Abstract
The intricate shared genetic architecture underlying halitosis and its related disorders—including salivary secretion disorders, chronic periodontitis, gastroesophageal reflux disease, dental caries, chronic sinusitis, helicobacter pylori infection, and porphyromonas genus abundance—remains incompletely characterized. Our study employed genomic structural equation modeling (Genomic SEM) to define the halitosis common factor (HCF) representing the shared genetic architecture of halitosis-related disorders. Coupled with diverse post-GWAS analytical methods, we aimed to discover susceptible loci and investigate genetic associations with external traits. Furthermore, we explored enriched genetic pathways, cellular layers, and genomic elements. Polygenic risk score analyses, leveraging our integrated GWAS data, were conducted to assess chromosomal-level risk associations for the HCF. A well-fitted genomic SEM integrated GWAS data, revealing the shared genetic architecture of halitosis-related disorders. We identified 23 independent genome-wide significant SNP loci, all previously unreported for this HCF relative to the input single-trait GWAS. Fine-mapping of variants and gene prioritization pinpointed numerous high-confidence putative causal variants and candidate susceptible genes. Subsequent analyses further illuminated the shared genetic architecture underlying HCF and multiple external traits, notably neuropsychiatric characteristics, cognitive function, and inflammatory or metabolic conditions. Notably, this study presents the first comprehensive genetic characterization of halitosis and its related disorders through a GWAS analysis of an unmeasured composite phenotype, providing novel insights into shared etiological pathways potentially linking oral health to systemic factors across these conditions.
Similar content being viewed by others
Introduction
Halitosis, characterized as an offensive breath odor, is a prevalent health concern affecting an estimated third of the global population1. This condition significantly impacts individual social interactions and overall quality of life2. The etiology of halitosis is complex. While most cases originate intra-orally, primarily linked to microbial activity associated with tongue coating, chronic periodontitis, and dental caries, extra-oral factors also contribute substantially. Such factors include otorhinolaryngologic conditions like chronic sinusitis (CRS), digestive system disorders, notably gastroesophageal reflux disease (GERD) and Helicobacter pylori infection, and salivary secretion disorders3,4. Despite extensive research into microbial and environmental determinants, the underlying host genetic susceptibility influencing halitosis and its biological mechanisms remains poorly understood5. Therefore, dissecting the shared genetic architecture of halitosis and its associated traits is crucial. Such analysis is expected not only to reveal complex biological mechanisms and common pathophysiological pathways6, but also holds significant public health relevance for identifying individuals at multiple risks and informing the development of more precise, effective prevention and intervention strategies7.
Previous studies into halitosis etiology relied heavily on observational epidemiology. These studies successfully identified associations between halitosis and various clinical factors, including chronic periodontitis, CRS, GERD, and specific microbial infections like helicobacter pylori infection and certain porphyromonas species8,9,10. However, observational approaches have inherent limitations in establishing causality, as identified associations may be confounded by unmeasured factors or subject to reverse causation. Concurrently, genetic investigations attempted to elucidate the role of host genetic background via candidate gene association studies, exemplified by analyses linking hTAS2R38 polymorphisms to halitosis susceptibility5. Parallel microbiological efforts concentrated on the specific roles of particular oral bacteria, such as porphyromonas gingivalis, Fusobacterium nucleatum, and Moraxella species, in the production of volatile sulfur compounds (VSCs) and the manifestation of halitosis11,12. However, these early research strategies had inherent limitations. Candidate gene methodologies are heavily reliant on prior biological hypotheses, rendering them susceptible to selection bias and incapable of capturing genetic effects across the entire genome. Similarly, studies focusing solely on specific microbes often neglected the holistic complexity of the oral microbiome as an ecosystem and the intricate interactions among its constituent species13. These limitations have impeded a comprehensive and systematic understanding of the genetic and microbial foundations underlying halitosis as a complex trait.
