Abstract
This study investigates the use of CT-based radiomics for predicting extrahepatic metastasis in hepatocellular carcinoma (HCC) following hepatectomy. We analyzed data from 374 patients from two centers (277 in the training cohort and 97 in an external validation cohort). Radiomic features were extracted from contrast-enhanced CT scans. Key features were identified using the least absolute shrinkage and selection operator (LASSO) to compute radiomics scores (radscore) for model development. A clinical model based on risk factors was also created. We developed a combined model integrating both radscore and clinical variables, constructing nomograms for personalized risk assessment. Model performance was compared via the Delong test, with calibration curves assessing prediction consistency. Decision curve analysis (DCA) was employed to assess the clinical utility and net benefit of the predictive models across different threshold probabilities, thereby evaluating their potential value in guiding clinical decision-making for extrahepatic metastasis. Radscore based on CT was an independent predictor of extrahepatic disease (p < 0.05). The combined model showed high predictive performance with an AUC of 87.2% (95% CI: 81.8%-92.6%) in the training group and 86.0% (95% CI: 69.4%-100%) in the validation group. Predictive performance of the combined model significantly outperformed both the radiomics and clinical models (p < 0.05). The DCA shows that the combined model has a higher net benefit in predicting extrahepatic metastases of HCC than the clinical model and radiomics model. The combined prediction model, utilizing CT radscore alongside clinical risk factors, effectively forecasts extrahepatic metastasis in HCC patients.
Similar content being viewed by others
Background
Hepatocellular carcinoma (HCC) is the fourth most prevalent cancer and the second leading cause of cancer-related mortality worldwide1,2. Surgical resection remains the primary treatment for HCC. Despite this, patients who develop extrahepatic metastasis post-surgery face a dismal prognosis. Median survival for advanced HCC patients with extrahepatic metastasis is less than one year3, compared to a five-year survival rate of approximately 70% for those without metastasis4,5. Extrahepatic metastasis is a critical prognostic factor, with 13.5–42% of patients exhibiting these metastases at diagnosis6,7,8. Common metastatic sites include the lungs, bones, adrenal glands, and brain9,10,11. Aggressive management of extrahepatic metastases can improve survival rates12,13. Thus, identifying reliable predictors of extrahepatic metastasis is essential for risk assessment and improving long-term survival.
Radiomics offers a sophisticated approach to tumor characterization by converting standard medical images into high-dimensional datasets, enabling detailed analysis of intra-tumor heterogeneity. It captures subtle imaging phenotypes beyond human visual perception, allowing comprehensive tumor profiling14,15,16,17. By analyzing these extensive data, radiomics holds promises for predicting various post-treatment outcomes, such as overall survival, tumor recurrence, and treatment response18,19,20. Studies indicate that radiomic features can effectively forecast these outcomes in HCC, as well as in other malignancies21,22,23. Specifically in hepatocellular carcinoma, radiomics has shown value in predicting microvascular invasion, early recurrence, and histopathological subtypes, facilitating personalized treatment planning24,25,26.
However, few studies have specifically explored the use of radiomics for predicting extrahepatic metastasis in HCC, particularly in the context of postoperative surveillance. To address this gap, our study aims to develop and validate a combined model that incorporates both CT-based radiomic features and clinical variables to predict extrahepatic metastasis after hepatectomy. By comparing this model with clinical and radiomics models, we aim to provide a more accurate, individualized, and non-invasive tool for early risk stratification in HCC patients.
Materials and methods
Patients
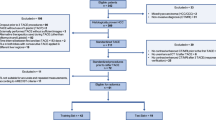
This study was approved by the Ethics Committees of Hospital 1 and Hospital 2 and informed consent was waived for this retrospective study. We included patients who underwent hepatectomy for HCC from January 2013 to November 2018. The selection criteria were: (1) pathologically confirmed HCC. (2) Newly diagnosed intrahepatic HCC lesions without any distant metastasis before surgery. (3) CT scans performed within two weeks prior to hepatectomy. Exclusion criteria included: (1) CT images were either unrecognizable or the lesion was less than three layers (scan layer thickness = 5 mm, pitch = 1.0); (2) incomplete clinical data; (3) preoperative anticancer treatments (n = 20); and (4) errors in feature extraction. Ultimately, 277 patients with complete data were selected from Hospital 1 and 97 from Hospital 2. Figure 1 depicts the patient selection flowchart.
