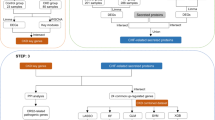
Abstract
Chronic kidney disease (CKD) can induce chronic heart failure (CHF), a condition referred to as type 4 cardiorenal syndrome (CRS4). The pathophysiological mechanisms remain unclear, and suitable early warning biomarkers for CHF in CKD patients are lacking. A total of 258 CHF key genes and 383 CKD-related secreted proteins were identified through differential expression analysis and WGCNA. PPI analysis revealed 81 genes as potential pathogenic genes related to CRS4. Enrichment analysis of these pathogenic genes highlighted pathways involved in cytokine activity, extracellular matrix remodeling, and immune response. Three machine learning algorithms identified two hub genes (MME and SERPINF1) as potential biomarkers for CHF, and a nomogram model was constructed. ROC analysis demonstrated that the model achieved an AUC greater than 0.80 in both the CHF merged dataset and two external cohorts. Furthermore, immune cell infiltration analysis indicated a correlation between these biomarkers and the infiltration scores of fibroblasts, CD8 T cells, and mast cells in CHF. Finally, our clinical cohort validated the expression patterns of these two biomarkers in serum, with the diagnostic model achieving an AUC of 0.880. CKD may promote the progression of CHF through proteins secreted by the kidneys and blood cells. MME and SERPINF1 may serve as potential biomarkers for CHF in CKD patients.
Similar content being viewed by others
Introduction
Chronic kidney disease (CKD) ranks among the top ten global disease burdens1. Epidemiological studies have revealed a significantly higher incidence of cardiovascular diseases (CVDs) in patients with CKD, with approximately 50% of end-stage renal disease (ESRD) patients succumbing to CVDs2. Among these, chronic heart failure (CHF) represents a common outcome of various CVDs, with a 10-year cumulative incidence of up to 20% in CKD patients, making it one of the leading causes of death in this population3. CHF resulting from CKD is referred to as type 4 cardiorenal syndrome (CRS4)4. However, the progression of CHF in CKD patients is often insidious, with a prolonged disease course and clinical features that overlap with the underlying manifestations of CKD. This is particularly true in the early stages of CHF, where symptoms are easily overlooked or underestimated in CKD patients. Therefore, early identification of high-risk CKD patients prone to CHF is of paramount importance.
Traditional heart failure biomarkers, such as high-sensitivity troponin (hs-TnT) and N-terminal B-type natriuretic peptide precursor (NT-proBNP), are influenced by renal function, and no universally accepted cutoff values exist for these markers in the CKD population. Moreover, their serum concentrations are often elevated only when overt clinical symptoms are already present, reflecting a delayed response. As a result, their clinical utility remains controversial5. Consequently, identifying novel and precise biomarkers for CHF in CKD patients is necessary.
In recent years, numerous studies have been published focusing on novel biomarkers for CHF based on the pathophysiological mechanisms in CKD patients. For instance, soluble suppression of tumorigenicity 2 (sST2) is a biomarker of myocardial fibrosis. A study by Gaggin et al. dynamically monitored changes in sST2 levels among CKD patients, revealing a positive correlation with the risk of developing CHF6. Galectin-3 (Gal-3), which promotes inflammation and fibrosis during kidney injury, can lead to remote myocardial damage and cardiac remodeling7. Research by Voroneanu et al. indicated that CKD patients with elevated Galectin-3 had a 1.11-fold increased risk of CHF8. However, current reports on novel CHF biomarkers often focus on a limited selection of candidate molecules of interest to researchers, lacking large-scale screening and comparative analyses, which poses significant limitations. Recently, studies have explored biomarker screening from the perspective of secreted proteins to investigate inter-organ interactions9,10,11. Secreted proteins, including various hormones, cytokines, and membrane proteins, are recognized for their role in mediating inter-organ communication and altering the pathophysiological states of distant organs. These proteins can be released into the bloodstream, making them suitable for clinical detection and offering unique advantages in disease biomarker screening.
Therefore, this study is based on gene expression datasets from the heart, kidneys, and blood cells, aiming to screen the most valuable potential novel biomarkers for CHF from 3947 secreted proteins and establish a diagnostic model, with the goal of helping clinicians to early identify high-risk CKD patients who are likely to develop CHF, enabling them to benefit from early treatment.
Materials and methods
Data collection and consolidation
Two CHF microarray datasets, GSE19303 and GSE21610, were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). In addition, raw expression profile datasets for kidney tissues from CKD patients (GSE66494) and whole blood (WB) (GSE37171) were retrieved from the GEO database. R software (version 4.4.0) was used to merge the CHF datasets using the “ComBat” function from the “SVA” package.
Differentially expressed genes (DEGs) analysis
Initially, the CHF merged dataset and the CKD-related dataset were subjected to gene symbol conversion and normalization. The “Limma” package in R was then used to identify DEGs in the CKD and CHF datasets. The criteria for screening DEGs were an adjusted p-value < 0.05 and a fold change > 1.5. Volcano plots were generated using the “ggplot2” package, and heatmaps were created with the “pheatmap” package to visualize the expression patterns of DEGs in R.