The completion of the Human Genome Project, enabled the advent of Genome-Wide Association Studies (GWAS), which rapidly emerged as the predominant paradigm for investigating the genetics of complex traits14. Indeed, GWAS has been widely and successfully employed to dissect the genetic basis of numerous individual diseases or traits relevant to halitosis etiology, including chronic periodontitis and dental caries, GERD, CRS, and susceptibility to Helicobacter pylori infection15,16. These efforts have identified a multitude of genetic susceptibility loci associated with these specific phenotypes. However, this approach inherently struggles to capture the potential shared genetic architecture and pleiotropic loci underlying these interrelated phenotypes. Consequently, this limitation hinders a comprehensive understanding of the holistic genetic basis of halitosis itself, recognized as a complex syndrome with multi-systemic and multi-factorial contributions7. A powerful strategy to overcome the limitations of single-trait analyses and the challenges of directly measuring a complex phenotype like halitosis in large cohorts is to model a latent common factor representing the shared genetic liability across its key etiological traits. Therefore, to overcome the constraints of single-trait analyses and fully leverage the wealth of existing GWAS data, there is a pressing need for the development and application of more sophisticated statistical genetic methodologies and integrative analytical strategies.
Genomic structural equation modeling (Genomic SEM) offers a robust statistical framework for integrating GWAS summary statistics across multiple traits, thereby enabling the construction and testing of complex models pertaining to their genetic architecture17. In the present study, this framework was leveraged using publicly available GWAS summary statistics specific to halitosis-related traits. Genomic SEM facilitates the elucidation of shared genetic underpinnings and putative causal relationships among traits by combining GWAS data with structural equation modeling principles, while rigorously accounting for sample overlap and pleiotropy. A key application of this methodinvolved estimating single nucleotide polymorphism (SNP) associations with a latent halitosis phenotype, effectively conducting a GWAS on this unmeasured construct. To further interrogate the genetic landscape, unexplained genetic variance potentially harboring novel loci associated with halitosis was investigated through complementary analyses informed by systems biology perspectives. While acknowledging that this genetic approach cannot fully capture the intricate interplay of genetic, environmental, and stochastic factors contributing to complex traits like halitosis, its application minimizes confounding from non-genetic factors often associated with direct biomarker measurements, thus permitting a robust analysis of challenging summary-level datasets. Subsequently, extensive causal inference analyses, utilizing the GWAS summary data, were performed to identify potential causal links between genetic variation and clinical outcomes. These analyses aim to provide predictive insights for clinicians and biologists, potentially informing preventative strategies and therapeutic interventions for patients.
Methods
GWAS summary statistics data sources for genomic SEM
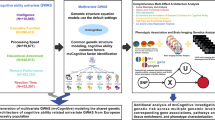
Figure 1 presents a schematic overview outlining the workflow employed in this study. For the Genomic SEM analysis, GWAS summary statistics were sourced from seven independent studies pertaining to halitosis-associated traits. These datasets originated from previously published GWAS investigations. The analysis encompassed traits including salivary secretion disorders, chronic periodontitis, GERD, dental caries, CRS, Helicobacter pylori infection, and porphyromonas genus abundance. Ethical approval was obtained from respective Institutional Review Boards for all contributing GWAS studies, and informed consent had been provided by all participants. Prior to our analysis, summary statistics were subjected to rigorous quality control procedures to ensure data integrity. Table S1 provides a detailed list of the GWAS datasets incorporated.
Rationale for trait selection
The selection of these seven traits was guided by the well-established multifactorial etiology of halitosis, which is broadly categorized into intra-oral and extra-oral origins. This framework aimed to construct a biologically coherent composite factor that captures the condition’s complex pathogenesis. Intra-orally, chronic periodontitis and dental caries were included as they are recognized primary pathological drivers of halitosis, creating anaerobic niches that facilitate the bacterial production of VSCs6,18. To capture the broader oral environment, salivary secretion disorders were included, as reduced salivary flow impairs oral clearance and promotes microbial proliferation, while the abundance of Porphyromonas genus was incorporated as a direct measure of the key VSC-producing microbiota19,20. For extra-oral contributions, GERD, Helicobacter pylori infection, and CRS were selected as they represent key, well-documented etiologic factors originating from the digestive and upper respiratory tracts that are linked to halitosis10,21,22. The final selection was determined by two primary criteria: first, a strong, evidence-based biological link to halitosis, and second, the crucial requirement of having publicly available, well-powered GWAS summary statistics from populations of European ancestry to ensure methodological consistency and statistical power. The final set of seven traits therefore represents the most comprehensive and biologically robust composite phenotype that could be constructed from the available genetic data.