Clinical endpoints and follow-up
The primary endpoint of this study was the development of liver cancer metastasis. Postoperative follow-up consisted of monthly liver ultrasounds for the first three months, transitioning to quarterly ultrasounds thereafter. Lung CT scans and enhanced CT or MRI of the liver were scheduled every three months for the initial two years. Following this period, imaging frequency was reduced to every six months to facilitate timely evaluation and intervention for potential disease recurrence or progression. During these follow-ups, liver function tests, including alanine aminotransferase (ALT), aspartate aminotransferase (AST), and alpha-fetoprotein (AFP), were conducted to monitor postoperative conditions and document any occurrences of extrahepatic metastases.
Image acquisition and tumor segmentation
CT imaging was performed using Siemens SOMATOM Definition Flash and GE Healthcare Discovery CT scanners at both hospitals. The imaging parameters were: 120 kV voltage, 200 to 350 mA current, 5 mm slice thickness and spacing, and a 512 × 512 matrix. A dual-head high-pressure injector administered contrast medium with 350 mg/mL iodine concentration, at 3.0 mL/s and 1.5 mL/kg dose. Enhancement phases were timed as follows: arterial phase at 30 s, portal venous phase at 60 s, and equilibrium phase at 120 s.
Feature extraction
Tumor regions of interest (ROIs) of the arterial, venous, and delayed phases were manually outlined by experienced radiologists using ITK-SNAP version 3.6.0 (http://www.itksnap.org). Image preprocessing and feature extraction of the three phases were conducted using the Radiomics package in 3D Slicer (https://www.slicer.org/). To evaluate the reproducibility of the segmentation results, ROI segmentation was repeated on CT images from 50 HCC patients. Additionally, another experienced radiologist independently performed ROI segmentation on these same patients. Intraclass and interclass correlation coefficients (ICCs) were calculated, retaining only features with ICCs greater than 0.75 for further analysis.
During the initial extraction phase, preprocessing was essential to enhance the differentiation of texture features. To address the batch effects arising from the use of different imaging equipment, all data underwent normalization via z-score standardization, adjusting the intensity values to have a mean of 0 and a standard deviation of 1. Additionally, the image slices were resampled to achieve a voxel size of 1 × 1 × 1 cm. A total of 1130 CT radiomic features were extracted from the tumor regions in each phase, including First order statistical features, Shape features, gray-level cooccurrence features, matrix-based features (GLCM), gray-level.
run-length matrix-based features (GLRLM), gray-level size zone matrix-based features (GLSZM), gray-level dependence matrix-based features (GLDM), Neighborhood Gray-Tone Difference Matrix (NGTDM) and Laplace wavelet changes. Figure 2 shows the flow diagram of feature selection and model construction.
Model and nomogram construction
To prevent overfitting the model, the Least Absolute Shrinkage and Selection Operator (LASSO) with 10 folds cross validation was used to further select key features and then compute the radiomic score (Radscore) and to build a radiomic model based on this score. Univariate and multivariate logistic regression analyses were conducted to identify clinical risk factors and independent predictors of extrahepatic metastasis in HCC patients, with p-values under 0.05 deemed statistically significant. The clinical risk factors determined through multivariate analysis were used to construct clinical models, for which corresponding nomograms were created. Additionally, a combined model was developed by integrating clinical risk factors with the Radscore, accompanied by a nomogram for this combined model. The predictive performance of the clinical, radiomic, and combined models in assessing the risk of liver cancer metastasis was evaluated using ROC curve analysis in both training and validation cohorts. Calibration curves were generated to assess model consistency. The DeLong test was employed to compare the predictive efficiency of the models, while decision curve analysis (DCA) was used to evaluate the net clinical benefit of predicting extrahepatic metastasis.
Statistical analysis
All statistical analyses were performed by R version 4.3.1 Categorical variables between the two groups were compared using the chi-square test or Fisher’s exact test, while continuous variables were analyzed using either the t-test or the Mann-Whitney U test. A p-value of less than 0.05 was considered statistically significant.
