Abstract
Transformer-based large language models (LLMs), such as ChatGPT-4, are increasingly used to streamline clinical practice, of which radiology reporting is a prominent aspect. However, their performance in interpreting complex anatomical regions from MRI data remains largely unexplored. This study investigates the capability of ChatGPT-4 to produce clinically reliable reports based on orbital MR images, applying a multimetric, quantitative evaluation framework in 25 patients with orbital lesions. Due to inherent limitations of current version of GPT-4, the model was not fed with MR volumetric data, but key 2D images only. For each case, ChatGPT-4 generated a free-text report, which was then compared to the corresponding ground-truth report authored by a board-certified radiologist. Evaluation included established NLP metrics (BLEU-4, ROUGE-L, BERTScore), clinical content recognition scores (RadGraph F1, CheXbert), and expert human judgment. Among the automated metrics, BERTScore demonstrated the highest language similarity, while RadGraph F1 best captured clinical entity recognition. Clinician assessment revealed moderate agreement with the LLM outputs, with performance decreasing in complex or infiltrative cases. The study highlights both the promise and current limitations of LLMs in radiology, particularly regarding their inability to process volumetric data and maintain spatial consistency. These findings suggest that while LLMs may assist in structured reporting, effective integration into diagnostic imaging workflows will require coupling with advanced vision models capable of full 3D interpretation.
Similar content being viewed by others
Introduction
Artificial intelligence is revolutionizing cognitive approaches to medical problems, including interpretation of clinical data, diagnostic images, and instrumental exams. In particular, large language models (LLMs) include a broad spectrum of architectures such as GPT-4, PaLM2, and LLaMA, and have become part of a daily application in clinical routine1.
Transformer-based models, an example of which is ChatGPT (OpenAI, San Francisco, CA, USA), are trained on billions of tokens across diverse textual corpora, exhibit unprecedented attentional capacities and are highly scalable2. As such, in the biomedical field, they have demonstrated increasingly promising abilities in question answering, clinical reasoning (for instance, in USMLE tests), and even simulation of real scenarios3. In light of these premises, LLMs are now being explored in radiology as potential tools for automating or augmenting the generation of radiology reports. The integration of LLMs language capabilities with architectures designed for 3D volumetric segmentation and image interpretation, including V-Net or Swin UNETR, represents an evolutionary perspective for radiology, and has been made widely available with ChatGPT. Moreover, from a textual point of view, radiology represents a challenging domain for LLM implementation. Traditionally, a radiological report follows a well-structured narrative format that captures the salient aspects of an image. Likewise, it is expected that this structure aligns well with the textual generative strengths of LLMs4,5.
Radiology reporting requires the extraction of organized features, including anatomical localization, definition of signal characteristics (in MR sequences) or Hounsfield Units (HU) density window (in CT scans), identification of pathological patterns, and, critically, avoidance of hallucinated content. Detrimental errors include laterality, mischaracterization of tissue signal, alteration of anatomical relationships, all factors which may have profound downstream consequences in diagnosis, treatment, and outcomes6.
Prior work has evaluated such models on traditional exams such as chest X-ray interpretation7 or orthopantograms8yet it is definitely more difficult to interpret volumetric data such as CT or MR. As the LLM is merely responsible for the generation of the textual report, which represents the ultimate endpoint assessed in this paper, the quality of AI radiological reports also depends on the effectiveness of architectures for image segmentation tasks, as well as vision transformers with attention mechanisms that encode the lesion features into structured tokens, which are then processed by the LLM9. In this respect, the authors decided to implement MR analysis in a highly selected anatomical region – the orbit. The orbit has a highly constant structure with clear boundaries between structures, including the eye globe, optic nerve, extraocular muscles, which are emphasized by the T1-hyperintense fat, thus representing an acceptable compromise between a reproducible segmentation and the heterogeneity of lesions. In fact, the orbital compartment might harbor a variety of lesions, benign, malignant, inflammatory, or syndromic, whose interpretation often requires the interpolation of tiny signal differences across T1, T2, fat-suppressed, and contrast-enhanced sequences. For this reason, orbital MRI interpretation provides a valuable benchmark to test the actual competence of transformed-based models10.
