Abstract
Acute respiratory distress syndrome (ARDS) is one of the most common and serious complications in the development of sepsis. Endoplasmic reticulum stress (ERS) plays an important role in the pathophysiologic process of sepsis-associated ARDS. The aim of this study was to identify and analyze hub genes related to ERS in sepsis-associated ARDS using bioinformatics and machine learning algorithms, which may serve as diagnostic markers and therapeutic targets. Based on the GSE32707 dataset from the GEO database, differentially expressed genes (DEGs) between patients with sepsis-associated acute respiratory distress syndrome (ARDS) and healthy controls were identified. A comprehensive evaluation was performed by integrating functional enrichment analysis, immune cell infiltration analysis, and weighted gene co-expression network analysis (WGCNA). By intersecting DEGs, key WGCNA module genes, and ERS-related genes(ERGs), ERS-associated differential genes in sepsis-related ARDS were obtained. Subsequently, three machine learning algorithms—least absolute shrinkage and selection operator (LASSO), random forest (RF), and support vector machine (SVM)—were used to further screen for hub ERS hub genes. The diagnostic value of these hub genes was assessed using receiver operating characteristic (ROC) curve analysis. Finally, their expression levels were validated in clinical samples using RT-qPCR. A total of 438 DEGs and five hub genes—STAT3, HSPB1, YWHAQ, LCN2, and SGK1—were identified.Diagnostic performance analysis demonstrated that all five genes had favorable discriminatory power, indicating their potential clinical utility.Further validation in clinical samples confirmed the reliability of the bioinformatics analysis. RT-qPCR results showed that STAT3 was significantly upregulated, while YWHAQ was significantly downregulated in sepsis-associated ARDS samples compared to healthy controls, with both differences reaching statistical significance. In conclusion, STAT3 and YWHAQ, as ERS-related key genes, not only play pivotal roles in sepsis-associated ARDS but also hold promise as diagnostic biomarkers and potential therapeutic targets.
Similar content being viewed by others
Introduction
Sepsis is characterized by organ dysfunction resulting from a dysregulated host response to infection1. Despite significant advancements in diagnosis and treatment, it remains a leading cause of mortality worldwide2, with an estimated 11 million deaths attributed to sepsis in 20173. ARDS is a frequent and severe complication of sepsis4. Sepsis-associated ARDS exhibits greater severity than non-sepsis-associated ARDS, resulting in higher morbidity and mortality rates, with a reported mortality of approximately 40%5. However, there is currently a lack of targeted and effective pharmacological treatments for sepsis-associated ARDS6. Therefore, the identification and analysis of key molecular biomarkers for early diagnosis and timely intervention are crucial to reducing the mortality associated with this condition.
In recent years, ERS has emerged as a prominent area of research interest. The endoplasmic reticulum (ER) is a vital organelle responsible for protein transport, folding, and post-translational modification7,8. Pathological conditions such as sepsis and ischemia can lead to the accumulation of unfolded or misfolded proteins, disrupting ER homeostasis and triggering ERS9,10,11,12. Prolonged or excessive ERS can overwhelm the unfolded protein response (UPR), ultimately leading to apoptosis, tissue damage, and organ dysfunction13. Several studies have demonstrated that ERS plays a significant role in the pathogenesis of sepsis-associated ARDS. Therefore, exploring effective molecular biomarkers related to ERS holds great potential for the early identification and targeted treatment of sepsis-associated ARDS.
In this study, we employed a range of bioinformatics approaches and machine learning algorithms to identify ERS-related hub genes in sepsis-associated ARDS, and validated their expression using RT-qPCR. A schematic overview of the study workflow is presented in Fig. 1.
Materials and methods
Data sources
The GSE32707 dataset, annotated with platform GPL10558, was downloaded from the Gene Expression Omnibus (GEO) database. This dataset comprises 144 human peripheral blood samples, from which 31 patients with sepsis-associated ARDS and 34 healthy controls were selected for analysis.