Weighted gene co-expression network analysis (WGCNA)
In the first step, a median absolute deviation of 0 was used to filter genes from each sample. The second step involved the use of the “goodSamplesGenes” function to detect missing values and exclude samples with over 20,000 values as outliers. In the third step, the optimal soft threshold was determined using cex1 = 0.85 to establish a scale-free co-expression gene network. After gene clustering, modules were obtained based on the criterion of MEDissThres = 0.25, and similar modules were merged. The fourth step involved creating heatmaps to illustrate the relationship between modules and traits. Lastly, we generated scatter plots depicting the MM-GS correlation for each module by calculating module membership (MM) and gene significance (GS) values.
Secreted protein gene set
The Human Protein Atlas database (https://www.proteinatlas.org/) is a public database that provides information on human encoded proteins. From the “SPOCTOPUS predicted of secreted proteins” category, 3947 genes encoding secreted proteins were identified.
Networks of protein–protein interactions (PPI)
This study explored the interactions between CHF key genes and CKD-related secreted proteins, leading to the construction of a PPI network. This network was implemented based on STRING database (https://cn.string-db.org/), applying a confidence score threshold of 0.4. The MCODE plugin in Cytoscape software (version 3.9.1, https://cytoscape.org/) was then used to evaluate important subsets and visualize the network. The top three subsets were identified as CRS4-related pathogenic genes and selected for further analysis and investigation.
Functional enrichment analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses are used to investigate the biological functions and mechanisms of CRS4-related pathogenic genes12. To this end, these genes are input into the SangerBox website (http://www.sangerbox.com/) for functional enrichment analysis, and bubble charts or circle diagrams are used for visualization.
cMAP (Connectivity Map) analysis
The cMAP database (https://clue.io) can reveal the associations between gene expression and small molecule compounds by analyzing differences in gene expression. The upregulated genes from the CRS4-related pathogenic genes are uploaded to the cMAP online repository for analysis to screen out the top ten small molecule drugs that can reverse the expression of the aforementioned genes.
Machine learning (ML) algorithms
This study combined three ML algorithms—Generalized Linear Model (GLM), Random Forest (RF), and Extreme Gradient Boosting (XGB)—to identify potential biomarkers for CHF. First, the intersection of CHF DEGs, CHF key module genes, and CKD-related secreted proteins was taken, resulting in 12 intersecting genes. Then, the XGB, RF, and GLM algorithms were used to perform CHF diagnostic importance ranking for these 12 intersecting genes within the CHF merged dataset. These three ML algorithms were realized using the “randomForest,” “kernlab,” “glmnet,” and “caret” packages in R 4.4.0. Subsequently, the intersection of the top six important genes identified by each algorithm was determined, and these intersecting genes are designated as hub genes, which are used to develop a CKD-related CHF diagnostic model. Violin plots are used to analyze the expression patterns of the hub genes in the CHF merged dataset.
Establishment and evaluation of the diagnostic nomogram model
Based on the two hub genes obtained, a CHF diagnostic nomogram model was established using the “rms” package in R software. The reliability of the model in diagnosing CHF were assessed using calibration curves. And the diagnostic performance of model for CHF was evaluated by generating decision curve analysis (DCA) and receiver operating characteristic (ROC) curves.
External validation of hub gene for CHF diagnosis
Two independent CHF datasets (GSE1145 and GSE141910) were obtained from the GEO database, which include CHF cases and control groups. Violin plots were used to examine hub gene expression patterns in the external datasets, and ROC analysis was used to evaluate the hub genes’ diagnostic performance.
Immune cell infiltration analysis
Two immune cell infiltration analysis algorithms, MCPcounter and single sample enrichment analysis (ssGSEA), were used to analyze the degree of immune cell and stromal cell infiltration in the CHF combined dataset. Box plots generated using the “ggplot2” package displayed the cell infiltration scores obtained from the two algorithms. Subsequently, Spearman’s rank correlation test was employed to analyze the correlation between the immune cell infiltration scores and the expression levels of hub genes, which were then presented using a correlation heatmap.
Patient sample collection
Participants were stratified into three groups: (1) Control group: healthy individuals lacking CKD, CHF, or major organic diseases; (2) CKD group: stage 3–5 CKD patients without CHF; (3) CRS4 group: stage 3–5 CKD patients with CHF diagnosed ≥ 3 months after initial CKD identification. CKD diagnosis complied with the 2024 CKD Clinical Practice Guidelines13, while CHF diagnosis followed the 2022 Heart Failure Management Guidelines14. Common exclusion criteria across all groups comprised: (i) receipt of any dialysis within 1 month; (ii) exposure to major stressful events (e.g., unemployment, bereavement) within 1 month; (iii) new-onset major organic diseases (e.g., stroke, severe infection) within 3 months; (iv) comorbidities affecting plasma MME or SERPINF1 levels (including malignancies, osteoarthritis); (v) pregnancy or lactation status.
