Abstract
Jute, a sustainable bast fibre crop from the Malvaceae family, has significant industrial applications and ecological benefits, making it an important focus of recent scientific research. This study investigates the genomic landscape of Miniature Inverted-Repeat Transposable Elements (MITEs), truncated derivatives of autonomous DNA transposons, in two cultivated jute species, Corchorus capsularis and Corchorus olitorius. Genome-wide analysis identified 51,369 and 50,829 MITEs, classified into 632 and 539 families in CC and C. olitorius, respectively. A notable proportion of MITEs were found in genic and near-genic regions, underscoring their potential as tools for gene function exploration. Additionally, 11,801 and 10,921 MITE-derived microRNAs were identified in CC and C. olitorius, emphasizing their roles in genetic and epigenetic regulation. Recognizing the scarcity of molecular markers in jute, this study developed an in-house database of genome-wide MITE insertional polymorphism (MIP) markers. These markers leverage MITEs’ unique features—high insertion polymorphism, genic region preference, and short sequence length—making them ideal for molecular breeding. Validation of thirty-eight randomly selected MIP markers across 30 jute accessions demonstrated their efficacy in genetic diversity analysis, grouping the accessions into five distinct clusters. This comprehensive study not only highlights the significance of MITEs in jute genomics but also provides valuable genomic resources for future breeding and improvement programs.
Similar content being viewed by others
Introduction
Jute, a member of the Malvaceae family, represents a versatile and environmentally sustainable bast fibre crop. Often referred to as “golden fibre” jute has been utilized for centuries. In recent years, it has attracted considerable scientific interest due to its myriad industrial applications and its potential role in sustainable development1. The two primary cultivated species, Corchorus olitorius (Tossa jute) and Corchorus capsularis (white jute), possess distinct traits and historical origins. C. olitorius (CO) is believed to have originated in Africa, whereas CC (CC) traces its roots to Asia2.
The crop is highly valued for its resilience, biodegradability, and minimal environmental impact, making it a preferred choice for various applications, including textiles, packaging, and composites. Furthermore, it significantly contributes to diminishing environmental carbon footprints by absorbing approximately 15 tons of CO2 from the atmosphere and releasing up to 11 tons of O2 per hectare3. However, the enhancement of jute quality and productivity has been constrained by the absence of well-characterized genetic markers and a limited understanding of the molecular mechanisms underlying crucial agronomic traits. To tackle these challenges, researchers have progressively utilized genomic tools, which possess significant potential for revealing the genetic capabilities of this vital fibre crop4. In recent years, significant advancements in genomics and genetic engineering have emerged, offering unprecedented opportunities to explore the genetic and molecular foundations of jute and facilitating more targeted crop improvement strategies1. A few draft genomes of jute, specifically tossa and white jute, have been reported5,6,7 offering insights into the organization and genetic information of the jute genome.
With the availability of jute genome data, it is now feasible to delve deeper into the genetic intricacies of this vital crop. Specifically, studying jute transposable elements will be highly beneficial. Understanding these elements can reveal insights into gene regulation8,9 genome architecture10, stress response and genome evolution11. One class of transposable elements, called Miniature Inverted-repeat Transposable Elements (MITEs), are a family of non-autonomous transposable elements that are notably more prevalent and active in plant genomes. These short DNA sequences, typically ranging from 70 to 800 base pairs in length, can transpose within the genome without encoding their own transposase enzyme. Instead, they rely on the transposase machinery of other autonomous transposable elements for their mobilization12,13. MITEs are characterized by their terminal inverted repeats and the presence of a target site duplication upon insertion. Their small size and lack of coding capacity render them less disruptive to the host genome compared to larger protein-coding transposable elements. Nevertheless, their high copy number and widespread distribution throughout the genome can still exert significant impacts on gene regulation and genome evolution. In several plant genomes, including rice and Arabidopsis, MITEs have amplified to a high copy number. For example, Rice has around 90,000 MITEs categorized into over 100 distinct families12,14,15. The preponderance of MITEs in these genomes is located inside the euchromatin arms of chromosomes16,17,18, as opposed to the heterochromatin. This preferential insertion into euchromatin areas indicates a possible function in affecting gene expression and maintaining genomic integrity.
Across diverse plant species, MITEs often insert near or within gene-coding regions, where they can influence gene expression by introducing alternative transcription start sites, disrupting splice sites, and providing new regulatory sequences such as promoters or enhancers. Furthermore, MITEs can generate new small RNAs, including microRNAs, which play crucial roles in regulating target genes at the transcriptional and post-transcriptional levels19. These MITE-derived microRNAs contribute to the diversification of gene regulatory networks and the evolution of new phenotypes. Understanding the prevalence and functional roles of MITE-derived microRNAs in jute is, therefore, a crucial area of research that can provide valuable insights into the regulation of important agronomic traits and the adaptive potential of this essential crop.
Given its significance in the global fibre market and its potential to support sustainable development, this research paper delves into the current understanding of jute MITEs, exploring their structural characteristics, genomic distribution, and potential implications for the plant’s development and adaptation. Additionally, the study of microRNAs in jute offers insights into gene expression regulation and new regulatory pathway evolution. The study’s significance lies in being one of the first to comprehensively identify and classify MITEs in the jute genome. While leveraging MITEs for genomic applications is well-documented in other crops, our work uniquely focuses on jute—a crop that has historically received limited genomic attention. By identifying jute-specific MITE markers and computationally predicting their potential roles in miRNA generation, this study fills a critical gap in the genomic toolkit available for jute.