DEGs analysis in sepsis-associated ARDS
The expression data from the two groups were standardized and normalized. DEGs between sepsis-associated ARDS patients and healthy controls were identified using the limma package in R14,with parameters set at | logFC |> 1 and a P-value < 0.05. The ggplot2 and pheatmap packages were employed to generate volcano plots and heatmaps, respectively, for result visualization.
Functional enrichment analysis of DEGs
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted on the DEGs using the clusterProfiler and org.Hs.eg.db packages15. GO enrichment analysis included three categories: molecular function (MF), biological process (BP), and cellular component (CC). KEGG pathway analysis was performed to identify significantly enriched signaling pathways. P-value < 0.05 was considered statistically significant.
Immunoinfiltration analysis
The CIBERSORT algorithm was applied to evaluate the relative abundance of 22 immune cell types in each sample16. Differences in immune cell infiltration between sepsis-associated ARDS patients and healthy controls were assessed and visualized using the ggpubr package’s ggboxplot function. Correlations among infiltrating immune cells were further analyzed using the corrplot package in R.
WGCNA and screening of ERS differential genes in sepsis-associated ARDS
WGCNA was conducted on the GSE32707 dataset17. Hierarchical clustering was first applied to detect and exclude outlier samples. Subsequently, an appropriate soft-thresholding power (β) was selected to construct a scale-free network. The weighted adjacency matrix was then transformed into a topological overlap matrix (TOM) to estimate network connectivity. Hierarchical clustering was used to construct a dendrogram of gene modules based on TOM dissimilarity. Modules showing the highest correlation with sepsis-associated ARDS were selected as key modules.
A total of 1301 ERGs with relevance scores > 7 were downloaded from the GeneCards database (https://www.genecards.org/). The intersection of DEGs, WGCNA key module genes, and ERGs was taken to identify ERS-related differential genes in sepsis-associated ARDS18.
Identification of hub genes for ERS in sepsis-associated ARDS
The ERS-related DEGs identified above were further analyzed using three machine learning algorithms: LASSO regression using the glmnet package, RF using the randomForest package19 and SVM20 Hub genes were identified as those commonly selected by all three algorithms.
Analysis of the diagnostic value of hub genes in sepsis-associated ARDS
The diagnostic performance of the identified hub genes was assessed using ROC curve analysis. The area under the curve (AUC) was calculated using the pROC package in R21, and AUC greater than 0.7 was considered indicative of good diagnostic performance.
Validation of ERS hub genes expression by RT-qPCR
A total of 19 patients with sepsis-associated ARDS hospitalized between May 2022 and August 2023 were enrolled as the test group. Nineteen healthy individuals undergoing routine physical examinations during the same period were recruited as the control group. Inclusion criteria: ① Test group: (i) met diagnostic criteria for sepsis-associated ARDS; (ii) age ≥ 18 years; (iii) provided informed consent. ② Healthy control group: (i) confirmed to be in good health by the hospital’s physical examination center; (ii) age ≥ 18 years; (iii) provided informed consent. Exclusion criteria: ① Age < 18 years;② Patients who died on admission or had incomplete clinical data;③ Patients with organic heart diseases, including ischemic cardiomyopathy, congenital heart disease, or myocarditis;④ Patients with end-stage chronic illnesses or malignancies;⑤ Individuals who refused to participate or did not provide informed consent;⑥ Patients with a history of psychiatric illness or cognitive dysfunction. Peripheral venous blood samples were collected, centrifuged at 4 °C, and the supernatant was aliquoted and stored at −80 °C. Informed consent was obtained from all participants, and the study was approved by the Clinical Research Ethics Committee of Shanxi Bethune Hospital (Approval No. SBQLL-2020-009).
Gene-specific primers were designed, and total RNA was extracted from serum samples of both groups. cDNA synthesis was performed using the HiScript II Q RT SuperMix kit. RT-qPCR was conducted using the Hieff qPCR SYBR Green Master Mix on a QuantStudio 6 Flex system. GAPDH was used as the internal control gene. The relative expression levels of ERS hub genes were calculated using the 2 − ΔΔCT method.