The objective of this study is to evaluate the capability of a large language model (ChatGPT 4o) to generate a clinically reliable radiology report based on standardized orbital MRI images. The focus is on free-text report generation to simulate a real-world reporting activity. Subsequently, each LLM-generated report is compared to the standard-of-care report composed by board-certified radiologist expert in orbital pathology. Validated computational metrics are employed to evaluate textual correspondence between LLM-generated reports and radiologist-generated reports, as well as the validity of extracted features. In this study, the authors aim to define both the capabilities and critical limitations of transformer-based models in clinical practice, defining a working frame applicable not only to orbital pathology, but generalizable to other anatomical regions.
Materials and methods
Study design
The aim of this pilot study is to evaluate the fidelity and clinical relevance of radiology reports generated by a LLM. Study design consists of a structured comparison between human-authored reports and reports generated by ChatGPT 4o using metrics validated for LLMs able to assess both textual correspondence, semantics and content. While LLM accounts only for the late step of generating the written report, the accuracy of report indirectly evaluates a stepwise computer-vision flow starting from visual pattern extraction using convolutional neural networks (CNN), specifically 2D U-Net architecture, which is particularly suitable for biomedical image segmentation due to its symmetric encoder-decoder structure with skip connections that preserve fine spatial details during downsampling and upsampling. Following segmentation, vision transformers (ViT) are used to encode into structured tokens features of each lesion, such as size, location, contrast enhancement, anatomical boundaries11. The LLM was tuned on radiology reports to generate a clinically coherent narrative incorporating domain-specific terms and lexicon. The sum of these steps relates to the accuracy of the AI-generated report, which thus measures not just the LLM performance, but also previous computer vision elaboration steps.
This evaluation was performed by screening clinical databases of two hub departments of maxillofacial surgery of Northern Italy, respectively the Academic Hospital of the University of Udine and San Paolo Hospital of the Milan University. Data eligible for this study included MR scans of orbital lesions with appropriate acquisition quality, absence of artifacts, T1 or T2, both contrast-enhanced or basal, with a radiology report written according to standardized guidelines12.
All procedures were performed in accordance with the relevant institutional guidelines and regulations, and the study adhered to the principles outlined in the Declaration of Helsinki. Ethical approval was granted by the Institutional Review Board of the University of Udine (approval number: IRB_45_2020). Written informed consent was obtained from all participants or their legal guardians. All radiological data were anonymized prior to analysis.
Integration of visual data in the transformer-based LLM
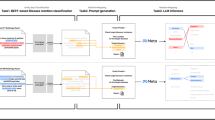
Although MR data was originally acquired and processed in volumetric NIfTI format to preserve spatial integrity and quantitative voxel information, the subsequent integration into the LLM was performed using selected two-dimensional (2D) key screen captures.The impossibility to implement LLM to interpret volumetric radiological data was driven by both technical limitations inherent to current LLM architectures and pragmatic considerations regarding standardization of interpretation. Using screenshots was deemed a more standardizable approach across cases, reducing variability introduced by different slice thickness, acquisition parameters, or specific artifacts that may affect volumetric data. Moreover, computer vision models integrated in GPT-4o or Gemini, while multimodal and capable of processing visual inputs, are currently programmed for 2D image understanding and lack the possibility to directly handle volumetric data in NIfTI or DICOM formats. In fact, a three-dimensional processing ability would require voxel-wise processing, sequential transformations and complex spatial reasoning across multiple planes, which nowadays would require a custom pipeline and the integration with external tools such as MONAI Label (Linux Foundation AI), 3D Slicer (Brigham and Women’s Hospital, USA) or nnU-Net. Figure 1 schematically illustrates the proposed analytical workflow.
Workflow for orbital MR images processing using a transformer-based model with image patching and tokenization and the resulting LLM radiology report. Quantitative evaluation to detect textual reliability with a human radiologist is performed using linguistic similarity indexes for LLM, while clinical completeness is quantified using RadGraph of CheXbert similarity index. Scores are visually represented in a confusion matrix.
Image preprocessing pipeline
Orbit MR datasets were exported from the institutional PACS archive as anonymized NIfTI files. For each patient, the region of interest (ROI) was centered on the lesion, and three orthogonal 2D slices (axial, coronal, sagittal) were extracted. In case of isotropic 3D acquisitions (voxel size ≤ 1 mm³, typically volumetric VIBE sequences), multiplanar reconstructions were generated from the same dataset; otherwise, dedicated planar coronal and sagittal sequences were used. Using 3DSlicer (Brigham and Women’s Hospital, Boston, Massachusetts, USA) all images were resampled to a uniform voxel size of 1 × 1 × 1 mm using trilinear interpolation, then cropped around the ROI and padded with black borders to a standard size of 256 × 256 pixels. To reduce differences between different MR scanners and sequences, intensity values were normalized (mean 0, standard deviation 1), and the final outputs were stored in PNG format for downstream analysis using transformer-based LLMs.