Materials and method
Identification and classification of jute mites
The genome sequences of CC var. ‘Huangma 179’ (HM179) and CO var. ‘Kuanyechangguo’ (KYCG) were obtained from https://bigd.big.ac.cn/gwh, accessed on 05.01.2025 with accession numbers GWHBCLC00000000 and GWHBCLB00000000, respectively7. In order to identify MITE elements in jute genomes the open-source software MITE Tracker20 was utilized with following parameters, Like element length between 50 and 800 bp, terminal inverted repeats (TIRs) of 10–30 bp, and flanking 2–10 bp target site duplications (TSDs)., minimum copy number threshold of 3. This software first identifies potential elements by detecting specific inverted repeat sequences and then computes the local composition complexity (LCC) score. After executing these initial operations, legitimate MITE candidates are recognized and subsequently grouped utilizing the VSEARCH tool with parameter --iddef 1 and --id 0.8, corresponding to a BLAST-like identity calculation and a minimum 80% similarity threshold20. MITE elements were further classified into known and unknown families by employing BLAST search with an e-value threshold of ≤ 1e−5 against plant-MITE database (P-MITE)21 which contain MITE sequences of different plant species.
To annotate all MITE elements in the jute genome, the putative MITE elements identified by the VSEARCH tool were used as a query and BLASTn search was performed against the jute genomes. The BLAST results were filtered in accordance with Chen et al., 201422 utilizing a standalone Perl script. The criteria encompassed a minimum element length of 50, a maximum element length of 800, a minimum query identity of 80%, a minimum threshold length ratio between query and subject of 0.85, a maximum threshold length ratio of 1.15, a minimum query coverage of 90%, and an overlap margin between query and subject of plus or minus 5 nucleotides.
MITE elements were subsequently categorized as either genic or near genic and intergenic according to their genomic location utilizing the BED tools intersect/window function. Elements were categorized as genic if they resided within boundary of gene. Furthermore, genic MITEs were classified according to their occurrence in coding or non-coding regions. The near-genic MITEs include sequences located within 1000 base pairs downstream or upstream of a gene whereas inter-genic were considered beyond the gene but within its 1000 bp flanking regions on both sides.
Genome‑wide analysis of unique and common mites
A comparative genome-wide analysis was performed to identify conserved and unique MITE sequences in the genomes of CO and CC. This analysis employed a sequence similarity method using the BLASTn program with an e-value threshold of ≤ 1e−5. The CO MITE sequences served as query sequence against the CC MITE sequences. This method allowed for the comparison of one or more nucleotide query sequences against a subject nucleotide sequence or a nucleotide sequence database, thereby facilitating the identification of common and unique MITE sequences within the genomes of these two jute species. In order to identify orthologus MITE elements between CC and CO, reciprocal best hit method was used.
Identification of MITE as MiRNA precursor
miRNA sequences from 271 organisms were obtained from the miRBase V.22 biological database23. miRNA sequences were searched against MITE elements using BLASTn program with criteria of alignment must be between 20 to 24nt and maximum of 3 mismatches (Axtell et al., 2018). The prediction of RNA secondary structure was performed using the ViennaRNA package v2.5.1 (http://www.tbi.univie.ac.at/~ivo/RNA/, accessed on 12.01.2025). Only predicted MITEs that demonstrated above mentioned hit with the small RNAs were classified as MITE-derived small RNAs.
In Silico mining of transcription factors associated with mites
Transcription factors containing MITE elements were identified transcription factor prediction tool of the Plant Transcription Factor Database (https://planttfdb.gao-lab.org/, accessed on 12.01.2025). The predicted transcription factors were subsequently validated through InterPro Scan to confirm presence of domain patterns.
Annotation of genes containing MITE elements
Annotations of MITE containing genes of both the species was completed using PANNZER web server24 which provides both Gene Ontology (GO) annotations and text description predictions. Function of 800 and 895 genes containing MITE elements, have been predicted through PANNZER2 web server in CC and CO respectively. Input file for WEGO web server was generated from PANNZER2 predicted GO terms and Gene ontology of both the species were graphically represented through WEGO web server (https://wego.genomics.cn/, accessed on 02.02.2025).
Development of MITE insertion polymorphic (MIP) markers
MITE elements along with 150 bp upstream and downstream sequences were fetched from the genome sequence by “blastdbcmd” function of the standalone BLAST tool. Then an in-house perl script “primer3_in.pl” was used to prepare input files for Primer 3 software. Standalone version of Primer 3 software was used to design a primer from flanking sequences of MITE elements with parameters annealing temperature:50–60 °C, GC content:40–60%, length 18–24 nucleotides, product length 100–900 bp. Primer3 produced multiple primer pairs for each MITE element. Hence, another in-house perl script named “extract_p3out.pl” was used to fetch a single pair of primers from each MITE element. In the present study, 38 randomly chosen primers were procured and genotyped in a panel of 30 jute accessions.
DNA extraction
Genomic DNA was isolated from fresh leaf tissues utilizing the CTAB method25. About 100 mg of leaf tissue was pulverized in Liquid nitrogen and reconstituted in 700 µL of CTAB extraction buffer (2% CTAB, 20 mM EDTA, 100 mM Tris-HCl, 1.4 M NaCl and 0.2% β-mercaptoethanol). Following a 30-minute incubation at 65 °C, the aqueous phase containing DNA was isolated utilizing dichloromethane. DNA was subsequently precipitated using isopropanol and rinsed with 70% ethanol.