Statistical analysis
All statistical analyses were performed using R software (version 4.3.1) and GraphPad Prism (version 9.0). Normally distributed continuous variables were expressed as mean ± standard deviation (X ± SD) and compared using independent samples t-tests. Non-normally distributed variables were presented as median and interquartile range [M (IQR)] and compared using the Mann–Whitney U test. The diagnostic efficacy of hub genes was evaluated by ROC curve analysis. A two-tailed P-value < 0.05 was considered statistically significant.
Results
DEGs in sepsis-associated ARDS
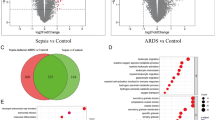
DEGs associated with sepsis-associated ARDS were identified using the limma package in R. A total of 438 DEGs were screened, including 226 upregulated and 212 downregulated genes (Fig. 2A). A heatmap was generated to visualize the top 25 upregulated and top 25 downregulated DEGs (Fig. 2B).
Functional enrichment analysis of DEGs
GO enrichment analysis revealed that the BP terms were primarily enriched in leukocyte activation and migration. The CC terms were mainly associated with specific granules, ribosomal subunits, and cytoplasmic ribosomes. The MF terms were predominantly enriched in structural constituents of ribosomes, haptoglobin binding, and antioxidant activity (Fig. 3A). KEGG pathway analysis indicated significant enrichment of several inflammatory signaling pathways, including the neutrophil extracellular trap (NET) formation pathway, the NOD-like receptor signaling pathway, and the NF-κB signaling pathway (Fig. 3B,C)22,23,24.
Immunoinfiltration analysis
Immunoinfiltration between the two groups was evaluated using the CIBERSORT algorithm based on the DEGs. Samples with a total immune cell abundance value of zero were excluded, resulting in the inclusion of 22 immune cell types. The distribution of immune cell infiltration across individual samples in both groups is shown in Fig. 4A. Compared to the healthy control group, the sepsis-associated ARDS group exhibited higher infiltration levels of activated CD4+T memory cells, γδ T cells, resting NK cells, and activated mast cells, while activated NK cells showed lower infiltration (Fig. 4B). Correlation analysis among immune cell populations revealed that eosinophils were positively correlated with activated dendritic cells and negatively correlated with neutrophils (Fig. 4C).
Immune cell subtype distribution based on DEGs. (A) Bar plot showing the relative proportion of immune cell infiltration across samples in the two groups. (B) Comparison of immune cell infiltration levels between the sepsis-associated ARDS and healthy control groups. (C) Heatmap of correlation analysis among immune cell types.*P < 0.05, **P < 0.01, ***P < 0.001; ns indicates no statistically significant difference.
WGCNA and identification of key ERS-related differential genes
The WGCNA R package was applied to perform clustering and establish a scale-free co-expression network, and 15 was selected as the soft threshold with a scale-free index of 0.85 for analysis (Fig. 5A,B). This analysis resulted in the identification of 11 distinct gene modules (Fig. 5C). The correlation between each module and sepsis-associated ARDS was assessed. The turquoise module, which showed the strongest positive correlation with sepsis-associated ARDS (r = 0.43, P = 3e−4), was selected as the key module and contained 2093 genes (Fig. 5D). To identify ERS-related differential genes, the intersection of three gene sets—DEGs, key module genes, and ERGs—was taken, yielding a total of 31 key ERS-associated differential genes (Fig. 5E).
WGCNA and identification of key ERS-related differentially genes. (A, B) Analysis of soft-thresholding power for scale-free topology in WGCNA. (C) Clustering dendrogram of gene modules identified by WGCNA. (D) Heatmap showing the correlation between gene modules and sepsis-associated ARDS. (E) Venn diagram showing the intersection of DEGs, WGCNA key module genes, and ERGs, resulting in 31 key ERS-related differential genes.