We collected clinical characteristics including baseline comorbidities, total cholesterol, low-density lipoprotein (LDL), NT-proBNP, cardiac troponin T (cTnT), left ventricular ejection fraction (LVEF), and estimated glomerular filtration rate (eGFR). The study protocol for human samples was approved by the Ethics Committee of the Shaoxing Second Hospital (Ethics batch number: 2024053), and all participants provided written informed consent. Additionally, the serum levels of membrane metalloendopeptidase (MME) and serpin family F member 1 (SERPINF1) were measured using enzyme-linked immunosorbent assay (ELISA) kits (Cloud-Clone Corp., Wuhan, China) following the manufacturer’s instructions.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 10.0.2 (GraphPad Software Inc., San Diego, CA, USA). Quantitative data were presented as mean ± standard deviation, and qualitative data were presented as number (percentage). The Kolmogorov–Smirnov test was employed for the normality test. The comparison of quantitative data between groups was performed using unpaired Student’s t-test or one-way analysis of variance, while the comparison of qualitative data between groups was performed using Chi-square test. P values less than 0.05 were considered statistically significant. For all figures, significance levels were defined as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
Identification of CHF key genes
The analytical workflow of this study is illustrated in Fig. 1. After batch correction, the CHF merged dataset included 61 CHF patients and 16 healthy controls. The box plot of mRNA expression showed that the batch effect between the two datasets was significantly reduced after batch effect elimination (Figs. 2A,B). Differential Expression Analysis (DEA) between the CHF and control groups identified a total of 364 DEGs, of which 227 genes were upregulated and 137 genes were downregulated. The expression characteristics of DEGs in the CHF merged dataset were visualized using volcano and heatmap (Figs. 2C,D). To further identify CHF key genes, we performed WGCNA to screen for gene modules highly correlated with CHF. Based on considerations of scale independence and mean connectivity, the soft-threshold power was set to 3. Using this threshold, nine modules were generated, and the clustering of module eigengenes is shown in Fig. 2E. We then analyzed the relationship between CHF and the nine gene modules, as depicted in Fig. 2F. The analysis revealed that the turquoise module had the strongest positive correlation with CHF (r = 0.42, p = 1 × 10−4). Additionally, the blue and yellow modules also showed statistical significance and, along with the turquoise module, were identified as CHF key modules. To identify CHF key genes, we intersected the DEGs with the genes in the CHF key modules, resulting in 258 CHF key genes (Fig. 2G).
Identification of CHF key genes. (A,B) Box plots of the expression profiles before (A) and after (B) batch correction of CHF datasets. (C) Volcano plot of differentially expressed genes in the CHF merged dataset. (D) Heatmap of the top 30 differentially expressed genes in the CHF merged dataset. (E) Gene dendrogram displaying the cleaved gene modules. (F) Heatmap showing the correlation between gene modules and traits. (G) Venn diagram illustrating the overlap between CHF differential genes and key module genes.
Identification of CKD-related secreted proteins
As illustrated by the volcano plot and heatmap, a total of 997 DEGs were identified in the WB samples of CKD patients (Fig. 3A,B), while 1,400 DEGs were identified in kidney tissue samples (Fig. 3C,D). Subsequently, the DEGs from WB (Fig. 3E) and kidney tissues (Fig. 3F) were intersected with the secreted proteins gene set. The union of these two intersections yielded a collection of CKD-related secreted proteins.
Identification of CKD-related secreted proteins. (A,B) Volcano plot (A) and heatmap (B) of differentially expressed genes in the GSE37171 dataset. (C,D) Volcano plot (C) and heatmap (D) of differentially expressed genes in the GSE66494 dataset. (E,F) Venn diagrams showing the overlap between the secreted protein gene set and differentially expressed genes in CKD whole blood (E) and kidney tissues (F).
PPI analysis, functional enrichment, and drug screening for CRS4-related pathogenic genes
To investigate the potential mechanisms of action of secreted proteins in CRS4, we utilized the STRING database to examine the interactions between CKD-related secreted proteins and CHF key genes. Using the MCODE plugin, three significant clusters comprising a total of 81 genes were identified as potential CRS4-related pathogenic genes. Gene Ontology (GO) enrichment analysis of these CRS4-related pathogenic genes revealed that biological processes (BP) such as “cellular response to cytokine stimulus” and “extracellular matrix organization” were among the top-ranked entries (Fig. 4A). Moreover, molecular function (MF) analysis indicated strong associations between the pathogenic genes and functions like “cytokine activity” and “endopeptidase activity” (Fig. 4A). KEGG pathway analysis demonstrated a robust correlation between CRS4-related pathogenic genes and key pathways, including the “renin-angiotensin system” and “hypertrophic cardiomyopathy” (Fig. 4B). Additionally, these genes were found to be enriched in several metabolic pathways, such as “arachidonic acid metabolism,” “glutathione metabolism,” and “phenylalanine metabolism.”
Furthermore, cMAP drug analysis identified the top 10 small molecule compounds capable of reversing the CRS4-related pathogenic genes’ expression, including dexamethasone, 3,3′-diindolylmethane, dimercaptosuccinic acid, voriconazole, dapsone, γ-homolinolenic acid, BRD-K97951054, orantinib, BRD-A61599461, and albendazole, which may serve as potential therapeutic agents for CRS4 (Fig. 4C). The targets or pathways affected by these small molecule compounds are depicted in Fig. 4D.