Quality control of input GWAS data
A stringent quality control (QC) pipeline, adhering to recommended filtering criteria, was implemented for all autosomal SNPs across the seven input GWAS datasets. To ensure consistency and compatibility, filtering was performed against the 1000 Genomes Project Phase 3 European (EUR) reference panel. Variants were excluded if they exhibited a minor allele frequency (MAF) < 0.01, reported a zero effect size estimate, presented reference panel mismatches, or possessed ambiguous allele assignments. Recognizing that the constituent GWAS datasets originated from diverse genomic repositories and study populations, potential sample overlap represented a critical methodological consideration. To address this, we utilized the multivariate extension of LDSC within the Genomic SEM framework. This statistical method inherently estimates the genetic covariance matrix while simultaneously calculating and adjusting for any sample overlap among the input GWAS summary statistics. This approach is designed to prevent test statistic inflation and enhance the robustness of subsequent Genomic SEM results by minimizing bias in effect size estimations.
Genomic SEM construction
Genomic SEM, implemented via the `GenomicSEM` R package (v0.0.5), was employed to investigate the shared genetic architecture underlying the selected halitosis-related traits. Genomic SEM provides a means of exploring the latent genetic structure connecting multiple phenotypes through the estimation of multivariate genetic models17. A key advantage of the Genomic SEM approach is its robustness to variations in sample overlap and sample size across input studies, thereby mitigating potential biases associated with these factors. The Genomic SEM analysis was conducted in two principal stages. The first step involved estimating the empirical genetic covariance matrix (S) and its corresponding sampling covariance matrix (V). To this end, QC-filtered GWAS summary statistics for the seven halitosis-related traits were compiled, and the multivariate extension of Linkage Disequilibrium Score Regression (LDSC) was applied. LDSC offers a powerful statistical framework that utilizes GWAS summary statistics and LD information. This approach distinguishes true polygenic signals from confounding factors such as cryptic relatedness and population stratification. Within our study, multivariate LDSC generated the empirical genetic covariance matrix for the seven traits. This matrix served as the input for the SEM model fitting in the subsequent stage. SNP-based heritability estimates (h²SNP) derived from LDSC for each individual trait are reported in Table S2. The second stage involved specifying and fitting a common factor SEM model. The primary goal was to identify a latent common genetic factor (s) underlying the seven halitosis-related traits by minimizing the discrepancy between the model-implied covariance structure and the empirical genetic covariance matrix derived from Stage 1. To assess model adequacy, multiple established fit indices were evaluated, including the Standardized Root Mean Square Residual (SRMR), the model chi-square test statistic (χ²), the Akaike Information Criterion (AIC), and the Comparative Fit Index (CFI) (Table S3 and S4). This common factor SEM specification provided a method for integrating individual autosomal SNP associations across the seven traits into a unified model. This integration facilitated a genome-wide association analysis for the identified latent common factor. To ensure consistent effect directions among SNPs significantly associated with the common factor, a heterogeneity test using Cochran’s Q statistic was performed for each genome-wide significant SNP; variants with a heterogeneity FDR-value < 0.05 were excluded.
Multi-level evaluation of the genomic SEM model
In addition to the standard model fit indices (SRMR, χ², AIC, CFI), supplementary evaluations were conducted to assess the stability and validity of the Genomic SEM results. Specifically, parameters such as the mean χ², genomic inflation factor (lambda GC, λGC), maximum χ², the overall h²SNP of the common factor, the LDSC intercept, and the attenuation ratio (calculated as (LDSC Intercept − 1) / (Mean χ² − 1)) were examined using LDSC based on the common factor GWAS summary statistics. Detailed controls for LDSC parameters included retaining SNPs with missing values, retaining SNPs with INFO scores < 0.9, retaining SNPs with MAF < 0.01, and excluding SNPs with p-values outside of the valid range or with unclear chain orientation.