Screening of ERS hub genes in sepsis-associated ARDS
Three machine learning algorithms—LASSO, RF, and SVM—were applied to identify hub genes related to ERS in sepsis-associated ARDS.LASSO regression analysis identified 12 candidate genes with non-zero coefficients (Fig. 6A,B). Random forest analysis selected 16 genes with a Gini index greater than 0.5 (Fig. 6C,D). The SVM algorithm identified 28 relevant genes based on recursive feature elimination (Fig. 6E,F). By intersecting the results from all three algorithms, five overlapping ERS-related hub genes were identified: HSPB1, LCN2, SGK1, STAT3, and YWHAQ (Fig. 6G).
Screening of ERS hub genes in sepsis-associated ARDS. (A) LASSO coefficient profiles of the 30 candidate genes. (B) Ten-fold cross-validation for LASSO regression with the optimal lambda value (λ = 0.0134), identifying 12 characteristic genes. (C) Error rate curve with confidence intervals for the random forest model (ntree = 500). (D) Variable importance plot showing the top genes ranked by mean decrease in Gini index in the random forest model. (E–F) Results of the SVM algorithm: the x-axis represents the number of selected features, with a cross-validation precision of 0.802 and error of 0.198. (G) Venn diagram showing the intersection of hub genes identified by LASSO, RF and SVM algorithms.
Analysis of ERS hub gene expression and diagnostic performance
The ggboxplot function in R was used to generate box plots illustrating the expression levels of endoplasmic reticulum stress-related hub genes in the sepsis-associated ARDS group versus healthy controls from the GSE32707 dataset. Among these, LCN2 and STAT3 were significantly upregulated in sepsis-associated ARDS, while HSPB1, SGK1, and YWHAQ were significantly downregulated (P < 0.05 for all; Fig. 7A–E). ROC curves were plotted using R to evaluate the diagnostic performance of each gene, and the AUC was calculated. The AUC values were as follows: HSPB1, 0.741; LCN2, 0.824; SGK1, 0.791; STAT3, 0.817; and YWHAQ, 0.798 (Fig. 7F).
Expression of ERS hub genes and their diagnostic efficacy in sepsis-associated ARDS. (A–E) Box plots showing the expression levels of HSPB1, LCN2, SGK1, STAT3, and YWHAQ in sepsis-associated ARDS patients and healthy controls from the GSE32707 dataset. (F) ROC curves illustrating the diagnostic performance of the five ERS-related hub genes.
Validation of ERS hub genes by RT-qPCR
To validate the expression of the identified ERS hub genes, RT-qPCR was performed on peripheral blood samples collected from patients with sepsis-associated ARDS and control samples. The results revealed that STAT3 expression was significantly elevated in the sepsis-associated ARDS samples (2.435 ± 1.147) compared to control samples (0.979 ± 0.319, P < 0.0001). Conversely, YWHAQ expression was significantly decreased [0.300 (0.170, 0.347)] compared to control samples [1.342 (0.470, 1.934), P < 0.001]. No statistically significant differences in expression were observed for HSPB1 [1.191 (0.641, 1.563) vs. 1.210 (0.649, 1.438)], SGK1 [1.587 (0.444, 2.284) vs. 1.289 (0.506, 1.221)], and LCN2 [0.764 (0.483, 1.054) vs. 1.243 (0.489, 1.594)] between the two groups (P > 0.05 for all)(Fig. 8A–E).
Expression of ERS hub genes in patients with sepsis-associated ARDS and healthy controls. (A–E) Relative mRNA expression levels of HSPB1, LCN2, SGK1, STAT3, and YWHAQ in peripheral blood samples measured by RT-qPCR.Data for STAT3 are presented as x ± SD, while data for HSPB1, LCN2, SGK1, and YWHAQ are expressed as median [interquartile range, IQR].Statistical significance: **P < 0.001, ***P < 0.0001; ns indicates no statistically significant difference.