Plant material
The genetic diversity of 16 CO (OEX-03, OEX-05, OEX-10, OIJ-001, OIJ-003, OIJ-009, OIN-0001, OIN-0002, OIN-0003, OIN-1082, OIN-1084, OIN-1091, OMU-07, OMU-09, OMU-17, OIM-18) and 14 CC(CIJ-001, CIJ-009, CIJ-055, CIJ-077, CIJ-085, CEX-001, CEX-004, CEX-055, CMU-001, CMU-023, CIM-001, CIN-001, CIN-038, CIN-040) germplasm maintained at ICAR-CRIJAF, Barrackpore, India was evaluated using 15 polymorphic MITE markers.
PCR amplification of MIPs markers
The PCR reaction (20 µL) included 50 ng DNA, 10 µL of 2x PCR master mix, 200 µM dNTPs, 1 µL each of primers, and nuclease-free water. Thermal cycler conditions: 94 °C for 5 min, 35 cycles of 94 °C for 45 s, 52–61 °C for 45 s, and 72 °C for 1 min, with a final extension at 72 °C for 7 min. PCR products were separated on a 2% agarose gel and visualized under UV light.
Construction of dendrogram
The bands of the amplified MITE insertion polymorphic markers were scored as binary data (i.e., ‘1’ for presence and ‘0’ for absence). Genetic similarity was determined using the Nei genetic similarity index26. The genetic similarity was analysed using NTsysPC software v2.1, and a dendrogram was constructed using the UPGMA method to illustrate the genetic relationships among the accessions.
Expression of genes containing MITE elements
To study expressions of genes which containing MITE elements within the exon region, publicly available RNA sequencing reads were retrieved from SRA database of NCBI. Details of the reads used in our study are given in the supplementary Table 1. Expression of the genes were studied in 7 different tissues viz. leaf, mature flowers, fruits, bast, stem, fibre, whole seedling. The RNA-seq data included in this investigation were obtained from the Sequence Read Archive (SRA). The quality control of raw readings was evaluated using FastQC (version 0.11.9). Subsequently, the Trimmatic (version 0.36)27 program was used via command line to eliminate adapters and low-quality readings from the raw data. After trimming raw reads, the quality of the filtered data was re-assessed using FastQC (version 0.11.9), and gene expression was measured by Kallisto28, which employs a pseudo-alignment method to expedite the alignment process. Kallisto program generated TPM (transcripts per million) data of each sample. Average value of replicated samples was log transformed, and heat map was generated using TB tool software.
Results
MITE identification in Corchorus genome
The MITE Tracker tool and Vsearch clustering algorithm identified 632 and 539 MITE families in the genomes of CC and CO, respectively, based on structural features and prevalence. The genome-wide annotation of MITE elements was performed utilizing the BLAST tool, leading to the identification of 51,369 candidate MITE elements in CC and 50,829 in CO. The identified MITE elements showed Target Site Duplication (TSD) lengths of 2 and 10 base pairs, and Terminal Inverted Repeat (TIR) lengths of at least 10 nucleotides.
A total of 15,143 sequences in CC and 13,291 sequences in CO were classified into established conserved MITE families from the identified potential MITE elements, whereas 36,226 novel MITEs in CC and 37,538 in CO which do not have similarity with any existing families, were also identified. The identified MITE families were subsequently categorized into 97 and 84 MITE families within the genomes of CC and CO, respectively. The study indicated that 48 families were common between CC and CO. Table 1 illustrates the comparative distribution of MITE families and superfamilies across the two jute species. Identified notable MITE superfamilies include hAT-like, Tc1/Mariner, Mutator-like, PIF/Harbinger, and CACTA. Interestingly no members were found from CACTA superfamily in CC. The DTM_Thc5 family (Mutator superfamily) contained the largest quantity of MITE sequences (2,220) in CC, whereas the DTA_Ric1 family (hAT superfamily) exhibited the highest number of MITE elements (1,661) in CO. A significant difference was observed in the distribution patterns of MITE families between the two species. For example, DTA_Ric1 family (hAT Superfamily) has 1661 candidates in CO whereas CC has only 136 candidates. The names and locations of all conserved MITE family candidates are provided in supplementary Table 2.
A homology search was performed to compare all identified MITEs in both genomes, identifying 43,018 MITE elements in CC with homologs in CO, and 47,800 MITE elements in CO with homologs in CC. As a result, 8,351 unique MITE elements were identified in the CC genome, while 3,029 unique MITE elements were found in the CO genome (Supplementary Table 3). We also identified 12,047 pairs of orthologous MITE elements using reciprocal best hit method, which are located on the same chromosomes of the two jute species (Supplementary Table 3, Supplementary Fig. 1). The substantial disparity in the number of unique MITE insertions—8,351 in C. capsularis (CC) versus 3,029 in C. olitorius (CO)—indicates differential transposable element (TE) activity and lineage-specific amplification or retention events. This imbalance may reflect underlying differences in genome size, rates of TE turnover, or the efficacy of host genome defense mechanisms. The lower unique MITE abundance in C. olitorius could be attributed to more effective silencing or purging of TEs, whereas the higher accumulation in C. capsularis may suggest relaxed purifying selection or historical bursts of MITE proliferation. These contrasting dynamics underscore the role of TEs in shaping genome architecture and evolutionary trajectories in the two Corchorus species.
Our analysis identified 5170 MITE elements located within 1488 genes in CC whereas 5405 MITES were located within 1658 different genes in CO, respectively. In CC, out of 5170 genic MITE elements, 1016 were in exons, while 4154 were found in intronic regions. In CO, 967 genic MITE elements were identified in exons, while 4438 were found in intronic regions. It was observed that there were instances where multiple MITE elements were inserted within the same gene. The maximum number of MITE insertions identified in a gene was 41, located within gene Cc.06G0032020 in CC. The highest number of MITE insertions within a gene in CO was identified as 30 in gene Co.06G0031700. Insertion of a single MITE element was identified in 540 genes of CO and 441 genes of CC, (Supplementary Table 4). As presence of MITE elements near the gene may influence expression of gene by altering promoter and other cis acting element’s activity, we have identified MITE elements which are located 1Kb upstream/downstream of the gene. 11,022 and 12,729 MITE elements were found to be located near genic region of CC and CO respectively.