Results
Patient characteristics
Table 1 summarizes the characteristics of patients in the training and validation cohorts. The training cohort consisted of 277 individuals, including 230 males (83.03%) and 47 females (16.97%). Among them, 35 patients had extrahepatic metastases, with 19 cases involving a single site: 7 lung metastases, 2 vertebral metastases, 8 lymph node metastases, 1 renal metastasis, and 1 diaphragm metastasis. Additionally, 16 patients experienced multiple metastases, primarily involving the lungs, bones, and lymph nodes. The validation cohort included 97 patients, with 83 males (85.57%) and 14 females (14.43%). Postoperative extrahepatic metastases were observed in 10 patients (10.31%), including 6 cases with a single site (4 lung metastases and 2 lymph node metastases) and 4 cases with multiple metastases.
Radiomics score construction
Feature selection was performed using the LASSO logistic regression algorithm (Fig. 2C), which identified the most relevant features. Ultimately, 9 features were selected from the training cohort, and a radscore was calculated. The radiomics model built based on a radscore showed that the AUC for the training group was 82.4% (95% CI: 75.8%−89.0%), with a sensitivity of 74.2%, a specificity of 78.6% and a F1-score of 74.8%. The AUC for validation group was 78.5% (95% CI: 62.9%−94.1%). The sensitivity was 80%, the specificity was 77.8% and the F1-score was 71.7%.
Radscore=
log-sigma-1-5-mm-3D_gldm_LargeDependenceHighGrayLevelEmphasis×0.188
+wavelet-HLL_gldm_SmallDependenceLowGrayLevelEmphasis×−0.062
+wavelet-HLH_glszm_LargeAreaHighGrayLevelEmphasis×0.067
+wavelet-HHL_glszm_ZoneEntropy×0.033
+log-sigma-0-5-mm-3D_glszm_SizeZoneNonUniformity×0.009
+log-sigma-1-0-mm-3D_glrlm_RunVariance×0.301
+wavelet-LHL_ngtdm_Contrast×−0.033
+wavelet-LHH_glcm_JointEnergy×−0.157
+wavelet-LLL_glcm_MaximumProbability×−0.499+(−2.157)
Development and assessment of clinical models
As shown in Table 2, univariate analysis identified BMI, histopathological grade, tumor diameter, MVI, PV, AST, ALB, PLT, and Cr as clinical risk factors for extrahepatic metastasis in liver cancer. Multivariate analysis further confirmed BMI, MVI, PV, and ALB as independent risk factors for postoperative extrahepatic metastasis. Using these independent predictors, we created a clinical prediction model. The AUC for the training set was 81.2% (95% CI: 74.7–87.7%), with sensitivity at 85.7%, specificity at 74.0% and F1-score at 79.4% (Fig. 3A). In the validation set, the AUC was 76.4% (95% CI: 57.5–95.4%), demonstrating a sensitivity of 80%, a specificity of 76.7% and a F1-score of 74.0% (Fig. 3B).
Development and evaluation of clinical prediction models. Clinical prediction models for training (A) and validation (B) cohorts. (C): Nomogram predicting extrahepatic metastasis. Calibration curves for the training (D) and validation (E) cohorts. Decision curve analyses for the training (F) and validation (G) cohorts.
A nomogram was constructed using these independent risk factors (Fig. 3C), and it visually represents the contribution of each predictor to the overall risk of postoperative extrahepatic metastasis. The calibration curves for both the training and validation sets showed good agreement between predicted probabilities and actual outcomes (Fig. 3D and E). Additionally, decision curve analysis (Fig. 3F and G) demonstrated that the nomogram provided a significant net benefit in predicting extrahepatic metastasis.
Development and assessment of combined models
According to Table 3, multivariate analysis integrating Radscore with clinical risk factors identifies MVI, PV, ALB, and Radscore as independent prognostic factors for predicting extrahepatic metastasis in HCC.