Discussion
In this study, we systematically screened and validated ERS-related key genes in sepsis-associated ARDS by integrating differential expression analysis, immunoinfiltration, WGCNA, and multiple machine learning algorithms using the GSE32707 dataset from the GEO database. Five ERS-related hub genes were ultimately identified: STAT3, HSPB1, YWHAQ, LCN2, and SGK1. Among them, STAT3 was significantly upregulated and YWHAQ significantly downregulated in patients, a finding further validated by RT-qPCR analysis. These results suggest that STAT3 and YWHAQ may serve as potential diagnostic biomarkers for sepsis-associated ARDS.
Differential expression analysis using R software identified a total of 438 DEGs, including 226 upregulated and 212 downregulated genes. Functional enrichment analysis indicated that these DEGs were significantly enriched in biological pathways related to leukocyte activation and migration, ribosomal subunits, antioxidant activity, and the nuclear factor kappa B (NF-κB) signaling pathway. These findings suggest that the biological roles of DEGs in sepsis-associated ARDS are closely related to inflammatory responses, ERS, and NF-κB pathway activation. Previous studies have demonstrated that 4-phenylbutyric acid (4-PBA) alleviates lipopolysaccharide (LPS)-induced lung inflammation by inhibiting ERS and suppressing activation of the NF-κB signaling pathway. Specifically, 4-PBA was shown to reduce the expression of unfolded protein response (UPR)-related proteins in the lungs of LPS-treated mice and to significantly decrease the number of inflammatory cells in bronchoalveolar lavage fluid (BALF), as well as lower the levels of inflammatory cytokines such as interferon-gamma (IFN-γ), tumor necrosis factor-alpha (TNF-α), interleukin-1 beta (IL-1β), and intercellular adhesion molecule 1 (ICAM-1)9. These results are consistent with the findings of the present study.
Immunoinfiltration analysis revealed a higher proportion of activated CD4⁺ T memory cells, γδ T cells, and mast cells in sepsis-associated ARDS compared to healthy controls. In addition, eosinophil infiltration showed a positive correlation with activated dendritic cells, and a negative correlation with neutrophil infiltration. Osorio et al. previously reported that the IRE1α–XBP1 axis of the UPR plays a crucial role in regulating dendritic cell function, and that deletion of XBP1 impairs antigen presentation by CD8α⁺ dendritic cells25. These findings suggest that ERS may influence the differentiation and functional regulation of both innate and adaptive immune cells involved in the pathogenesis of sepsis-associated ARDS.
WGCNA analysis was performed to obtain the gene modules with the highest association with sepsis-associated ARDS, containing a total of 2093 genes. The intersection of DEGs, key module genes, and ERGs yielded 31 key ERS-related differential genes. Five ERS hub genes, STAT3, HSPB1, YWHAQ, LCN2 and SGK1, were identified through three distinct machine learning algorithms. ROC analysis was performed to evaluate diagnostic efficacy, revealing that the AUC for all five hub genes was greater than 0.74, suggesting favorable diagnostic potential. The expression levels of the identified hub genes were validated in clinical samples using RT-qPCR. Compared with the healthy control group, the expression of STAT3 was significantly upregulated, while YWHAQ was significantly downregulated in the sepsis-associated ARDS group. In contrast, the expression levels of HSPB1, SGK1, and LCN2 showed no statistically significant differences between the two groups.
STAT3 is a member of the STAT protein family. The JAK/STAT pathway is a major signaling pathway for various key cytokines and initiates the expression of NF-κB, IL-1β, TNF-α, IL-6, and others. It is widely involved in various biological processes, including inflammation and apoptosis26,27,28. In animal models of sepsis-induced lung injury, STAT3 expression is positively correlated with the expression of these inflammatory factors and the severity of lung damage. Tofacitinib, a JAK inhibitor, blocks JAK-STAT3 pathway activation, markedly alleviating lung tissue injury and pulmonary edema severity in sepsis models29. In addition, methotrexate (MTX), an anti-inflammatory agent, inhibits the JAK2/STAT3 signaling pathway and has been proven to reduce systemic inflammation and pulmonary injury in CLP-induced sepsis rat models30. Forkhead box protein A2 (FOXA2) mitigates LPS-induced ERS in part by suppressing the p38/STAT3 signaling cascade31.