Case selection
Only one MR sequence was selected for each case between T1-weighted or T2-weighted, depending on the individual features of the lesions. In general, masses with static fluids, like those with a fluid content, were studied in T2 sequences. Vascular sequences, including time-of-flight (TOF) and 3D phase-contrast (PC) imaging, were excluded due to their selective enhancement of vascular structures, which limits the visualization of surrounding anatomical landmarks. Similarly, diffusion-weighted imaging (DWI) was considered unsuitable given its inherently low spatial resolution and poor anatomical delineation.
Inclusion criteria were the following: presence of a soft tissue orbital lesion, at least 256 × 256 voxel matrix, visualization of at least two of the following anatomical structures: eye globe, optic nerve, inferior rectus, superior rectus, medial rectus, lateral rectus, superior obliquus, possibility to identify the lesion boundaries. Exclusion criteria were: low quality images, image with a artifacts, lesions whose localization prevented the visualization of at least two of the aforementioned structures. The study did not include MR of patients for which a report from a board certified radiologist was not available. Radiology reports were translated to English using DeepL (DeepL SE, Köln, Germany) and checked by a native English speaker to decrease potential biases.
LLM prompt structure
Textual prompts for ChatGPT-4 followed a standardized input structure that than be resumed in the following points: (1) instructions directing the LLM to perform as a board-certified radiologist and avoid statements unsupported by image analysis; (2) always specifying to consider the radiological convention of flipped side to correctly locate the side of disease; (3) providing case-specific data, including the definition of standardized 2D MRI views (axial, coronal, sagittal), sequence type, presence of contrast enhancement, and a brief clinical context; and (4) output constraints, requiring a structured report with sections on anatomical relationships, lesion characteristics in terms of radiological features, such as contrast enhancement, regional hypo- or hyperintensity, and conclusive diagnostic impression. The same structure was used across all prompts for each case.
Computer vision pipeline
For the purposes of this study, segmentation and feature extraction tasks were entirely processed end-to-end by GPT-4o’s native multimodal architecture rather than through custom-built pipelines based on external libraries. For each case, the model received three lesion-centered, standardized MRI screenshots (axial, coronal, sagittal; 256 × 256 pixels) from T1- or T2-weighted sequences. Using its integrated visual encoder, GPT-4o performed direct parsing of the orbital anatomy, automatically segmenting key structures (globe, optic nerve, extraocular muscles, periorbital fat) and the lesion itself. According to the textual prompt, the model estimated lesion size, boundaries, signal intensity patterns, and anatomical relationships with nearby structures. Such visual features were automatically tokenized by GPT-4o, to be then handled by the transformer-based language module to compose a structured, textual, radiology report, including sections on anatomy, lesion description, and diagnostic impression. This fully end-to-end workflow, relying exclusively on OpenAI’s built-in computer vision and language capabilities without external pipelines, removed the need for separate CNNs, vision transformers, or external feature encoders, ensuring a uniform and reproducible process across all cases.
LLM performance evaluation
LLM textual reliability
For LLM analysis, ChatGPT 4o was used to compare the performance of the LLM-generated report with a board-certified radiologist expert in orbital imaging. The evaluation was carried out across two levels: linguistic similarity between LLM and human-generated reports and medical report recognition.
Linguistic similarity between LLM-generated and expert reports was first quantified using three validated natural language processing (NLP) metrics, which are briefly summarized as follows:
-
The BLEU-4 (Bilingual Evaluation Understudy) score measured n-gram overlap (up to four words in sequence) between the computer-generated and human-reference texts. While BLEU is sensitive to exact word choice and order—thus capturing lexical precision—it is known to penalize clinically acceptable paraphrases and rephrasings13,14.
$$\:BLEU=BP\cdot\:exp\left(\sum\:_{n=1}^{N}{w}_{n}log{\:p}_{n}\right)$$-
\(\:{p}_{n}\): n-grams precision.
-
\(\:BP\): brevity penalty (penalizes texts that are too short compared with the ground truth).
-
\(\:{w}_{n}\): weight for every n-gram (generally uniform).