A combined model was developed using these factors, resulting in an AUC of 87.2% (95% CI: 81.8%−92.6%) for the training cohort, showing a sensitivity of 88.6%, a specificity of 70.1% and a F1-score of 81.6% (Fig. 4A). In the validation cohort (Fig. 4B), the AUC was 86.0% (95% CI: 69.4%−100%), achieving a sensitivity of 80%, a specificity of 85.1% and an F1-score of 78.2%. As shown in Fig. 4C, the nomogram that combines radscore and clinical risk factors provides a clear assessment of the risk of postoperative extrahepatic metastasis. Additionally, it plays a significant role in personalized medicine and clinical decision-making aimed at improving overall patient outcomes. The calibration curves in Fig. 4D and E demonstrate excellent consistency in predicting extrahepatic metastasis after surgery. Decision curve analysis for both the training and validation cohorts further confirmed that the nomogram offered substantial net clinical benefit in predicting extrahepatic metastasis (Fig. 4F and G).
The performance of Combined prediction model. ROC curves for the combined model in the training (A) and validation (B) cohorts. C: Nomogram combining radscore and clinical risk factors to predict extrahepatic metastasis. Calibration curves for the combined model in the training (D) and validation (E) cohorts shows a high degree of agreement between the predicted probabilities and actual outcomes, indicating that the combined model is well-calibrated and performs reliably when applied to unseen data. Decision curves for the combined model in the training (F) and validation (G) cohorts demonstrate that the combined model provides the highest net clinical benefit across a wide range of threshold probabilities, compared with the radiomics-only and clinical-only models.
Comparative predictive performance of clinical, radiomics, and combined models
In the training cohort (Fig. 5A), the combined model achieved the highest predictive performance with an AUC of 87.2% (95% CI: 81.8%−92.6%), followed by the radiomics model at 82.4% (95% CI: 75.8%−89.0%), and the clinical model with the lowest AUC of 81.2% (95% CI: 74.7%−87.7%). The combined model significantly outperformed both the radiomics model (DeLong test P = 0.028) and the clinical model (DeLong test P = 0.008), while no significant difference was found between the radiomics and clinical models (DeLong test P = 0.739). Additionally, the decision curve analysis indicated that the combined model with Radscore offered the highest clinical net benefit compared to the other models in the threshold range of 0-0.4 (Fig. 5C).
Comparison of the performance of three prediction models. Comparative analysis of ROC curves of radiomic, clinical, and combined models in training (A) and validation (B) cohorts. The ROC curves reveal that the combined model achieves the highest AUC, outperforming both the clinical and radiomic models alone. Decision curves for the radiomics, clinical, and combined models were compared in the training (C) and validation (D) cohorts. This indicates that the combined model is more effective in supporting clinical decision-making.
In the validation cohort (Fig. 5B), the combined model again demonstrated the highest predictive accuracy, achieving an AUC of 86.0% (95% CI: 69.4%−100%). It was followed by the clinical model at 76.4% (95% CI: 57.5%−95.4%) and the radiomics model at 78.5% (95% CI: 62.9%−94.1%). While no significant differences in predictive power were observed among the three models (DeLong test P > 0.05), decision curve analysis consistently showed that the combined model offered the greatest clinical net benefit compared to the other two models in the threshold range of 0-0.3 (Fig. 5D).
Discussion
Previous research has explored radiomics models for predicting tumor prognosis using contrast-enhanced CT27. To the best of our knowledge, this is the first study to utilize enhanced CT sequence data specifically for assessing postoperative extrahepatic metastasis in HCC patients.
Radiomics has garnered significant attention in cancer research due to its ability to extract a vast array of feature data from standard medical images, thereby potentially enhancing clinical decision-making28. By employing advanced imaging analysis techniques, radiomics can identify subtle patterns and features within tumors, offering a cost-effective, and reproducible method for characterizing tumor phenotypes related to internal heterogeneity. This approach enhances our understanding of tumor biology and may contribute to the development of more personalized treatment strategies29. These features are derived from imaging modalities such as CT, MRI, and PET scans30,31,32, encompassing various aspects of tumor morphology, texture, and intensity, which collectively offer a comprehensive understanding of tumor biological behavior. Radiomic feature scoring, when integrated with clinical and pathological risk factors, can improve diagnostic accuracy, predict therapeutic responses, and monitor disease progression33,34, ultimately advancing the field of precision oncology. As research progresses, radiomics has been applied to liver tumors for diverse purposes, including diagnosis, prognosis, pathological grading, and microvascular invasion (MVI)35,36,37,38. A comprehensive summary of recent radiomics studies related to hepatocellular carcinoma and other malignancies in Supplementary Table S1.