Identification of MITE derived MiRNA
MITE sequences identified from both genomes were searched for homology using 48,885 miRNA sequences as a query. Small RNA sequences that exactly matched to the MITE sequences were aggregated as MITE-derived sequences. The analysis indicated that 1246 and 1368 MITE elements could putatively produce miRNAs in CC and CO, respectively (Supplementary Table 5).
MITE derived transcription factor
MITEs found within the genes of both jute genomes were searched against the Plant Transcription Factor database (plantTFDB), which encompasses transcription factors (TFs) from multiple families. A comparative analysis of transcription factor families in CC and CO indicated notable differences in the distribution of MITE insertions (Fig. 1). In CO, highest number of MITE insertions were found in MYB family (11) followed by NAC (10). However, in CC highest MITE insertion was found in bHLH family (9) followed by MIKC_MADS (7). Various transcription factor families, including Whirly, Dof, YABBY, CAMTA, and HD-ZIP, exhibited MITE insertions solely in CO, whereas families such as NF-YB, HB-other, HB-PHD, and C2H2 demonstrated insertions exclusively in CC.
Gene ontology analysis of genes containing mites
The functional annotation of genes containing MITEs may elucidate the involvement of MITE elements in molecular functions, biological processes and biological pathways. Gene Ontology (GO) annotation of the genic MITEs in the genomes of CC (1,488 sequences) and CO (1,658 sequences) was conducted utilizing the PANNZER2 web server. A total of 800 (53.7%) and 895 (53.9%) MITE containing genes from the CC and CO genomes, respectively, were annotated with at least one Gene Ontology (GO) term related to the Cellular Component, Molecular Function, or Biological Process categories (Fig. 2). In the Cellular Component domain, CO and CC display a significant proportion of genes associated with general cellular structures, specifically “cell part” (GO:0044464) and “cell” (GO:0005623), accounting for 57.2% and 56.5% of their respective gene compositions. Significant components include “organelle” (GO:0043226), which accounts for 41.7% in CO and 39% in CC, and “membrane” (GO:0016020), representing approximately 55% in both species. Minor categories such as “supramolecular complex” (GO:0099080) and “membrane-enclosed lumen” (GO:0031974) constitute a smaller proportion of genes.
In the Molecular Function domain, both species exhibit a significant prevalence of genes associated with “catalytic activity” (GO:0003824), with 64.1% in CO and 65.5% in CC. Over 55% of genes in both species are associated with “binding” (GO:0005488). Functional categories like “transporter activity” (GO:0005215) and “transcription regulator activity” (GO:0140110) comprise a smaller percentage of genes (2–8%), whereas “antioxidant activity” (GO:0016209) and “molecular function regulator” (GO:0098772) are even less common.
In the biological process domain, both species demonstrate a significant number of genes linked to extensive biological functions, including “cellular process” (GO:0009987) and “metabolic process” (GO:0008152), with each representing over 50% of the total genes. Processes such as “developmental process” (GO:0032502), “biological regulation” (GO:0065007), and “response to stimulus” (GO:0050896) are also significant. Rare biological processes include “immune system process” (GO:0002376), “reproduction” (GO:0000003), and “detoxification” (GO:0098754). Gene Ontology (GO) enrichment analysis of genes containg MITE elements revealed distinct patterns of functional specialization between Corchorus olitorius and Corchorus capsularis. In olitorius, significant enrichment was observed in secondary metabolite biosynthetic processes, including carotenoid biosynthesis (GO:0016104), flavone biosynthesis (GO:0051553), and oxidosqualene cyclase activity (GO:0031559), highlighting its potential role in specialized metabolism and defense signaling pathways (supplementary Fig. 2.a). Additionally, the response to salicylic acid (GO:0009751) was notably enriched, suggesting an active role in hormonal regulation and stress adaptation. In contrast, capsularis showed strong enrichment in primary metabolic processes such as lipid metabolism (GO:0006629, GO:0044255) and vitamin biosynthesis pathways, including pantothenate (GO:0019742) and C5-branched dibasic acid biosynthesis (GO:0019745) (supplementary Fig. 2.b). Although carotenoid biosynthesis was also enriched in capsularis, the fold enrichment and significance were comparatively lower than in olitorius.
Chromosome distribution of MITE elements in CC and CO
In CC, the distribution of MITE elements is heterogeneous across chromosomes, with chromosome 4 exhibiting the highest count of 8,849 elements, while the scaffold region contains the lowest number at 140 elements. The distribution across other chromosomes is relatively balanced, with values ranging from 6,197 to 7,231 elements. This suggests a significant abundance of MITE transposable elements throughout the genome, potentially influencing genomic plasticity and adaptation.
In CO, the distribution of MITE elements across chromosomes exhibits variability, presenting distinct differences when compared to CC. Chromosome 2 contains the highest quantity of MITE elements, totalling 7,446, whereas chromosome 1 has the lowest count at 5,861. The scaffold region in CO exhibits a significantly higher quantity of MITE elements (3,445) in contrast to CC, which has only 140 MITE elements. This indicates that CO potentially possesses a larger proportion of MITE elements in non-assembled chromosomal regions. This may suggest variations in genome structure or assembly completeness between the two species. Figure 3 presents a comparative distribution of MITEs in CC and CO across seven chromosomes (Chr01–Chr07) and scaffolds.