-
-
To address such limitations, we integrated the ROUGE-L (Recall-Oriented Understudy for Gisting Evaluation, Longest common subsequence) score, which evaluates similarity based on the longest shared subsequence (LCS) of words. Unlike BLEU, ROUGE-L accounts for structural coherence even in the presence of word reordering and rephrasing, making it particularly useful for assessing sentence-level alignment between clinical descriptions15.
$$\:ROUGE-L=\:\frac{(1+{\beta\:}^{2})\cdot\:R\cdot\:P}{R+{\beta\:}^{2}\cdot\:P}$$-
R = \(\:\frac{LCS}{reference\:text\:length}\).
-
P = \(\:\frac{LCS}{generated\:text\:length}\).
-
\(\:\beta\:\) is usually set to 1 (equal importance recall and precision).
-
-
Acknowledging the limitations of purely lexical metrics, textual evaluation was enhanced using the BERT (Bidirectional Encoder Representations from Transformers) Score, which computes is able to compare tokens semantically. From a computational perspective, in BERTScore, the similarity between two sentences is evaluated as the sum of the cosine similarities between their tokens, enabling to detect paraphrases. This metric allows for a finer understanding of the semantical approximation of two texts. In radiology for instance, similar terms are possible, such as “vascular mass” and “hemangioma” and in this respect BERTScore provides a more significant assessment of interpretive equivalence16.
$$\:BERTScoreF1=\frac{2\cdot\:P\cdot\:R}{P+R}$$-
P: semantic precision (the extent the AI report includes the ground truth report).
-
R: semantic recall (the extent the ground truth report is represented in the AI text).
-
LLM conceptual reliability
Radiological report structure and recognition of medical concepts was based on different evaluation metrics, which included:
-
RadGraph F1, initially developed for natural language processing (NLP) in radiology, assesses the correct identification of clinical entities by the radiology report, such as lesions and anatomical targets, as well as the relationships between them. Therefore, this metric evaluates the saliency of medical content rather than textual form17.
$$\:F1=2\cdot\:\frac{Precision\cdot\:Recall}{Precision+Recall}$$-
$$\:Precision=\frac{True\:Positives}{True\:Positives+False\:Positives}$$
-
$$\:Recall=\frac{True\:Positives}{True\:Positives+False\:Negatives}$$
-
-
Another proxy metric, the CheXbert label similarity, namely an adaptation from a transformer-based labeler originally trained on chest X-rays, was implemented to provide an estimation of the content alignment between reports based on keywords which recur throughout reports18. Despite being developed for chest X-ray, CheXbert score was repurposed in this study as a proxy metric to assess content alignment between LLM-generated and ground-truth radiology reports. No additional model retraining was performed, relying on its capability to identify recurring clinical entities, such as anatomical structures and lesion types, through keyword-based matching and contextual embeddings. Basically, yet not optimized for orbital imaging, it was used as a coarse measure of conceptual consistency between reports. For this reason, CheXbert was strictly considered as a quantitative proxy metric in addition to RadGraph F1 for evaluating content completeness, rather than serving as a diagnostic classifier for orbital lesions.
$$\:CheXbert\:Similarity=\:\frac{1}{8}\sum\:_{i=1}^{8}1({l}_{i}^{gen}={l}_{i}^{ref})$$-
\(\:{l}_{i}^{gen}\): predicted label from LLM report for the i-condition.
-
\(\:{l}_{i}^{ref}\): extracted label from ground-truth report
-
\(\:1\left(\cdot\:\right)\): is the indicator function (1 if true, 0 if false)
-
Clinician evaluation
In addition to automated evaluations, three additional parameters reviewed by expert clinicians were developed:
-
Clinical completeness: evaluating whether the generated report included all essential anatomic, pathologic, and topographic elements necessary to formulate a correct report.
$$\:Clinical\:Completeness=\frac{Sum\:of\:correct\:and\:partially\:correct\:items}{Total\:expected\:items\:\left(from\:reference\:report\right)}$$ -
Diagnostic accuracy assessed whether the LLM’s output correctly conveyed the primary diagnosis formulated.
$$\:Diagnostic\:accuracy=\frac{Number\:of\:correctly\:identified\:diagnoses}{Total\:number\:of\:cases}$$ -
Clinical utility was conducted on a multidimensional perspective, integrating clarity, completeness, and the absence of misleading data/hallucinations. This was essential to serve as an overall surrogate of completeness and usefulness of the report. Clinical utility was calculated as follows:
$$\:Clinical\:Utility=0.7\:\cdot\:Diagnostic\:accuracy+0.3\cdot\:Clinical\:Completeness$$
To simplify the evaluation, such parameters were semantically categorized in the following classes: very low (0-0.3); low (0.3–0.5); moderate (0.5–0.7), high (0.7-1). Each case was independently assessed by five maxillofacial surgeons for clinical completeness, diagnostic accuracy, and clinical utility. Inter-observer agreement was quantified using Cohen’s kappa, which demonstrated substantial concordance (κ = 0.83, 0.87, and 0.84, respectively). Thus, no formal adjudication was performed, as the high level of agreement obviated the need for a third observer consensus review.