YWHAQ, a member of the 14-3-3 protein family, plays key roles in processes such as protein transport, signal transduction, and cell apoptosis. The encoded protein 14-3-3θ is a negative regulator of apoptosis signal-regulating kinase 1 (ASK1). Experimental evidence further suggests that regulated IRE1-dependent decay (RIDD) in the IRE1α pathway induces apoptosis by downregulating 14-3-3θ expression and activating the ASK1/JNK signaling pathway32. 14-3-3 proteins can inhibit NF-κB activation mediated by Toll-like receptor 2 (TLR2), while enhancing the activation of transcription factors dependent on TLR433. In addition, 14-3-3 proteins can bind to exosomes and play a vital role in sepsis-associated acute lung injury (ALI)34.
HSPB1, a prominent member of the heat shock protein (HSP) family, plays a vital role in the host response to various pathophysiological stresses, including injury, stress, hypoxia, and infection35. In cecal ligation and puncture (CLP) models, HSPB1 knockout (HSPB1−/−) mice exhibited nearly twice the mortality rate compared to wild-type (WT) mice.SGK1, a member of the protein kinase subfamily, is a serine/threonine kinase that serves as a key regulatory node in multiple signaling pathways and phosphorylation processes. It plays essential roles in cell proliferation, ion channel regulation, and signal transduction and is considered a critical player in inflammatory responses36. It plays essential roles in cell proliferation, ion channel regulation, and signal transduction and is considered a critical player in inflammatory responses37. LCN2, a member of the lipocalin family, is a key regulator of iron metabolism, oxidative stress, and inflammation in mammals. Experimental data indicate that LCN2 levels in plasma-derived extracellular vesicles (EVs) are significantly elevated in sepsis-associated ARDS patients compared to those without ARDS38.
However this study inevitably has some limitations. First, the clinical sample size was small, and the limited sample size may affect the stability and statistical efficacy of the results. Second, although the hub genes of ERS were identified and analyzed, the exact mechanism of the roles of STAT3 and YWHAQ in sepsis-associated ARDS was not elucidated. Therefore, further in-depth exploration is needed in the future to provide an important theoretical basis for translational clinical applications.
In summary, this study systematically identified five ERS-related hub genes—STAT3, HSPB1, YWHAQ, LCN2, and SGK1—in sepsis-associated ARDS through integrated bioinformatics analysis and multiple machine learning algorithms. Diagnostic efficacy analysis demonstrated favorable discriminatory power for all five genes. Subsequent RT-qPCR validation further confirmed that STAT3 was significantly upregulated and YWHAQ was downregulated in samples. These findings suggest that STAT3 and YWHAQ play pivotal roles in the pathogenesis of sepsis-associated ARDS and may serve as promising molecular markers for early diagnosis and potential targets for precision intervention.
Data availability
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) datasets (http://www.ncbi.nlm.nih.gov/geo/). All data are available from the corresponding author on reasonable request.
References
Singer, M. et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). Jama 315, 801–810. https://doi.org/10.1001/jama.2016.0287 (2016).
Chiu, C. & Legrand, M. Epidemiology of sepsis and septic shock. Curr. Opin. Anaesthesiol. 34, 71–76. https://doi.org/10.1097/aco.0000000000000958 (2021).
Rudd, K. E. et al. Global, regional, and National sepsis incidence and mortality, 1990–2017: Analysis for the global burden of disease study. Lancet (London England). 395, 200–211. https://doi.org/10.1016/s0140-6736(19)32989-7 (2020).