MITE density across chromosomes
A heatmap illustrated MITE density across chromosomes of jute, where green signified low density and red indicated high density (Fig. 4). In CC, significant variation in MITE density is evident among chromosomes. Cc_Chr01, Cc_Chr03, and Cc_Chr07 display extensive high-density regions, whereas Cc_Chr05 and Cc_Chr06 present a more uniform distribution with lower densities. This demonstrates notable clustering of MITEs in specific regions, particularly in Cc_Chr01 and Cc_Chr03. A comparable variation is noted in CO. Co_Chr03 and Co_Chr06 exhibit extensive high-density regions, while Co_Chr02 and Co_Chr05 demonstrate more dispersed patterns with reduced density. The distribution patterns of MITEs in CO exhibit similarities with those in CC; however, the specific density patterns and locations differ between the two species. This comparison elucidates the genomic distribution of MITEs in the two Corchorus species, indicating species-specific variations in the accumulation and dispersion of transposable elements, potentially impacting genome evolution and function.
MIPs marker analysis
Transposition of MITE elements within genome is random and it can be utilised a valuable resource to develop molecular markers. An in-house database of MITE insertion polymorphic markers was developed by designing flanking primers of each MITE elements. In supplementary Table 6 genome-wide distributed MIP markers sequence information is available which can be readily accessed and utilised in jute molecular breeding. Thirty-eight MITE inserting polymorphic markers were genotyped in a panel of 30 jute accessions including both CO and CC. Out of 38 markers 15 markers were found to be polymorphic and data derived from these polymorphic MITE markers was used for further statistical analysis. A total of 50 alleles were found, with the number of alleles per locus ranging from 2 to 3, yielding an average of 2.66 alleles per locus. The PIC varied from 0.14 to 0.35 per locus, with an average of 0.28. The computed PIC was greatest for many primers, including MITE_7 (0.35) and MITE_5 (0.338), MITE_9 (0.336) and lowest for the primer MITE_6 (0.143) and MITE_15 (0.23) (Table 2). The observed heterozygosity for each primer varied between 0.28 and 0.70. The number of alleles detected per marker and the genetic diversity of markers are contingent upon the number of genotypes analysed, which were relatively less in the current research.
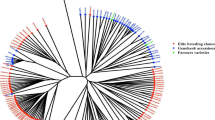
UPGMA dendrogram using MIPs marker
The UPGMA dendrogram was generated utilizing molecular markers derived from MITE elements (Fig. 5). Fifteen markers were utilized to categorize 30 genotypes, including both CO and CC, into five separate clusters with a Nei genetic distance threshold of 0.6. Cluster II was identified as the largest cluster, containing 12 genotypes, while Cluster III included 10 genotypes. Cluster IV comprised one genotype (CIN-040). Cluster I comprised five genotypes, including four CO (OIN-01, OIN-02, OIN-03, OEX-03) and one CC(CIJ-009). Cluster II comprised eight genotypes of CO and four of CC, with a sub-cluster exclusively containing five CO genotypes (OIJ-001, OEX-10, OIJ-003, OEX-05, OIM-18). Clusters IV and V consisted solely of CC genotypes.
Expression profiles of genes containing mites
We analysed gene expression profiles across seven distinct tissue types (Bast, Mature Flowers, Leaf, Fruits, Stem, Fiber Cell, and Whole Seedlings) utilizing TPM values to evaluate expression of genes containing MITE insertions within coding region. (Supplementary Fig. 2 A and 2B and supplementary Table 7). The objective of this study was to see influence of MITE elements insertion in exon on expression pattern of the gene.
In CO, several genes were identified that display tissue-specific expression patterns with elevated TPM values (> 100) in particular tissues. Genes GWHTBCLB005438 and GWHTBCLB005637 exhibited high expression levels in leaf and fruit tissues, respectively, suggesting specialized functions in these organs. Furthermore, GWHTBCLB008763 and GWHTBCLB004374 exhibited elevated expression levels in Fiber Cell and Stem tissues, respectively, suggesting their role in structural functions. Conversely, we identified seven genes (GWHTBCLB001706, GWHTBCLB016055, GWHTBCLB016158, GWHTBCLB021677, GWHTBCLB023542, GWHTBCLB023875, and GWHTBCLB024778) exhibiting consistently high expression (TPM > 100) across all tissues, suggesting essential roles in fundamental cellular processes, including metabolism, gene regulation, and structural maintenance. The dataset identified 84 genes exhibiting no expression across all tissues (TPM < 1) and 278 genes expressed in at least one tissue (TPM > 1). We identified 178 genes consistently expressed across all seven tissues (TPM > 1), offering valuable insights for future genetic and functional studies focused on tissue-specific or universal plant genes.
In Capsularis, 180 genes were consistently expressed across all tissues (TPM > 1). Four genes (GWHTBCLC000830, GWHTBCLC013106, GWHTBCLC013711, and GWHTBCLC017610) demonstrated elevated expression levels (TPM > 100) in all tissues, indicating their essential roles in basic cellular functions. Furthermore, 46 genes exhibited elevated expression levels (TPM > 100) tissues, underscoring their potential specialization and functional significance in those tissues. A total of 313 genes were identified as expressed in at least one tissue (TPM > 1), while 45 genes were found to be not expressed in any tissue (TPM < 1).
Discussion
MITEs constitute 4.58% in CC and 3.81% in CO of the total jute genome, compared to 3.3% in Arabidopsis thaliana29, 5.24% in rice13 and 0.32% in maize31. We identified 68.54% of MITEs in CC and 64.39% in CO within intergenic regions, while 10.06% in CC and 10.66% in CO were in genic regions. The remaining 21.45% in CC and 25.04% in CO were near genic regions. This distribution is consistent with other species like Arabidopsis thaliana32, Oryza sativa33, and Brassica34. Among genic MITEs, 19.78% in CC and 30.88% in CO were located in exon regions, potentially altering gene expression or function.