Functional enrichment, and drug screening for CRS4-related pathogenic genes. (A) The lollipop chart displays the results of the GO enrichment analysis, including the BP, CC, and MF terms associated with the CRS4-related pathogenic genes. (B) The circle diagram shows the pathways that are significantly enriched in the KEGG analysis for CRS4-related pathogenic genes. (C) Heatmap of the top 10 small-molecule drug enrichment fractions obtained by cMAP analysis. (D) The Sankey diagram presents the target descriptions of the top 10 drugs.
Biomarker screening and diagnostic model construction for CKD-related CHF
CKD related secreted proteins play a critical role in promoting the onset and progression of CHF. We further intersected CHF DEGs, genes of CHF key modules, and CKD-related secreted proteins, resulting in the identification of 12 intersecting genes (Fig. 5A). These genes may serve as unique biomarkers for early warning in CKD-related CHF. To narrow down the scope of potential biomarkers and enhance their clinical applicability, we employed three ML algorithms: GLM, RF, and XGB, to rank the importance scores of the 12 intersecting genes (Figs. 5B–D). Notably, after identifying the common genes from all three ML algorithms, only two hub genes—MME and SERPINF1—were consistently present in all three subsets (Fig. 5E).
Compared to the control group, the mRNA expression levels of MME and SERPINF1 were significantly elevated in the CHF group (Figs. 5F,G). ROC analysis showed that the area under the curve (AUC) values for both hub genes exceeded 0.8 (Fig. 5H,I). To improve diagnostic accuracy and predictive capacity, a nomogram model incorporating these two hub genes was constructed (Fig. 5J). Calibration curves indicated that the predicted probabilities from the nomogram model closely aligned with those from the ideal model (Fig. 5K). DCA demonstrated that the nomogram model provides clinical benefit for the diagnosis of CHF (Fig. 5L). In the ROC analysis, the nomogram’s AUC reached 0.892, demonstrating excellent diagnostic performance (Fig. 5M).
Biomarker screening and diagnostic model construction for CKD-related CHF. (A) Venn diagram showing the intersection of CHF DEGs, CHF key module genes, and CKD-related secreted proteins. (B–D) Importance rankings of the 12 overlapping genes using three ML algorithms: GLM (B), RF (C), and XGB (D). (E) Venn diagram of the intersection of the top 6 important genes ranked by each of the three algorithms. (F,G) Violin plots displaying the expression patterns of MME (F) and SERPINF1 (G) in the CHF merged dataset. (H,I) ROC analysis for diagnosing CHF using MME (H) and SERPINF1 (I). (J) Nomogram model for diagnosing CHF using MME and SERPINF1. (K) Calibration curve of the nomogram model. (L) DCA curve for diagnosing CHF using the nomogram model. (M) ROC analysis of the nomogram model for diagnosing CHF.
External verification of the expression patterns and diagnostic performance of two biomarkers
Datasets GSE1145 and GSE141910 served as two independent external CHF datasets for validation. Violin plots demonstrated that both biomarkers were significantly upregulated in the CHF groups of GSE1145 (Fig. 6A,B) and GSE141910 (Fig. 6C,D). ROC analysis indicated that the two biomarkers exhibited strong diagnostic performance for CHF in the external datasets (Fig. 6E,G). Additionally, the AUC values for the nomogram model reached 0.939 and 0.833 for the GSE1145 and GSE141910 datasets, respectively (Fig. 6F,H).
External verification of the expression patterns and diagnostic performance of two biomarkers. (A,B) The violin plots display the expression patterns of MME (A) and SERPINF1 (B) in the GSE1145 dataset. (C,D) The violin plots show the expression patterns of MME (C) and SERPINF1 (D) in the GSE141910 dataset. (E,F) ROC analysis demonstrates the diagnostic performance for CHF of individual biomarkers (E) and the model (F) in the GSE1145 dataset. (G,H) ROC analysis presents the diagnostic performance for CHF of individual biomarkers (G) and the model (H) in the GSE141910 dataset.
Immune cell infiltration analysis of CHF merged dataset
Enrichment analysis revealed a strong association between immune processes and CRS4, prompting us to perform two immune cell infiltration analyses. The enrichment ratio of 8 kinds of immune cells and 2 kinds of stromal cells was calculated in MCPcounter algorithm, while the enrichment fraction of 16 kinds of immune cells was calculated in ssGSEA algorithm. In the MCPcounter algorithm, the proportion of CD8 T cells, cytotoxic lymphocytes and fibroblasts increased significantly in the CHF group (Fig. 7A). And SERPINF1 expression was positively correlated with the proportion of CD8 T cells, cytotoxic lymphocytes and fibroblasts (Fig. 7B). In the ssGSEA algorithm, CD8T cells and mast cells rose significantly in the CHF group (Fig. 7C). And SERPINF1 expression was positively correlated with the proportion of CD8 T cells and mast cells (Fig. 7D). In both algorithms, the expression of MME was significantly positively correlated with CD8T cells (Fig. 7B,D).