Li, W. et al. Classic signaling pathways in alveolar injury and repair involved in sepsis-induced ALI/ARDS: New research progress and prospect. Disease Markers 2022, 6362344 https://doi.org/10.1155/2022/6362344 (2022).
Sheu, C. C. et al. Clinical characteristics and outcomes of sepsis-related vs non-sepsis-related ARDS. Chest 138, 559–567. https://doi.org/10.1378/chest.09-2933 (2010).
Liu, C., Xiao, K. & Xie, L. Advances in the regulation of macrophage polarization by mesenchymal stem cells and implications for ALI/ARDS treatment. Front. Immunol. 13, 928134. https://doi.org/10.3389/fimmu.2022.928134 (2022).
Sano, R. & Reed, J. C. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta. 1833, 3460–3470. https://doi.org/10.1016/j.bbamcr.2013.06.028 (2013).
Senft, D. & Ronai, Z. A. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 40, 141–148. https://doi.org/10.1016/j.tibs.2015.01.002 (2015).
Kim, H. J. et al. Inhibition of Endoplasmic reticulum stress alleviates lipopolysaccharide-induced lung inflammation through modulation of NF-κB/HIF-1α signaling pathway. Sci. Rep. 3, 1142. https://doi.org/10.1038/srep01142 (2013).
Ron, D. & Walter, P. Signal integration in the Endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529. https://doi.org/10.1038/nrm2199 (2007).
Minamino, T., Komuro, I. & Kitakaze, M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circul. Res. 107, 1071–1082. https://doi.org/10.1161/circresaha.110.227819 (2010).
Kim, I., Xu, W. & Reed, J. C. Cell death and Endoplasmic reticulum stress: Disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 7, 1013–1030. https://doi.org/10.1038/nrd2755 (2008).
Li, L., Peng, X., Guo, L., Zhao, Y. & Cheng, Q. Sepsis causes heart injury through Endoplasmic reticulum stress-mediated apoptosis signaling pathway. Int. J. Clin. Exp. Pathol. 13, 964–971 (2020).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 https://doi.org/10.1093/nar/gkv007 (2015).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics: J. Integr. Biol. 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. https://doi.org/10.1186/1471-2105-9-559 (2008).
Bardou, P., Mariette, J., Escudié, F., Djemiel, C. & Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 15, 293. https://doi.org/10.1186/1471-2105-15-293 (2014).
Izmirlian, G. Application of the random forest classification algorithm to a SELDI-TOF proteomics study in the setting of a cancer prevention trial. Ann. N. Y. Acad. Sci. 1020, 154–174. https://doi.org/10.1196/annals.1310.015 (2004).
Huang, S. et al. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteom. 15, 41–51. https://doi.org/10.21873/cgp.20063 (2018).
Robin, X. et al. pROC: An open-source package for R and S + to analyze and compare ROC curves. BMC Bioinform. 12, 77. https://doi.org/10.1186/1471-2105-12-77 (2011).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–d677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. Public. Protein Soc. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Osorio, F. et al. The unfolded-protein-response sensor IRE-1α regulates the function of CD8α + dendritic cells. Nat. Immunol. 15, 248–257. https://doi.org/10.1038/ni.2808 (2014).
Zhang, J. et al. Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS. Ther. Adv. Respir. Dis. https://doi.org/10.1177/1753466619879840 (2019).
Chang, X. et al. Influence of Roflumilast on sepsis mice through the JAK/STAT signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 1335–1341. https://doi.org/10.26355/eurrev_201902_17028 (2019).
Yan, X. et al. HO-1 overexpressed mesenchymal stem cells ameliorate sepsis-associated acute kidney injury by activating JAK/stat3 pathway. Cell. Mol. Bioeng. 11, 509–518. https://doi.org/10.1007/s12195-018-0540-0 (2018).
Xu, F. et al. Inhibition of gp130 alleviates LPS-induced lung injury by attenuating apoptosis and inflammation through JAK1/STAT3 signaling pathway. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 72, 493–507. https://doi.org/10.1007/s00011-022-01686-9 (2023).