MITEs are often located next to or within genes and have a role in gene regulation35. For instance, an insertion in the wheat Vrn-A1 gene promoter removes vernalization needs for flowering36. Similarly, a small tourist family MITE in the 3′-UTR of TaHSP16.9–3 A in wheat regulates heat shock protein gene expression37. In this study, we identified genome-wide distributed MITE elements in genic as well as near genic region of two jute species, which may provide a valuable resource for identifying functions of genes through further studies. Our study also identified exonic as well as intronic MITE insertions which can have profound functional consequences, such as disrupting coding sequences, introducing premature stop codons, or causing frameshift mutations, potentially leading to loss-of-function alleles or altered protein products. In contrast, insertions in intronic regions may influence splicing efficiency, alternative exon usage, or transcript stability by affecting cis-regulatory elements or splice sites. Additionally, both exonic and intronic insertions can impact gene expression through epigenetic modifications (e.g., DNA methylation) or by serving as sources of small RNAs that mediate gene silencing. Thus, the positional context of MITE insertions is critical for understanding their functional and regulatory implications.
Upon the detection of potential MITEs, they were categorized into various families and superfamilies. In CC, the Tc1/Mariner and Mutator superfamilies were predominant, while in CO, the Hat-like and Tc1/Mariner superfamilies were more common. Previous studies have similarly identified Tc1-like elements in several angiosperms, including Oryza sativa, Brassica rapa, Cannabis sativa, and Triticum urartu38. Unclassified MITEs made up 70.5% and 73.8% in CC and CO, respectively, suggesting distinct functionality and distribution than previously classified MITE elements. Guo et al.31 found similar observation in Arabidopsis thaliana where 80.7% of total MITE elements identified were unclassified.
The distribution of MITE elements is comparable in terms of chromosomal occurrence across both species. CO exhibited a higher prevalence of these elements in scaffolds and specific chromosomes, such as Chr02 and Chr03. Whereas CC has higher prevalence of MITE elements on Chr. 04. This suggests species-specific differences in transposable element activity and genomic organization. Such variation could reflect evolutionary divergence in the regulatory roles and genomic impact of MITE elements between the two species, with potential implications for their adaptability and genome stability.
Our study identified 11,801 and 10,921 MITE elements producing miRNAs in CC and CO, respectively, which might have a role in the genetic and epigenetic regulation of gene expression. Most miRNA-generating MITEs were detected in intergenic regions (69.12% in CC and 77.25% in CO), while 30.88% and 22.75% were in genic and near-genic regions. Similar findings were observed in rice where 51.8% of small RNA were generated from intergenic MITE elements39. A comparison of experimentally determined microRNAs with repeat databases revealed that 6.5% of Arabidopsis and 35% of rice microRNAs co-localized with TEs, the majority of which were MITEs40,41. The present investigation also showed MITE elements of both the jute species is a rich reservoir of miRNA biogenesis which through their rapid rate of rearrangement or decay represent a constantly evolving source of new microRNA genes.
Insertion of MITE elements within genic region may influence gene expression through transcriptional, post-transcriptional or translational regulation. Previous reports show that MITE insertions in transcription factor genes can negatively regulate their expression. For instance, MITE insertion in the WRKY45-1 gene intron produces siRNA (TE-siR815), which negatively regulates ST1 expression42. Similarly, a MITE in the ZmNAC111 promoter correlates with maize drought tolerance variation by inhibiting this gene’s expression through RdDM and H3K9 dimethylation43. Our study identified 22 and 26 different transcription factor family in CC and CO respectively where MITE elements were inserted within genic region. Genome-wide analysis showed a high number of MITE insertions in the bHLH family, crucial for growth and development, suggesting evolutionary pressures influenced by MITE activity. In CO, the NAC family had the highest MITE insertion frequency, indicating its role in species-specific adaptations. The MYB family also showed more insertions in CO than in CC, suggesting increased regulatory flexibility in CO related to stress responses and development. Some families, such as NF-YB and HB-PHD, had insertions only in CC, while others, like Whirly, Dof, YABBY, CAMTA, and HD-ZIP, had insertions only in CO, indicating evolutionary divergence and functional specialization between the two species. The bHLH family, crucial for numerous growth and developmental processes, displayed a significant number of MITE insertions in both species, indicating that this family may have experienced conserved evolutionary pressures shaped by MITE activity.
MITEs’ unique attributes, including prevalence, short sequence length, non-autonomous nature, high insertion polymorphism, and preferential insertion into genic regions, make them suitable as molecular markers. MITE-derived markers have been used in crops like citrus44, brassica33, rice45,46,47, and maize48. Three major MITE-based marker systems have been created and are employed in diverse molecular plant breeding studies include MITE- Inter-MITE polymorphism (IMP), Display (MD), and MITE insertional polymorphism (MIP). MD and IMP markers are dominant and multi-locus, limiting their use in plant breeding. MIP markers, being single-locus and co-dominant, are ideal for genetic studies like diversity analysis and QTL mapping35. Despite their advantages, no MIP markers in jute have been reported until now. In this study, we identified genome-wide MITE elements in two jute species and converted them into MIP markers. We amplified 38 MIP markers in 30 jute accessions, with 15 polymorphic markers, demonstrating their utility in genetic diversity analysis. MIP markers have also been used in rice48, tea49, and maize50. A significant number of MITE elements were located within or near genes, making MIP markers valuable for gene mapping and tagging through techniques like MAP-based cloning or insertional mutagenesis.