Results
A total of 25 lesions were deemed appropriate for this study. Table 1 reports clinical and radiological features of selected patients, including antomical localization, MR sequence, and selected screenshots (Table 1).
For each couple of LLM-generated report and ground-truth report, language similarity metrics and conceptual recognition metrics were calculated using ChatGPT and verified in Matlab (MathWorks Inc, Natick, MA, USA). Results are extensively reported in Supplementary materials 1 and statistically described in Table 2.
Among all the LLM metrics, BERTscore resulted the most accurate in terms of language similarity (0.6856 ± 0.1198), while RadGraphF1 was the most performing to evaluate content completeness (0.4856 ± 0.2013). Performance of LLM metrics is graphically represented in Fig. 2 as radar plot, while Fig. 3 represents a confusion matrix in which darker colors are associated with increased performance of all metrics.
In addition, given the superior performance of BERTscore in predicting the language similarity between the LLM report and the ground-truth, a confusion matrix was created to evaluate the amount of cases in which a satisfactory diagnostic accuracy was achieved using a BERTscore threshold value of at least 0.7 to define reliability (Fig. 4).
Concerning clinician evaluation of LLM performance, inter-observer bias was reduced having five surgeons calculating scores of clinical completeness, diagnostic accuracy and clinical utility for each of the 25 cases. Cohen’s kappa was measured to estimate variability of measures between different clinicians, with a value of 0.83 for clinical completeness, 0.87 for diagnostic accuracy, and 0.84 for clinical utility. As shown by Table 3, overall, almost half of cases were described with a low or very low clinical utility metric. Figure 5 provides a bar plot to illustrate values of clinician-derived scores. All data generated or analysed during this study are included in this published article and reported in Supplementary materials 2.
To investigate whether LLM performance was influenced by lesion complexity, cases were stratified into well-circumscribed lesions (sharp margins) and infiltrative lesions, with poorly defined margins. Supplementary Materials 3 provides a detailed table indicating the allocation of each of the 25 cases to the simple or infiltrative lesion category. As shown by Table 4, across all evaluation metrics, LLM performed significantly better in well-circumscribed lesions, with mean BERTScore, RadGraph F1, and CheXbert scores of 0.75, 0.55, and 0.50, respectively, compared to 0.62, 0.42, and 0.35 for infiltrative lesions (p < 0.05, Mann–Whitney U test). Likewise, ratings provided by surgeons for clinical completeness, diagnostic accuracy, and clinical utility were consistently higher in the well-circumscribed group (0.60, 0.55, 0.58) versus the infiltrative group (0.40, 0.35, 0.38; p < 0.05). Such findings support the hypothesis that the model’s relative strength is superior in characterizing single, encapsulated masses, as well as its limitations in segmenting and interpreting infiltrative pathologies with poorly defined margins and complex anatomical relationships (Fig. 6).
Discussion
The importance of this study is to provide a quantitative and multimetric evaluation of ChatGPT 4o, a widely available, commercial, transformer-based model, in assessing features of diagnostic MR images of orbital lesions. The choice of the orbit for this study was based on a relatively constant anatomy, with easy identifiable anatomical structures as they are surrounded by fat that can be discerned by MR sequences. Thus, it represented an optimal benchmark for a preliminary evaluation of the diagnostic capabilities of LLM in the field of maxillofacial surgery. Despite the larger availability of CT scan, in this study we decided not to evaluate CT owing to the substantially inferior contrast of structures, which might result challenging to discern by computer vision operations.
The evaluation was conducted across multiple levels: linguistic similarity, content completeness, and human perception. The first two modalities are quantitative and mathematically designed, while the latter is based on clinician judgement and related to the perception of the LLM utility in clinical practice.