Immune cell infiltration analysis of CHF merged dataset. (A) The box plot displays the differences in immune-related cells infiltration scores between CHF and the control group as obtained by the MCPCounter algorithm. (B) The heatmap shows the correlation between immune-related cells and two biomarkers in the MCPCounter algorithm. (C) The box plot presents the differences in immune cell infiltration scores between CHF and the control group as obtained by the ssGSEA algorithm. (D) The heatmap illustrates the correlation between immune cells and two biomarkers in the ssGSEA algorithm.
Clinical serological validation of the diagnostic performance of two biomarkers
To further validate the clinical significance of these two biomarkers, this study collected serum samples to measure protein concentrations. The clinical characteristics of the study cohort are shown in Table 1. No significant differences were observed in age or sex among the three groups. Compared with the Control group, the two disease groups (CKD group and CRS4 group) exhibited significantly reduced renal function indices. Except for cardiac function indices (LVEF and NT-proBNP), no other significant differences were observed between the CKD group and the CRS4 group. These results indicate consistency between the clinical characteristics and disease grouping.
Compared with the healthy control group and the CKD group, the concentrations of the two biomarkers were significantly elevated in the CRS4 group, with statistically significant differences (Fig. 8A,B). ROC analysis revealed that the AUC values for diagnosing CRS4 were 0.811 and 0.790 for MME and SERPINF1, respectively (Fig. 8C,D). As shown in Fig. 8E, a nomogram model incorporating these two biomarkers was constructed. Analysis of the calibration curve indicated that the predicted probabilities generated by the nomogram closely matched those of the ideal model (Fig. 8F). Furthermore, DCA was performed to evaluate the performance of the nomogram model, demonstrating its advantage in identifying CRS4 (Fig. 8G). Additionally, ROC analysis of the nomogram model yielded an AUC value of 0.880, indicating excellent diagnostic performance (Fig. 8H).
Clinical serological validation of the diagnostic performance of two biomarkers. (A,B) Bar graphs showing the protein concentrations of MME (A) and SERPINF1 (B) in serum. (C,D) ROC analysis demonstrating the diagnostic efficacy of MME (C) and SERPINF1 (D) for CRS4. (E) A nomogram model for diagnosing CRS4 based on MME and SERPINF1. (F) Calibration curve of the nomogram model used for diagnosing CRS4. (G) DCA curve of the nomogram model for diagnosing CRS4. (H) ROC analysis showing the efficacy of the nomogram model in diagnosing CRS4.
A comprehensive diagnostic model established based on MME, SERPINF1 and NT-proBNP
NT-proBNP and cTnT are established cardiac biomarkers. In this cohort, serum cTnT levels were significantly elevated in both CKD and CRS4 groups compared to control group (Fig. 9A), while serum NT-proBNP levels showed greater elevation in the CRS4 group versus both control and CKD patients (Fig. 9B). ROC analysis revealed AUC values of 0.682 and 0.865 for cTnT and NT-proBNP in diagnosing CRS4, respectively (Fig. 9C,D). Given the superior diagnostic value of NT-proBNP, we developed a nomogram integrating MME, SERPINF1, and NT-proBNP (Fig. 9E). Analysis of the calibration curve indicated that the predicted probabilities generated by the nomogram closely matched those of the ideal model (Fig. 9F). DCA confirmed the clinical utility of the nomogram for CRS4 identification (Fig. 9G). The model achieved an AUC of 0.942 in ROC analysis, indicating excellent diagnostic performance (Fig. 9H).
A comprehensive diagnostic model established based on MME, SERPINF1 and NT-proBNP. (A,B) Bar graphs showing the protein concentrations of cTnT (A) and NT-proBNP (B) in serum. (C,D) ROC analysis demonstrating the diagnostic efficacy of cTnT (C) and NT-proBNP (D) for CRS4. (E) A nomogram model for diagnosing CRS4 based on MME, SERPINF1, and NT-proBNP. (F) Calibration curve of the nomogram model used for diagnosing CRS4. (G) DCA curve of the nomogram model for diagnosing CRS4. (H) ROC analysis showing the efficacy of the nomogram model in diagnosing CRS4.
Discussion
This study investigates the relationship between CKD and the subsequent development of CHF through bioinformatics analysis. The potential humoral mechanisms through which CKD induces CHF may involve cytokine activity, adhesion molecule binding, and immune- and metabolism-related signaling pathways. ML algorithms facilitated the development of a diagnostic nomogram that integrates two hub genes, MME and SERPINF1, for CHF risk prediction. ROC curve analysis demonstrates that these two hub genes and the model exhibit strong performance in predicting CHF. Validation of MME and SERPINF1 upregulation patterns in two external GEO datasets confirmed their consistency with findings from this study. Additional validation with clinical cohort further confirmed the predictive value of MME and SERPINF1 as biomarkers for CHF in CKD patients.