In this study, we utilized enhanced CT imaging data from patients with liver cancer for tumor region segmentation and feature extraction. We assessed the consistency of the segmentation results using ICC to ensure that the extraction of tumor characteristics is highly stable and reproducible across different time points and observers. This evaluation method is crucial for enhancing the reliability of imaging features in clinical research, as it ensures the accuracy and consistency of the research data and minimizes variations introduced by observer39,40. Additionally, features with high consistency contribute to improving the predictive capability of models, thereby enhancing the quality of diagnostic and therapeutic decision-making.
We employed the LASSO algorithm to identify features most relevant to extrahepatic metastasis, utilizing ten-fold cross-validation to mitigate overfitting. The LASSO algorithm is effective for analyzing large feature sets with relatively small sample sizes, reducing data noise interference and enhancing model reliability41. Imaging features linked to extrahepatic metastasis were selected through LASSO regression, and Radscore values were computed. Our analysis confirmed that BMI, MVI, PV, ALB and Radscore are independent predictors of HCC prognosis, aligning with previous findings42,43,44.
We developed predictive models for postoperative extrahepatic metastasis in HCC patients, including a radiomics model, a clinical model, and a combined model integrating clinical factors and Radscore. In both the training and validation cohorts, the combined model that includes Radscore, MVI, PV, and ALB exhibited better AUC performance than both the radiomics and clinical models (P < 0.05). There was no significant difference between the clinical and radiomics models in predicting extrahepatic metastasis (P > 0.05), which aligns with previous research findings45,46. The clinical net benefit of patients was analyzed according to the decision curve. Whether in the training group or the validation group, the net benefit of patients in the combined model was generally higher, indicating that the model was safe. Our study validated the use of enhanced CT imaging tools that can predict postoperative extrahepatic metastases in patients with liver cancer, further demonstrating the stability and reproducibility of radiomics in prognostic assessment of liver cancer47,48.
However, this study has limitations: manual 2D ROI delineation was time-consuming, suggesting that automated lesion segmentation should be a focus of future research. In future studies, the performance of automatic and semi-automatic segmentation methods can be further improved through algorithm optimization, integration of multimodal imaging data (such as CT and MRI), and enhancement of user interactivity. Developing interactive semi-automatic tools may help balance segmentation accuracy and clinical usability. Furthermore, the complex relationship between radiomics features and biological behaviors remains challenging to fully elucidate. Integrating radiomics with genomics or pathology data may help elucidate the biological basis of the radiomic features, thus enhancing model interpretability and clinical utility.
Conclusions
Our findings indicate that radscore is an independent prognostic indicator for predicting extrahepatic metastasis in HCC. By integrating radscore with clinical risk factors, we developed a non-invasive radiomics model that provides a data-driven tool for the early prediction of extrahepatic metastasis after hepatectomy. This model may assist in formulating personalized follow-up strategies and enabling timely interventions. Future research directions include validating this model in larger, multicenter, and prospective cohorts to ensure generalizability.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Cao, W., Qin, K., Li, F. & Chen, W. Comparative study of cancer profiles between 2020 and 2022 using global cancer statistics (GLOBOCAN). J. Natl. Cancer Cent. (4 (2), 128–134. https://doi.org/10.1016/j.jncc.2024.05.001 (2024). -06-01.
Yang, J. D. et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. NAT REV GASTRO HEPAT. 2019-10-01; 16 (10): 589–604. https://doi.org/10.1038/s41575-019-0186-y
Altekruse, S. F., McGlynn, K. A. & Reichman, M. E. Hepatocellular carcinoma incidence, mortality, and survival trends in the united States from 1975 to 2005. J. CLIN. ONCOL. 27 (9), 1485–1491. https://doi.org/10.1200/JCO.2008.20.7753 (2009). -03-20.