Metrics designed for NLP systems evaluation represent the most objective and quantitative method to measure performance of AI compared to the radiologist ground-truth. Prior to such metrics, the assessment of LLM outputs depended on human evaluators, rendering it a laborious and time-consuming process. It was thus necessary for evaluators to develop scores like the BLEU score, which was the first one to be created13. Such metrics analyze the tokenization process and are designed to compare n-grams or longest common textual subsequences to identify similarities in sentence structure or semantics19.
Among scores for textual resemblance analysis, BERTScore is considered superior to traditional and older metrics such as BLEU and ROUGE since it assesses text similarity based on contextual embeddings from pretrained language models20. Moreover, BERT score performs more efficiently as, rather than binary yes/no matching, it uses soft alignment functions by computing cosine similarity between all token embeddings of the LLM and the ground-truth. This allows to capture semantic meaning rather than relying solely on exact word or n-gram matches. As a result, BERTScore is more resistant to lexical variation and synonyms and can better reflect human judgment, performing efficiently in particular in tasks involving paraphrasing or domain-specific language such as medical or scientific texts21. Its capacity to perform soft token alignment allows to capture meaning rather than form, making it especially suited for evaluating outputs from LLM16,22.
These assumptions confirm our results showing that BERTscore achieves higher performances compared to BLEU and ROUGE-L, with a mean BERT value of 0.6856, and mean BLEU and ROUGE-L value of respectively 0.4140 and 0.4988. Thus, it must be acknowledged that poorer scores are in part attributable to inadequate metrics to judge between the LLM report and the ground truth report, rather than incapacity of the LLM itself.
Concerning metrics to evaluate clinical completeness, RadGraph F1 and CheXbert should be understood in relation of their structural differences to correctly interpret data. RadGraph F1 schematically assesses the alignment between the LLM and ground-truth reports by comparing structured representations of extracted clinical entities and their interconnections. Thus, this metric not only detects clinical findings, but also their relations, allowing for instance to infer the anatomical localization, achieving factual consistency. Conversely, CheXbert is derived from pretrained classifiers or labelers, and quantifies report accuracy only based on the correct detection of items from a fixed semantic set (e.g., “optic nerve”, “hemangioma”, “adenoma”), emphasizing exact diagnostic correspondence23. Thus, CheXbert provides a simpler, point-by-point evaluation of textual accuracy, while RadGraph F1 is more sophisticated and relates with clinical reasoning and radiologist annotation practices. According to our results, RadGraph F1 yielded higher scores (mean: 0.4856) compared to CheXbert (mean: 0.4124), confirming that also for content completeness results should be interpreted considering the appropriateness of available metrics, which might not capture all the clinical meaning.
In relation to clinician assigned scores, there was a substantial agreement between observers as confirmed by the Cohen’s kappa coefficient. Results confirmed an almost equally distributed clinical completeness score between high-moderate and low-very low classes.
Limitations
Aside from the appropriateness of metrics, there are multiple reasons which make current applications of available transformer-based models not fully ready to interpret MR images of the orbit.
First, a substantial technological limitation of currently available models like ChatGPT is the incapacity to handle volumetric datasets. As previously explained, ChatGPT can not currently interpret NIfTI files without external code execution, owing to the absence of voxel-wise transformers, thus it has a substantially limited interpretation of volumetric features of lesions. To decrease the impact of this limitation, we provided multiplanar screenshots by placing the localizer in the center of the lesion, allowing LLM to integrate missing data24,25.
In addition, from a methodological standpoint, the use of key representative screenshots - carefully selected by an expert clinician to capture the most diagnostically relevant features - simulates the traditional approach in clinical reporting, where static images are routinely used to summarize complex volumetric findings for interdisciplinary discussions, surgical planning, or documentation. The radiologist then produces a narrative report by describing salient features of image analysis. In this setup, the AI model is employed exclusively for the reporting task, relying on images manually selected by human experts. This makes the process more comparable to conventional clinical workflows, as opposed to a fully automated system capable of independently navigating the entire DICOM dataset. However, automatic interpretation of volumetric data surely represents the evolutionary perspective in AI-powered radiology, at the same time needing new parameters to evaluate not only the language output related to the LLM report, but also the efficiency of three-dimensional segmentation and vision transformers, for which metrics such as BERTScore are not adequate.
In addition, LLMs for radiology must be trained with some preliminary assumptions, first of all the radiologic convention of flipped side, as the LLM arbitrarily defines the right part of the screen as the right of the patient. In this study, this needed to be preliminarily declared, but in a real setting adoption of LLMs the absence of this convention might lead to detrimental results in clinical practice26.