The primary characteristics of heart damage in CRS4 include myocardial hypertrophy, interstitial fibrosis, and a subsequent decline in diastolic and contractile function. The pathogenesis of CRS4 is complex and challenging to fully elucidate. It is currently believed that the primary mechanism by which CRS4 induces heart damage involves activation of the sympathetic nervous system, the renin-angiotensin-aldosterone system (RAAS), the release of inflammatory factors, and the accumulation of uremic toxins in end-stage CKD patients4,15. This study investigates the pathogenesis of CRS4 from the perspective of secreted proteins. KEGG enrichment analysis of CRS4-related pathogenic genes revealed important risk factors for heart failure, such as the RAAS system and hypertrophic cardiomyopathy. GO enrichment analysis identified entries related to secreted proteins, including “cellular response to cytokine stimulus” and “cytokine activity.” Moreover, the top-ranked BP entries included “extracellular matrix remodeling,” while the top-ranked CC and MF entries also involved extracellular matrix-related terms. This evidence suggests that CKD may interact with the heart through various cytokines secreted by kidney and circulatory blood cells, induce myocardial hypertrophy and myocardial fibrosis, and accelerate the progression of heart failure.
It is well established that CHF is a chronic inflammatory disease, and increasing evidence suggests that immune cell dysfunction plays a critical role in its pathogenesis16. In this study, both the MCPcounter and ssGSEA algorithms were applied to a combined CHF dataset to identify key immune cell alterations. The MCPcounter algorithm revealed a significant enrichment of fibroblasts in the hearts of CHF patients, consistent with prior findings. The ssGSEA algorithm identified an elevated mast cell infiltration score in the hearts of CHF patients. Previous studies have demonstrated that in the hearts of mice with experimental hypertension, myocardial infarction, or chronic cardiac volume overload, there is a significant increase in mast cell numbers. These cells store and release various active mediators, including TNF-α and proteases such as elastase, renin, and matrix metalloproteinases, which contribute to heart remodeling and the progression of heart failure17. Inhibition of mast cell proteases or degranulation has been shown to prevent the progression of heart fibrosis and improve left ventricular dysfunction18,19. Interestingly, both algorithms consistently identified a significant upregulation of CD8 T cell infiltration in the hearts of CHF patients. Previous studies have suggested that CD8 T cells are activated in hypertensive mice and release perforin, which promotes TGFβ1 release and heart fibrosis20. Further research has shown that in mice with myocardial remodeling following myocardial infarction or left ventricular dysfunction, CD8 T cells are recruited and activated, releasing granzyme B to induce inflammation and myocardial cell apoptosis, thus promoting heart failure progression. Moreover, CD8 T cell exhaustion significantly alleviates the progression of heart failure. Additionally, recruitment of CD8 T cells and elevated expression of granzyme B have been observed in pig models of ischemia-reperfusion and in human patients with myocardial infarction21,22. These findings suggest that CD8 T cell activation and cytokine release play a pivotal role in the progression of heart failure. Furthermore, GO and KEGG analyses of the CRS4 pathogenic genes in this study were enriched in the “Toll-like receptor signaling pathway.” Toll-like receptors (TLRs) are a family of pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). TLRs play a critical role in innate immune responses, leading to both acute and chronic inflammation23. Recent evidence has shown that myocardial TLRs, including TLR224, TLR325, TLR426, and TLR927, among others, contribute to the progression of CHF through innate immune activation and inhibiting TLRs has emerged as a promising therapeutic approach for CHF. These findings suggest that CKD-related secreted proteins may contribute to the onset and progression of CHF through various immune pathways, including the recruitment of mast cells and CD8 T cells, and activation of TLRs.
Despite the widespread occurrence of CHF, diagnosing it in CKD populations remains a significant challenge. Since CHF is diagnosed clinically based on characteristic medical history and physical examination, its accurate diagnosis depends on symptoms and signs of volume overload and increased cardiac filling pressure. These symptoms and signs are also observed in CKD patients, particularly as they approach end-stage, complicating the distinction between CHF and systemic volume overload due to CKD28. Therefore, identifying new early biomarkers of CHF in CKD patients is crucial. This study, based on secreted proteins from CKD patients, identifies two CHF biomarkers and establishes a CHF diagnostic model using three machine learning algorithms. RNA detection in the external cohort and serum ELISA detection in the clinical cohort of this study suggest that these two biomarkers have high diagnostic value for CHF. MME encodes neprilysin, a protein that inactivates multiple peptide hormones, including enkephalin, substance P, and neurotensin, inducing vasoconstriction and playing a key role in the progression of CHF. Its inhibitor, sacubitril/valsartan, has been extensively studied and shown to improve the long-term prognosis of CHF patients29. SERPINF1, a member of the non-inhibitory serpin family, promotes myocardial cell apoptosis, thereby inducing the progression of CHF. Current research has shown that its serum concentration is significantly associated with the incidence of adverse events in patients with advanced heart failure30. Moreover, correlation studies have identified a significant positive correlation between these biomarkers and CD8 T cells, mast cells, or fibroblasts, suggesting that these markers may mediate the progression of CHF in CKD patients through an immunological pathway.
