-0284-4 (2008).
De Falco, F. et al. Bovine delta papillomavirus E5 oncoprotein interacts with TRIM25 and hampers antiviral innate immune response mediated by RIG-I-like receptors. Front. Immunol. 10, 658762. https://doi.org/10.3389/fimmu.2021.658762 (2021).
De Falco, F. et al. Bovine delta papillomavirus E5 oncoprotein negatively regulates the cGAS-STING signaling pathway in cattle in a spontaneous model of viral disease. Front. Immunol. 13, 937736. https://doi.org/10.3389/fimmu.2022.937736 (2022).
Brandt, S., Haralambus, R., Schoster, A., Kirnbauer, R. & Stanek, C. Peripheral blood mononuclear cells represent a reservoir of bovine papillomavirus DNA in sarcoid-affected equines. J. Gen. Virol. 89, 1390–1395. https://doi.org/10.1099/vir.0.83568-0 (2008).
Brandt, S. et al. A subset of equine sarcoids harbours BPV-1 DNA in a complex with L1 major capsid protein. Virology 375, 433–441. https://doi.org/10.1016/j.virol.2008.02.014 (2008).
De Falco, F. et al. Possible etiological association of ovine papillomaviruses with bladder tumors in cattle. Virus Res. 328, 199084. https://doi.org/10.1016/j.virusres.2023.199084 (2023).
Acknowledgements
We would like to thank Dr. G. Altamura, Department of Veterinary Medicine and Animal Productions, Uni¬versity of Naples Federico II, (Naples, Italy) and Dr. A.Venuti, IRCSS Regina Elena National Cancer Institute, (Rome, Italy) for providing BPV1- and BPV-2-positive samples as a kind gift. Furthermore, we wish to thank Dr S. Morace, Uni¬versity of Catanzaro “Magna Graecia”, Loredana De Vita from the Istituto Zooprofilattico Sperimentale del Mezzogiorno, and La Rizza Maria Antonia for their technical assistance.
Funding
This research was partially supported by the Istituto Zooprofilattico Sperimentale del Mezzogiorno, Portici (NA). The funder of the work did not influence the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
A.C., G.F.: methodology; acquisition and analysis of the data; A.B., R.P., M.N.: visualization and statistical analysis; S.B. and S.R.: conceptualization, writing (original draft) S.R.: supervision, validation, funding acquisition, writing (review & editing). All authors reviewed the results; they approved the final version of the manuscript and declared that the content has not been published elsewhere.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declaration
The animal collection and handling and tissue sampling procedures were performed in accordance with the ethics guidelines of the Veterinary University Vienna and Austrian Law. Animal studies performed in Naples were approved by the Institutional Animal Care and Use Committee (Protocol PG/2024/0023599, Naples University Federico II). Permission to collect samples was obtained from the animals’ owners who were previously informed and in agreement with the purpose and methods used.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cutarelli, A., Buonavoglia, A., Fusco, G. et al. Accurate identification of bovine deltapapillomavirus in equine sarcoids by ddPCR. Sci Rep 15, 29414 (2025). https://doi.org/10.1038/s41598-025-15353-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-15353-6