Another limitation of this study is that it postulates that the radiology report is the ground-truth. However, if radiology report is not created following the standard reporting method, this might be the source of additional bias, as the LLM report, which is highly standardized, would be compared with a suboptimal ground-truth, resulting in a confounding effect27. In fact, official radiology reports may themselves include variability or occasional inconsistencies in style, completeness, or diagnostic interpretation. This inherent variability would unavoidably influence the comparative metrics, and some discrepancies attributed to the LLM may reflect limitations of the ground-truth itself rather than a model error.
Likewise, our preliminary results revealed that the LLM performs more efficiently in the interpretation of well-demarcated, unifocal lesions, and shows poorer performance in assessing more infiltrative lesions, such as neoplastic or inflammatory, for which it might misevaluate contours and anatomical boundaries28.
Transformer-based LLMs applied to computer vision and CNNs represent a powerful and ever-evolving branch of AI radiology and exhibit increasing effectiveness in various domains of knowledge, including their generative and discriminative capabilities. However, yet ChatGPT implements basic CNN and ViT models to encode images in textual tokens, its core architecture is fundamentally distinct from certified AI platforms currently used in radiological image analysis. ChatGPT code is written predominantly on a transformer-based LLM, optimized for the semantic understanding and generation of textual content, including radiology reports and clinical documentation29. In contrast, most FDA-cleared AI software for diagnostic radiology, examples of which are such as Aidoc (Israel), Viz.ai (USA), Gleamer (France), and Arterys (USA), heavily rely on CNNs and ViT architectures, and are explicitly programmed to process DICOM-based medical images30. Such models have been validated to perform specific tasks, including lesion detection, segmentation, and triage, while NLP components, if present, are typically auxiliary and used to structure final reports. Likewise, as mentioned, to accomplish similar tasks, ChatGPT needs to be integrated with external open-source platforms such as MONAI and 3D Slicer which similarly prioritize volumetric image processing and annotation, with minimal or no use of NLP modules.
Future work
In conclusion, while ChatGPT is becoming a daily tool in clinical practice for its reliable guidance on clinical decision making, also in radiology its use is highly promising. With current architecture, while NLP models such as ChatGPT can assist in downstream creation of the final report, they do not natively analyze pixel data and thus cannot replace more advanced image-based AI models. However, this work represents a pilot experience in interrogating ChatGPT for radiology report generation, and its robustness lies in the evaluation of model performance through appropriate, quantitative metrics, to assess the fidelity of automated report creation. To date, no application of such models in the orbital region was reported. With examined limitations, this paper anticipates a possible implementation of LLM models to create radiology reports in the next future, paving the way for a novel reporting methodology amongst radiologists.
Data availability
All data generated or analysed during this study are included in this published article and reported in **Supplementary materials 2** .
References
Naveed, H. et al. A comprehensive overview of large Language models. ACM Trans. Intell. Syst. Technology (2023).
Vaswani, A. et al. Polosukhin, L.I. Attention is all you need. Advances in neural information processing systems, 30 (2017).
Singhal, K. et al. Toward expert-level medical question answering with large Language models. Nat. Med. 31, 943–950 (2025).
Chai, S. et al. Ladder fine tuning approach for Sam integrating complementary network. Procedia Comput. Sci. 246, 4951–4958 (2024).
Kao, J. P. & Kao, H. T. Large Language models in radiology: A technical and clinical perspective. Eur. J. Radiol. Artif. Intell. 2, 100021 (2025).
Sun, C. et al. Generative large Language models trained for detecting errors in radiology reports. Radiology May;315 (2), e242575 (2025).
Lee, R. W., Lee, K. H., Yun, J. S., Kim, M. S. & Choi, H. S. Comparative analysis of M4CXR, an LLM-Based chest X-Ray report generation model, and ChatGPT in radiological interpretation. J. Clin. Med. 13, 7057 (2024).
Dasanayaka, C. et al. AI and large Language models for orthopantomography radiology report generation and Q&A. Appl. Syst. Innov. 8, 39 (2025).
Wang, S. et al. Interactive computer-aided diagnosis on medical image using large Language models. Commun. Eng. 3, 133 (2024).
Brown, R. A. et al. Deep learning segmentation of orbital fat to calibrate conventional MRI for longitudinal studies. Neuroimage 208, 116442 (2020).