Furukawa, K. et al. Survival benefit of hepatic resection for hepatocellular carcinoma beyond the Barcelona clinic liver cancer classification. J HEPATO-BIL-PAN SCI. 2017-04-01; 24 (4): 199–205. https://doi.org/10.1002/jhbp.436
Sala, M., Forner, A., Varela, M. & Bruix, J. Prognostic prediction in patients with hepatocellular carcinoma. SEMIN LIVER DIS. 25 (2), 171–180. https://doi.org/10.1055/s-2005-871197 (2005-01-01).
Katyal, S. et al. Extrahepatic metastases of hepatocellular carcinoma. Radiology 216 (3), 698–703. https://doi.org/10.1148/radiology.216.3.r00se24698 (2000).
Shuto, T. et al. Treatment of adrenal metastases after hepatic resection of a hepatocellular carcinoma. Dig. Surg. 18 (4), 294–297. https://doi.org/10.1159/000050155 (2001).
Hsu, C. Y. et al. Metastasis in patients with hepatocellular carcinoma: prevalence, determinants, prognostic impact and ability to improve the Barcelona clinic liver cancer system. Liver Int. 38 (10), 1803–1811. https://doi.org/10.1111/liv.13748 (2018).
Hanazaki, K. et al. Survival and recurrence after hepatic resection of 386 consecutive patients with hepatocellular carcinoma. J. Am. Coll. Surg. 191 (4), 381–388. https://doi.org/10.1016/s1072-7515(00)00700-6 (2000).
Yan, W. T. et al. Predictors and long-term prognosis of early and late recurrence for patients undergoing hepatic resection of hepatocellular carcinoma: a large-scale multicenter study. Hepatobiliary Surg. Nutr. 12 (2), 155–168. https://doi.org/10.21037/hbsn-21-288 (2023).
Sasaki, A. et al. Receptor activator of nuclear factor-kappaB ligand (RANKL) expression in hepatocellular carcinoma with bone metastasis. Ann. Surg. Oncol. 14 (3), 1191–1199. https://doi.org/10.1245/s10434-006-9277-4 (2007).
Tomimaru, Y. et al. The significance of surgical resection for pulmonary metastasis from hepatocellular carcinoma. Am. J. Surg. 192 (1), 46–51. https://doi.org/10.1016/j.amjsurg.2005.12.006 (2006).
Poon, R. T., Fan, S. T., O’Suilleabhain, C. B. & Wong, J. Aggressive management of patients with extrahepatic and intrahepatic recurrences of hepatocellular carcinoma by combined resection and locoregional therapy. J. Am. Coll. Surg. 195 (3), 311–318. https://doi.org/10.1016/s1072-7515(02)01226-7 (2002).
Barry, N. et al. Evaluating the impact of the radiomics quality score: a systematic review and meta-analysis. Eur. Radiol. 2025-03-01; 35 (3): 1701–1713. https://doi.org/10.1007/s00330-024-11341-y
Kocak, B., Keles, A., Kose, F. & Sendur, A. Quality of radiomics research: comprehensive analysis of 1574 unique publications from 89 reviews. Eur. Radiol. 35 (4), 1980–1992. https://doi.org/10.1007/s00330-024-11057-z (2025). -04-01.
Alabi, R. O., Elmusrati, M., Leivo, I., Almangush, A. & Mäkitie, A. A. Artificial Intelligence-Driven radiomics in head and neck cancer: current status and future prospects. INT J MED INFORM. 2024-08-01; 188 105464. https://doi.org/10.1016/j.ijmedinf.2024.105464
Zhou, H. Y. et al. A systematic review and Meta-Analysis of MRI radiomics for predicting microvascular invasion in patients with hepatocellular carcinoma. Curr. Med. Imaging. 01-01, 20 1–11. https://doi.org/10.2174/0115734056256824231204073534 (2024).
Chen, Y., Zeng, J., Guo, P., Zeng, J. & Liu, J. Prognostic significance of Platelet-to-Lymphocyte ratio (PLR) in extrahepatic metastasis of hepatocellular carcinoma after curative resection. Cancer Manag Res. 13, 1395–1405 (2021).
Limkin, E. J. et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 28, 1191–1206 (2017).
Brenet Defour, L. et al. Hepatocellular carcinoma: CT texture analysis as a predictor of survival after surgical resection. Eur. Radiol. 29, 1231–1239 (2019).