Conclusion
The comprehensive genomic analysis of Miniature Inverted-Repeat Transposable Elements (MITEs) in jute highlights their significant roles in gene regulation, genome architecture, and breeding applications. By identifying and characterizing MITE families, miRNA precursors, and insertional polymorphism markers, this study provides invaluable resources for advancing jute breeding and genetic research. The developed MITE-based molecular markers demonstrate strong potential in genetic diversity analysis and molecular breeding. These findings not only deepen our understanding of MITE dynamics in plant genomes but also lay a robust foundation for future innovations in jute crop improvement, emphasizing sustainable agriculture and industrial applications. This work sets a precedent for leveraging genomic tools to address challenges in fibre crop development. However, there is significant scope for further research, including the application of MIP markers for identifying genes associated with agronomically important traits and the functional validation of MIP-derived microRNAs to confirm their roles in gene regulation.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Majumder, S., Saha, P., Datta, K. & Datta, S. K. Fiber crop, jute improvement by using genomics and genetic engineering. Adv. Crop Improv. Tech. 363–383. https://doi.org/10.1016/B978-0-12-818581-0.00022-X (2020).
Kundu, B. C. Origin of jute. Indian J. Genet. Plant. Breed. 11, 95–99 (1951).
Palit, P. & Meshram, J. H. Physiology of jute yield and quality. In Jute and Allied Fibre Updates (eds Karmakar, P. G. et al.) 112–124 (Central Research Institute for Jute and Allied Fibres, 2008).
Tao, A. et al. Construction of a full-length cDNA library and analysis of expressed sequence tags in white jute (Corchorus capsularis L). Afr. J. Biotechnol. 14, 1928–1935. https://doi.org/10.5897/AJB2015.14619 (2015).
Sarkar, D. et al. The draft genome of Corchorus olitorius cv. JRO-524 (Navin). Genomics Data. 12, 151–154. https://doi.org/10.1016/j.gdata.2017.05.007 (2017).
Islam, M. S. et al. Comparative genomics of two jute species and insight into fibre biogenesis. Nat. Plants. 3, 1–7. https://doi.org/10.1038/nplants.2016.223 (2017).
Zhang, L. et al. Reference genomes of the two cultivated jute species. Plant. Biotechnol. J. 19, 2235–2248. https://doi.org/10.1111/pbi.13652 (2021).
Hirsch, C. D. & Springer, N. M. Transposable element influences on gene expression in plants. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 157–165. https://doi.org/10.1016/j.bbagrm.2016.05.010 (2017).
Judd, J. & Feschotte, C. Transposons take remote control. eLife 7, e40921. https://doi.org/10.7554/eLife.40921 (2018).
Roessler, K., Bousios, A., Meca, E. & Gaut, B. S. Modeling interactions between transposable elements and the plant epigenetic response: a surprising reliance on element retention. Genome Biol. Evol. 10, 803–815. https://doi.org/10.1093/gbe/evy043 (2018).
Hosaka, A. & Kakutani, T. Transposable elements, genome evolution and transgenerational epigenetic variation. Curr. Opin. Genet. Dev. 49, 43–48. https://doi.org/10.1016/j.gde.2018.02.012 (2018).
Jiang, N. et al. An active DNA transposon family in rice. Nature 421, 163–167. https://doi.org/10.1038/nature01214 (2003).
Jiang, N., Feschotte, C., Zhang, X. & Wessler, S. R. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). Curr. Opin. Plant. Biol. 7, 115–119. https://doi.org/10.1016/j.pbi.2004.01.004 (2004).
Feschotte, C., Swamy, L. & Wessler, S. R. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163, 747–758 (2003).
Juretic, N., Bureau, T. E. & Bruskiewich, R. M. Transposable element annotation of the rice genome. Bioinformatics 20, 155–160. https://doi.org/10.1093/bioinformatics/bth019 (2004).
Feng, Q. et al. Sequence and analysis of rice chromosome 4. Nature 420, 316–320. https://doi.org/10.1038/nature01183 (2002).
Santiago, N., Herráiz, C., Goñi, J. R., Messeguer, X. & Casacuberta, J. M. Genome-wide analysis of the emigrant family of mites of Arabidopsis Thaliana. Mol. Biol. Evol. 19, 2285–2293. https://doi.org/10.1093/oxfordjournals.molbev.a004052 (2002).
Wright, S. I., Agrawal, N. & Bureau, T. E. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis Thaliana. Genome Res. 897–1903. https://doi.org/10.1101/gr.1281503 (2003).
Portnoy, V., Huang, V., Place, R. F. & Li, L. C. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev. RNA. 2, 748–760. https://doi.org/10.1002/wrna.90 (2011).
Crescente, J. M., Zavallo, D., Helguera, M. & Vanzetti, L. S. MITE tracker: an accurate approach to identify miniature inverted-repeat transposable elements in large genomes. BMC Bioinform. 19, 348. https://doi.org/10.1186/s12859-018-2376-y (2018).
Chen, J., Hu, Q., Zhang, Y., Lu, C. & Kuang, H. P-MITE: a database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 42, D1176–D1181. https://doi.org/10.1093/nar/gkt1000 (2014).
Zavallo, D. et al. Genomic re-assessment of the transposable element landscape of the potato genome. Plant. Cell. Rep. 39, 1161–1174. https://doi.org/10.1007/s00299-020-02554-8 (2020).
Kozomara, A. & Griffiths-Jones, S. MiRBase: annotating high confidence MicroRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. https://doi.org/10.1093/nar/gkt1181 (2014).