Bringué, J. et al. Methotrexate ameliorates systemic inflammation and septic Associated-Lung damage in a cecal ligation and puncture septic rat model. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22179612 (2021).
Xue, Z., Li, Y., Xiao, S., Zhang, H. & Xu, J. FOXA2 attenuates lipopolysaccharide–induced pneumonia by inhibiting the inflammatory response, oxidative stress and apoptosis through blocking of p38/STAT3 signaling. Exp. Therapeutic Med. 26, 469. https://doi.org/10.3892/etm.2023.12168 (2023).
Gómora-García, J. C., Gerónimo-Olvera, C., Pérez-Martínez, X. & Massieu, L. J. C. D. D. IRE1α RIDD activity induced under ER stress drives neuronal death by the degradation of 14-3-3 θ mRNA in cortical neurons during glucose deprivation. Cell Death Discov. 7, 131. https://doi.org/10.1038/s41420-021-00518-9 (2021).
Liu, J. et al. The role of 14-3-3 proteins in cell signalling pathways and virus infection. J. Cell. Mol. Med. 25, 4173–4182. https://doi.org/10.1111/jcmm.16490 (2021).
Wang, D., Zhang, M. & Pei, H. Advances in the application of exosomes in sepsis Chinese critical care medicine. Chin. Crit. Care Med. 30, 377–380. https://doi.org/10.3760/cma.j.issn.2095-4352.2018.04.019 (2018).
Breed, E. R. et al. The small heat shock protein HSPB1 protects mice from sepsis. Sci. Rep. 8, 12493. https://doi.org/10.1038/s41598-018-30752-8 (2018).
Li, Q., Zheng, H. & Chen, B. Identification of macrophage-related genes in sepsis-induced ARDS using bioinformatics and machine learning. Sci. Rep. 13, 9876. https://doi.org/10.1038/s41598-023-37162-5 (2023).
Li, J. et al. Melatonin attenuates sepsis-induced acute lung injury through improvement of epithelial sodium channel-mediated alveolar fluid clearance via activation of SIRT1/SGK1/Nedd4-2 signaling pathway. Front. Pharmacol. 11, 590652. https://doi.org/10.3389/fphar.2020.590652 (2020).
Ye, R. et al. Plasma-derived extracellular vesicles prime alveolar macrophages for autophagy and ferroptosis in sepsis-induced acute lung injury. Mol. Med. (Cambridge Mass). 31, 40. https://doi.org/10.1186/s10020-025-01111-x (2025).
Acknowledgements
The authors are grateful for the data provided by the GEO database(GSE32707).
Funding
This work was supported by the Shanxi Province Overseas Students Science and Technology Activities Selection Funding Project (20200030). 2023 Talent Introduction Support Program of Shanghai Pudong New Area Zhoupu Hospital (ZP-XK-2023B-4). 2023 Public Institution People’s Livelihood Research Special Project, Pudong New Area Science and Technology Development Fund (PKJ2023-Y12). 2024 Teaching Research Project for Faculty, Shanghai University of Medicine & Health Sciences (CFDZ20240025P).
Author information
Authors and Affiliations
Contributions
LG conceived and designed the study. LG and TT analyzed data and wrote the manuscript. YX revised the manuscript and make final approval of the version. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The research was conducted in full compliance with the guidelines outlined in the Declaration of Helsinki. Third Hospital of Shanxi Medical University approved the study protocol ethics committee (Ethical Approval No.SBQLL-2020-009). Approval of ethics and agreement to participate.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, L., Liu, T. & Li, X. Identification and analysis of the endoplasmic reticulum stress hub genes in sepsis-associated ARDS. Sci Rep 15, 31007 (2025). https://doi.org/10.1038/s41598-025-16644-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-16644-8
Keywords
This article is cited by
-
Comparison between eDNA and traditional morphological methods for fish diversity monitoring in rivers
Scientific Reports (2026)