Jia, P. F., Li, Y. R., Wang, L. Y., Lu, X. R. & Guo, X. Radiomics in esophagogastric junction cancer: A scoping review of current status and advances. EUR J RADIOL. 2024-08-01; 177 111577. https://doi.org/10.1016/j.ejrad.2024.111577
Lee, J. H. et al. Preoperative prediction of early recurrence in resectable pancreatic cancer integrating clinical, radiologic, and CT radiomics features. CANCER IMAGING. 24 (1), 6. https://doi.org/10.1186/s40644-024-00653-3 (2024-01-08).
Zheng, H., Jian, L., Li, L., Liu, W. & Chen, W. Delta-Radiomics based on dynamic Contrast-Enhanced MRI for predicting lymphovascular invasion in invasive breast cancer. ACAD RADIOL. 2024-05-01; 31 (5): 1762–1772. https://doi.org/10.1016/j.acra.2023.11.017
Huang, Z. et al. Decision fusion model for predicting microvascular invasion in hepatocellular carcinoma based on Multi-MR habitat imaging and Machine-Learning classifiers. ACAD. RADIOL. 32 (4), 1971–1980. https://doi.org/10.1016/j.acra.2024.10.007 (2025-04-01).
Mirza-Aghazadeh-Attari, M., Srinivas, T., Kamireddy, A., Kim, A. & Weiss, C. R. Radiomics features extracted from Pre- and postprocedural imaging in early prediction of treatment response in patients undergoing transarterial radioembolization of hepatic lesions: A systematic review, Meta-Analysis, and quality appraisal study. J. AM. COLL. RADIOL. 21 (5), 740–751. https://doi.org/10.1016/j.jacr.2023.12.029 (2024-05-01).
Wang, Q. et al. Radiomics models for preoperative prediction of the histopathological grade of hepatocellular carcinoma: A systematic review and radiomics quality score assessment. EUR. J. RADIOL. 166, 111015. https://doi.org/10.1016/j.ejrad.2023.111015 (2023-09-01).
He, Y. et al. A novel multimodal radiomics model for predicting prognosis of resected hepatocellular carcinoma. Front. Oncol. 12, 745258. https://doi.org/10.3389/fonc.2022.745258 (2022).
Lambin, P. et al. Radiomics: the Bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14, 749–762. 10.1038/ nrclinonc.2017.141 (2017).
Davnall, F. et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical Practice?Insights imaging 3:573–589. (2012). https://doi.org/10.1007/s13244-012-0196-6
Xu, Y. et al. A CT-based radiomics approach to predict intra-tumoral tertiary lymphoid structures and recurrence of intrahepatic cholangiocarcino’ma. Insights Imaging. 14 (1), 173. https://doi.org/10.1186/s13244-023-01527-1 (2023-10-15).
Stefano, A. Challenges and limitations in applying radiomics to PET imaging: possible opportunities and avenues for research. COMPUT. BIOL. MED. 179, 108827. https://doi.org/10.1016/j.compbiomed.2024.108827 (2024).
Feng, Q. et al. An MRI-Based radiomics nomogram for differentiation of benign and malignant vertebral compression fracture. ACAD. RADIOL. 31 (2), 605–616. https://doi.org/10.1016/j.acra.2023.07.011 (2024-02-01).
Wang, T., Li, Y. Y., Ma, N. N., Wang, P. A. & Zhang, B. A MRI radiomics-based model for prediction of pelvic lymph node metastasis in cervical cancer. World J. Surg. Oncol. 2024–02–17; 22 (1): 55. https://doi.org/10.1186/s12957-024-03333-5
Yang, G. et al. Enhancing recurrence risk prediction for bladder cancer using multi-sequence MRI radiomics. Insights Imaging. 15 (1), 88. https://doi.org/10.1186/s13244-024-01662-3 (2024-03-25).
Guo, J. et al. Diagnostic performance of Contrast-Enhanced multidetector computed tomography and Gadoxetic acid Disodium-Enhanced magnetic resonance imaging in detecting hepatocellular carcinoma: direct comparison and a Meta-Analysis. Abdom. Radiol. (NY). 41, 1960–1972. https://doi.org/10.1007/s00261-016-0807-7 (2016).