Chen, J. et al. TransUNet: rethinking the U-Net architecture design for medical image segmentation through the lens of Transformers. Med. Image Anal. 97, 103280 (2024).
dos Santos, D. P., Kotter, E., Mildenberger, P. & Martí-Bonmatí, L. European society of radiology (ESR). ESR paper on structured reporting in radiology—update 2023. Insights into Imaging. 14, 199 (2023).
Papineni, K., Roukos, S., Ward, T. & Zhu, W. J. BLEU: a method for automatic evaluation of machine translation. in Proceedings of the 40th Annual Meeting on Association for Computational Linguistics - ACL ’02 311Association for Computational Linguistics, Philadelphia, Pennsylvania, (2001). https://doi.org/10.3115/1073083.1073135
Badshah, S. & Sajjad, H. D. A. F. E. LLM-Based Evaluation Through Dynamic Arbitration for Free-Form Question-Answering. arXiv preprint arXiv:2503.08542, (2025).
Chin-Yew, L. ROUGE: A package for automatic evaluation of summaries. In Text Summarization Branches Out, 74–81, Barcelona, Spain. Association for Computational Linguistics (2004).
Zhang, T., Kishore, V., Wu, F., Weinberger, K. Q. & Artzi, Y. BERTScore: Evaluating Text Generation with BERT. Preprint at (2020). https://doi.org/10.48550/arXiv.1904.09675
Jain, S., Agrawal, A., Saporta, A., Truong, S. Q., Duong, D. N., Bui, T., … Rajpurkar,P. (2021). Radgraph: Extracting clinical entities and relations from radiology reports.arXiv preprint arXiv:2106.14463.
Smit, A. et al. CheXbert: Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT. Preprint at (2020). https://doi.org/10.48550/arXiv.2004.09167
de Inácio, A., Lopes, H. S. & S. & Evaluation metrics for video captioning: A survey. Mach. Learn. Appl. 13, 100488 (2023).
Liu, Y. et al. RoBERTa: A Robustly Optimized BERT Pretraining Approach. Preprint at (2019). https://doi.org/10.48550/arXiv.1907.11692
Shor, J. et al. Association for Computational Linguistics, Toronto, Canada,. Clinical BERTScore: An Improved Measure of Automatic Speech Recognition Performance in Clinical Settings. in Proceedings of the 5th Clinical Natural Language Processing Workshop 1–7 (2023). https://doi.org/10.18653/v1/2023.clinicalnlp-1.1
Morgan, A., BERTScore For, L. L. M. & Evaluation Comet (2024). https://www.comet.com/site/blog/bertscore-for-llm-evaluation/
Jain, S., Agrawal, A., Saporta, A. & Truong, S. Q. RadGraph: Extracting Clinical Entities and Relations from Radiology Reports.
Bradshaw, T. J. et al. Large Language models and large multimodal models in medical imaging: A primer for physicians. J. Nucl. Med. 66, 173–182 (2025).
Tian, D., Jiang, S., Zhang, L., Lu, X. & Xu, Y. The role of large Language models in medical image processing: a narrative review. Quant. Imaging Med. Surg. 14, 1108–1121 (2024).
Kathait, A. S. et al. Assessing laterality errors in radiology: comparing generative artificial intelligence and natural Language processing. J. Am. Coll. Radiol. 21, 1575–1582 (2024).
Koçak, B. et al. Bias in artificial intelligence for medical imaging: fundamentals, detection, avoidance, mitigation, challenges, ethics, and prospects. Diagn. Interventional Radiol. https://doi.org/10.4274/dir.2024.242854 (2025).
Hatamizadeh, A. et al. UNETR: Transformers for 3D Medical Image Segmentation. Preprint at (2021). https://doi.org/10.48550/arXiv.2103.10504
GPT-4. (2024). https://openai.com/index/gpt-4-research/.
Health, C. D. and R. Artificial Intelligence and Machine Learning in Software as a Medical Device. FDA (2025).
Funding
This study did not receive any funding.
Author information
Authors and Affiliations
Contributions
All Authors contributed substantially to the manuscript.A.T: ideation, writing, dataF.Bo: ideation, dataL.M: dataF.Bi: final approvalM.R: final approval.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tel, A., Bolognesi, F., Michelutti, L. et al. Assessment of ChatGPT performance in orbital MRI reporting with multimetric evaluation of transformer based language models. Sci Rep 15, 35654 (2025). https://doi.org/10.1038/s41598-025-19669-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-19669-1