Providing appropriate CHF treatment for CKD patients can be challenging once diagnosed. Traditional CHF treatments, such as excessive use of diuretics, can decrease blood volume and worsen CKD progression. ACE inhibitors (ACEI) or angiotensin II receptor blockers (ARBs) may cause hyperkalemia, requiring cautious use in patients with severely reduced renal function. Emerging CHF treatments, such as sacubitril/valsartan and SGLT2 inhibitors, show great potential for CKD patients, but more evidence from clinical studies is needed. This study performed a drug sensitivity analysis based on CKD-induced upregulated pathogenic genes, identifying ten small-molecule drugs that significantly reverse their expression, with dexamethasone ranked first. Dexamethasone, a synthetic glucocorticoid with strong anti-inflammatory and immunosuppressive effects, is commonly used to treat CKD. In a mouse experiment, the leukemia drug ponatinib induced strong inflammation and significant cardiotoxicity. However, dexamethasone intervention nearly completely reversed the cardiac dysfunction caused by ponatinib31. Another study on rats with CHF found that dexamethasone improved symptoms by inhibiting the RAAS system and significantly increasing urine output32. Additionally, dexamethasone enhances NPRA expression in the renal collecting ducts, improving kidney sensitivity to ANP, promoting sodium excretion, and reducing left ventricular end-diastolic pressure33. It should be noted that short-term use of dexamethasone may prevent cardiac injury by exerting anti-inflammatory effects, inhibiting immune cells, and promoting diuresis, but long-term dexamethasone treatment may also lead to opposite results, similar to cardiovascular complications in patients with Cushing’s syndrome34. Therefore, rational use of dexamethasone could offer dual benefits for both CHF and CKD.
This study has several limitations: the single-center design and modest sample size limit the statistical power, while the restricted cohort precluded subgroup analyses across CKD stages. Additionally, the study could not follow up with the clinical cohort to further explore the predictive value of the two biomarkers for CHF progression and prognosis in CKD patients. Further research is also needed to understand the mechanisms by which these biomarkers contribute to CRS4 and to explore their potential as therapeutic targets.
Data availability
The public datasets were downloaded and analyzed in this study including GEO data repository (accession numbers: GSE19303, GSE21610, GSE37171, GSE66494, GSE1145, and GSE141910).
Abbreviations
- CKD:
-
Chronic kidney disease
- CVDs:
-
Cardiovascular diseases
- ESRD:
-
End-stage renal disease
- CHF:
-
Chronic heart failure
- CRS4:
-
Type 4 cardio-renal syndrome
- NT-proBNP:
-
N-terminal B-type natriuretic peptide precursor
- sST2:
-
Soluble suppression of tumorigenicity 2
- Gal-3:
-
Galectin-3
- WB:
-
Whole blood
- DEG:
-
Differentially expressed gene
- WGCNA:
-
Weighted gene co-expression network analysis
- MM:
-
Module membership
- GS:
-
Gene significance
- PPI:
-
Protein–protein interaction
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- BP:
-
Biological processes
- CC:
-
Cellular component
- MF:
-
Molecular function
- cMAP:
-
Connectivity map
- ML:
-
Machine learning
- RF:
-
Random forest
- XGB:
-
eXtreme gradient boosting
- GLM:
-
Generalized linear model
- DCA:
-
Decision analysis curve
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- TLRs:
-
Toll-like receptors
- PRRs:
-
Pattern recognition receptors
- RAAS:
-
Renin–angiotensin–aldosterone system
- ME:
-
Module eigengene
- ssGSEA:
-
Single sample enrichment analysis
- ELISA:
-
Enzyme-linked immunosorbent assay
References
Diseases, G. B. D. & Injuries, C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the global burden of disease study 2019. Lancet. 396 (10258), 1204–1222 (2020).
Li, X. & Lindholm, B. Cardiovascular risk prediction in chronic kidney disease. Am. J. Nephrol. 53 (10), 730–739 (2022).
House, A. A. et al. Heart failure in chronic kidney disease: conclusions from a kidney disease: improving global outcomes (KDIGO) controversies conference. Kidney Int. 95 (6), 1304–1317 (2019).
Ronco, C., Haapio, M., House, A. A., Anavekar, N. & Bellomo, R. Cardiorenal syndrome. J. Am. Coll. Cardiol. 52 (19), 1527–1539 (2008).
Aimo, A. et al. Prognostic value of high-sensitivity troponin T in chronic heart failure: an individual patient data Meta-Analysis. Circulation 137 (3), 286–297 (2018).
Gaggin, H. K. et al. Head-to-head comparison of serial soluble ST2, growth differentiation factor-15, and highly-sensitive troponin T measurements in patients with chronic heart failure. JACC Heart Fail. 2 (1), 65–72 (2014).
Bansal, N. et al. Cardiac biomarkers and risk of incident heart failure in chronic kidney disease: the CRIC (Chronic renal insufficiency Cohort) study. J. Am. Heart Assoc. 8 (21), e012336 (2019).
Voroneanu, L. et al. Prospective validation of a screening biomarker approach combining amino-terminal pro-brain natriuretic peptide with Galectin-3 predicts death and cardiovascular events in asymptomatic Hemodialysis patients. Angiology. 69 (5), 449–455 (2018).
Zhu, E. et al. Screening of immune-related secretory proteins linking chronic kidney disease with calcific aortic valve disease based on comprehensive bioinformatics analysis and machine learning. J. Transl. Med. 21 (1), 359 (2023).
Zhang, C. et al. Screening of secretory proteins linking major depressive disorder with heart failure based on comprehensive bioinformatics analysis and machine learning. Biomolecules. 14(7). (2024).