Kim, H. Y. et al. Gadolinium ethoxybenzyl diethylenetriamine pentaacetic Acid-Enhanced magnetic resonance imaging predicts the histological grade of hepatocellular carcinoma only in patients with Child-Pugh class A cirrhosis. Liver Transpl. 18, 850–857. https://doi.org/10.1002/lt.23426 (2012).
Huppertz, A. et al. Enhancement of focal liver lesions at Gadoxetic Acid-Enhanced MR imaging: correlation with histopathologic findings and spiral CT–Initial observations. Radiology 234, 468–478. https://doi.org/10.1148/radiol.2342040278 (2005).
Sumie, S. et al. Microvascular invasion in patients with hepatocellular carcinoma and its predictable clinicopathological factors. Ann. Surg. Oncol. 15, 1375–1382. https://doi.org/10.1245/s10434-008-9846-9 (2008).
Li, Y., Tan, G., Vangel, M., Hall, J. & Cai, W. Influence of feature calculating parameters on the reproducibility of CT radiomic features: a thoracic Phantom study. QUANT. IMAG. MED. SURG. 10 (9), 1775–1785. https://doi.org/10.21037/qims-19-921 (2020-09-01).
Fiset, S. et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. RADIOTHER ONCOL. 2019-06-01; 135107–135114. https://doi.org/10.1016/j.radonc.2019.03.001
Wang, G., Wang, W. L., Liu, Y. Q., Dong, H. M. & Hu, Y. X. Positioning error and expanding margins of planning target volume with kilovoltage cone beam computed tomography for prostate cancer radiotherapy. Onco Targets Ther. 11, 1981–1988. https://doi.org/10.2147/OTT.S152915 (2018).
Wang, Y. et al. Deciphering intratumoral heterogeneity of hepatocellular carcinoma with microvascular invasion with radiogenomic analysis. J. Transl Med. 2023-10-18; 21 (1): 734. https://doi.org/10.1186/s12967-023-04586-6
He, Y. et al. A novel multimodal radiomics model for predicting prognosis of resected hepatocellular carcinoma. Front. Oncol. 12 (745258). https://doi.org/10.3389/fonc.2022.745258 (2022-01-01).
Tian, Y. et al. Prognostic value of systemic immune-inflammation index/ albumin for transcatheter arterial chemoembolization treatment. Heliyon 9 (4), e15156. https://doi.org/10.1016/j.heliyon.2023.e15156 (2023-04-01).
Zhou, Y. et al. CT-Based radiomics signature: A potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom. Radiol. (NY). 42, 1695–1704. https://doi.org/10.1007/s00261-017-1072-0 (2017).
Zheng, B. H. et al. Radiomics score: A potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer. 18, 1148. https://doi.org/10.1186/s12885-018-5024-z (2018).
Liu, P. et al. Prediction of microvascular invasion in solitary hepatocellular carcinoma ≤ 5 cm based on computed tomography radiomics. World J. Gastroenterol. 27 (17), 2015–2024. https://doi.org/10.3748/wjg.v27.i17.2015 (2021).
Feng, S. T. et al. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur. Radiol. 29 (9), 4648–4659. https://doi.org/10.1007/s00330-018-5935-8 (2019).
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 82293660/82293665), National Natural Science Foundation of China (Grant No. 82370890), Taishan Scholars Program of Shandong Province (grant number tsqn202312382), Natural Science Foundation of Shandong Province (grant number ZR2023MH243), Shandong Higher Education Young Science and Technology Support Program (grant number 2020KJL005 and 2023KJ224).
Author information
Authors and Affiliations
Contributions
CY and CZ conceived and designed the study. BD, BH, XH and NX collected and analyzed the data. YH wrote the draft manuscript. QD revised the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Informed consent was waived by Ethics Committees of Affiliated Hospital of Qingdao University (Hospital 1) and Qilu Hospital of Shandong University (Hospital 2). All methods were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, Y., Dong, B., Hu, B. et al. Radiomics-based machine-learning method to predict extrahepatic metastasis in hepatocellular carcinoma after hepatectomy: a multicenter study. Sci Rep 15, 29784 (2025). https://doi.org/10.1038/s41598-025-15406-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-15406-w