Törönen, P. & Holm, L. PANNZER—a practical tool for protein function prediction. Protein Sci. 31, 118–128. https://doi.org/10.1002/pro.4193 (2022).
Doyle, J. J. (ed Doyle, J. L.) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987 11–15 (1987).
Nei, M. Genetic distance between populations. Am. Nat. 106, 283–292. https://doi.org/10.1086/282771 (1972).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. https://doi.org/10.1038/nbt.3519 (2016).
Hu, T. T. et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481. https://doi.org/10.1038/ng.807 (2011).
Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis Thaliana. Nature 408, 796–815. https://doi.org/10.1038/35048692 (2000).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. https://doi.org/10.1126/science.1178534 (2009).
Guo, C. et al. Genome-wide comparative analysis of miniature inverted-repeat transposable elements in 19 Arabidopsis Thaliana ecotype accessions. Sci. Rep. 7, 2634. https://doi.org/10.1038/s41598-017-02855-1 (2017).
Oki, N. et al. A genome-wide view of miniature inverted-repeat transposable elements (MITEs) in rice, Oryza sativa ssp. Japonica. Genes Genet. Syst. 83, 321–329 (2008).
Sampath, P. & Yang, T. J. Miniature inverted-repeat transposable elements (MITEs) as valuable genomic resources for the evolution and breeding of brassica crops. Plant. Breed. Biotech. 2, 322–333. https://doi.org/10.9787/PBB.2014.2.4.322 (2014).
Venkatesh, N. B. Miniature inverted-repeat transposable elements (MITEs), derived insertional polymorphism as a tool of marker systems for molecular plant breeding. Mol. Biol. Rep. 47, 3155–3167. https://doi.org/10.1007/s11033-020-05365-y (2020).
Yan, L. et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. https://doi.org/10.1126/science.1094305 (2004).
Li, J., Wang, Z., Peng, H. & Liu, Z. A MITE insertion into the 3′-UTR regulates the transcription of TaHSP16.9 in common wheat. Crop J. 2, 381–387. https://doi.org/10.1016/j.cj.2014.07.001 (2014).
Liu, Y. & Yang, G. Tc 1-like transposable elements in plant genomes. Mob. DNA. 5, 17. https://doi.org/10.1186/1759-8753-5-17 (2014).
Lu, C. et al. Miniature inverted–repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol. Biol. Evol. 29, 1005–1017. https://doi.org/10.1093/molbev/msr282 (2012).
Lisch, D. & Bennetzen, J. L. Transposable element origins of epigenetic gene regulation. Curr. Opin. Plant. Biol. 14, 156–161. https://doi.org/10.1016/j.pbi.2011.01.003 (2011).
Piriyapongsa, J. & Jordan, I. K. Dual coding of SiRNAs and MiRNAs by plant transposable elements. RNA 14, 814–821. https://doi.org/10.1261/rna.916708 (2008).
Zhang, H. et al. Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants. 2, 1–8. https://doi.org/10.1038/nplants.2016.16 (2016).
Mao, H. et al. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6, 8326. https://doi.org/10.1038/ncomms9326 (2015).
Liu, Y. et al. Comparative analysis of miniature inverted–repeat transposable elements (MITEs) and long terminal repeat (LTR) retrotransposons in six Citrus species. BMC Plant. Biol. 19, 101. https://doi.org/10.1186/s12870-019-1757-7 (2019).
Jiang, N. & Wessler, S. R. Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant. Cell. 13, 2553–2564. https://doi.org/10.1105/tpc.010235 (2001).
Monden, Y. et al. High potential of a transposon mPing as a marker system in Japonica× Japonica cross in rice. DNA Res. 16, 131–140. https://doi.org/10.1093/dnares/dsp004 (2009).
Komori, T. & Nitta, N. High frequency of sequence polymorphism in rice mites and application to efficient development of PCR-based markers. Breed. Sci. 53, 85–92 (2003).
Casa, A. M. et al. The MITE family Heartbreaker (Hbr): molecular markers in maize. Proc. Natl. Acad. Sci. USA 97, 10083–10089, (2000). https://doi.org/10.1073/pnas.97.18.10083
Rohilla, M. et al. Genome-wide identification and development of miniature inverted-repeat transposable elements and intron length polymorphic markers in tea plant (Camellia sinensis). Sci. Rep. 12, 16233. https://doi.org/10.1038/s41598-022-20400-7 (2022).
Zerjal, T. et al. Maize genetic diversity and association mapping using transposable element insertion polymorphisms. Theor. Appl. Genet. 124, 1521–1537. https://doi.org/10.1007/s00122-012-1807-9 (2012).
Acknowledgements
Authors are thankful to ICAR-CRIJAF for funding inhouse project titled “Development of molecular marker panel for DNA fingerprinting of jute”.
Author information
Authors and Affiliations
Contributions
RB: Writing– review & editing, Writing– original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. KNC: Writing– review & editing, Formal analysis, Data curation. JKM: Writing– review & editing, Supervision. KM: Writing– original draft, Data analysis. JM: Supervision, Writing– review & editing. GK: Supervision, Writing– review & editing. TR: Conceptualization, Methodology, Writing– review & editing, Writing– original draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Authors are thankful for ICAR-CRIJAF for funding inhouse project titled “Development of molecular marker panel for DNA fingerprinting of jute”.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bhowmick, R., Chourasia, K.N., Meena, J.K. et al. Exploring the genomic landscape of mites and harnessing MITE insertion polymorphism markers in jute (Corchorus sp.). Sci Rep 15, 35419 (2025). https://doi.org/10.1038/s41598-025-18966-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18966-z