Deng, Y. T. et al. Atlas of the plasma proteome in health and disease in 53,026 adults. Cell. (2024).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Kidney Disease. Improving global outcomes CKDWG. KDIGO 2024 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. 105 (4S), S117–S314 (2024).
Heidenreich, P. A. et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: A report of the American college of cardiology/american heart association joint committee on clinical practice guidelines. J. Am. Coll. Cardiol. 79 (17), e263–e421 (2022).
Kumar, U., Wettersten, N. & Garimella, P. S. Cardiorenal syndrome: pathophysiology. Cardiol. Clin. 37 (3), 251–265 (2019).
Zhang, Y., Bauersachs, J. & Langer, H. F. Immune mechanisms in heart failure. Eur. J. Heart Fail. 19 (11), 1379–1389 (2017).
Janicki, J. S., Brower, G. L. & Levick, S. P. The emerging prominence of the cardiac mast cell as a potent mediator of adverse myocardial remodeling. Methods Mol. Biol. 1220, 121–139 (2015).
Ricciotti, E. & FitzGerald, G. A. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31 (5), 986–1000 (2011).
Willoughby, S., Holmes, A. & Loscalzo, J. Platelets and cardiovascular disease. Eur. J. Cardiovasc. Nurs. 1 (4), 273–288 (2002).
Brassington, K. et al. Crosstalk between cytotoxic CD8 + T cells and stressed cardiomyocytes triggers development of interstitial cardiac fibrosis in hypertensive mouse hearts. Front. Immunol. 13, 1040233 (2022).
Santos-Zas, I. et al. Cytotoxic CD8(+) T cells promote granzyme B-dependent adverse post-ischemic cardiac remodeling. Nat. Commun. 12 (1), 1483 (2021).
Wang, D. et al. CD8 T cells promote heart failure progression in mice with preexisting left ventricular dysfunction. Front. Immunol. 15, 1472133 (2024).
Zhang, Y., Wu, J., Dong, E., Wang, Z. & Xiao, H. Toll-like receptors in cardiac hypertrophy. Front. Cardiovasc. Med. 10, 1143583 (2023).
Shishido, T. et al. Toll-like receptor-2 modulates ventricular remodeling after myocardial infarction. Circulation 108 (23), 2905–2910 (2003).
Chen, C. et al. Role of extracellular RNA and TLR3-Trif signaling in myocardial ischemia-reperfusion injury. J. Am. Heart Assoc. 3 (1), e000683 (2014).
Yang, Y. et al. The emerging role of Toll-like receptor 4 in myocardial inflammation. Cell. Death Dis. 7 (5), e2234 (2016).
Oka, T. et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature 485 (7397), 251–255 (2012).
Tuegel, C. & Bansal, N. Heart failure in patients with kidney disease. Heart 103 (23), 1848–1853 (2017).
Bayes-Genis, A. et al. Prognostic value and kinetics of soluble Neprilysin in acute heart failure: A pilot study. JACC Heart Fail. 3 (8), 641–644 (2015).
Rychli, K. et al. Prognostic value of pigment epithelium-derived factor in patients with advanced heart failure. Chest 138 (3), 656–664 (2010).
Tousif, S. et al. Ponatinib drives cardiotoxicity by S100A8/A9-NLRP3-IL-1beta mediated inflammation. Circ. Res. 132 (3), 267–289 (2023).
Liu, C., Ge, N., Zhai, J. L. & Zhang, J. X. Dexamethasone-induced diuresis is associated with Inhibition of the renin-angiotensin-aldosterone system in rats. Kaohsiung J. Med. Sci. 32 (12), 614–619 (2016).
Liu, C. et al. Glucocorticoids improve renal responsiveness to atrial natriuretic peptide by up-regulating natriuretic peptide receptor-A expression in the renal inner medullary collecting duct in decompensated heart failure. J. Pharmacol. Exp. Ther. 339 (1), 203–209 (2011).
Kanzaki, A., Kadoya, M., Katayama, S. & Koyama, H. Cardiac hypertrophy and related dysfunctions in Cushing syndrome Patients-Literature review. J. Clin. Med. 11, 23 (2022).
Funding
This research was supported by the Zhejiang Province Medicine and Health Science and Technology Project (2023KY377) and Keqiao District Science and Technology Project (2024KZ29).
Author information
Authors and Affiliations
Contributions
Conceptualization: Z.N.W., Z.Y.W., and O.T.; Methodology: H.S., X.D., J.W., and Z.N.W.; Investigation: Z.N.W., Z.Y.W., and Y.H.; Visualization: Z.N.W. and Z.Y.W.; Data curation: Z.N.W.; Project administration: Y.C. and O.T.; Writing—original draft preparation: Z.N.W.; Writing—review and editing: Y.C. and O.T.; Funding acquisition: Z.N.W. and J.W. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Shaoxing Second Hospital (2024053).
Consent for publication
All authors have thoroughly reviewed the manuscript and consent to its publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, Z., Wu, Z., Sun, H. et al. Identification of early warning biomarkers for type 4 cardio-renal syndrome based on bioinformatics analysis and secreted proteins. Sci Rep 15, 35071 (2025). https://doi.org/10.1038/s41598-025-16651-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-16651-9