Identification of significant and novel genomic loci
We utilized FUMA (Functional Mapping and Annotation; https://fuma.ctglab.nl/) to systematically identify genomic risk loci associated with the halitosis common factor (HCF) derived from the Genomic SEM23. Independent significant SNPs were defined as those reaching genome-wide significance (P < 5 × 10⁻⁸). Lead SNPs within each locus were designated based on the lowest P-value and independence from other lead SNPs (r² < 0.1). In order to ascertain novelty, a ‘GWAS-by-Subtraction’ approach was additionally employed. This involved contrasting loci identified via the Genomic SEM (P < 5 × 10⁻⁸) with those reaching genome-wide significance in any single-trait input GWAS (P < 5 × 10⁻⁸). Further comparisons were made against previously published associations (P < 5 × 10⁻⁸) in the GWAS Catalog to evaluate potential pleiotropy.Risk locus annotation and prioritization for genome-wide significant variants (P < 5 × 10⁻⁸) from the Genomic SEM common factor GWAS were subsequently performed using FUMA. Post-GWAS analyses were conducted using MAGMA to investigate gene-level associations with the HCF.
Fine-mapping of association signals
In order to pinpoint the most probable causal variants within the identified loci, a Bayesian fine-mapping approach was implemented using FINEMAP, executed via the `echolocatoR` R package (v2.0.3)24. For each independent significant signal, a 250 kb window centered on the lead SNP was analyzed. Posterior probability (PP) of causality for each variant within the region is calculated by FINEMAP, accounting for LD structure (1000 Genomes EUR reference). 95% credible sets were defined for each signal, encompassing the minimal set of variants whose PP ≥ 0.95. Variants within these credible sets were considered putative causal variants.
Transcriptome-Wide association study
Given that association signals may be mediated through gene expression, and recognizing that fine-mapping solely based on SNP proximity can be limited, a Transcriptome-Wide Association Study (TWAS) was performed25. The sCCA-TWAS method was employed across multiple tissues to identify genes whose predicted expression levels associate with the HCF. This analysis leveraged pre-computed tissue-specific eQTL weights for 37,920 genes derived from the Genotype-Tissue Expression (GTEx) project (v8) dataset. Genes exhibiting a significant TWAS association after FDR correction (FDR < 0.05) were selected for further analysis. In order to refine TWAS findings and assess the likelihood of causal effects versus LD-induced correlation, FOCUS (Fine-mapping Of Causal gene Sets) was applied to significant TWAS genes within each locus. The FOCUS framework calculates the posterior inclusion probabilities (PIP) for each gene being the causal mediator. This Bayesian approach integrates GWAS summary statistics and eQTL weights, adjusting for LD and potential colocalization. A PIP threshold > 0.8 was used to identify genes with strong evidence supporting a causal role.
Gene set and pathway enrichment analysis
Gene set and pathway enrichment analyses were performed to elucidate the biological functions and pathways potentially underlying the genetic associations. Genes implicated by MAGMA served as the input set. These analyses were conducted using tools such as FUMA’s gene2func module, querying canonical pathways and functional categories from the Molecular Signatures Database (MsigDB).
Cell type annotation and partitioned heritability analysis
To identify specific cell types potentially relevant to the etiology of the HCF, the CELLECT pipeline was employed26. This method provides a means of integrating cell-type expression specificity profiles from single-cell RNA sequencing (scRNA-seq) data with GWAS summary statistics. Pre-processed expression specificity likelihood scores, computed via CELLEX using the Tabula Muris dataset27, were utilized. Stratified LD Score Regression (S-LDSC) was subsequently applied to test for enrichment of the common factor heritability within genomic regions specific to each cell type. Furthermore, partitioned heritability analysis using S-LDSC was performed to estimate the contribution of different functional genomic annotations (e.g., coding regions, enhancers) to the overall heritability. This analysis allows for the assessment of heritability enrichment within specific genomic categories, offering insights into the functional context of the associated variants.
Correlation and causal inference with external traits via Mendelian randomization
In order to explore potential causal relationships between various exposures and the HCF, two-sample Mendelian Randomization (MR) was performed. The IEU OpenGWAS database, encompassing data for 50,033 phenotypes, was utilized as a comprehensive source for potential exposure traits. Instrumental variables (IVs) for each exposure were selected based on genome-wide significance (P < 5 × 10⁻⁸) and independence (r²< 0.001, kb = 10,000). The inverse variance weighted (IVW) method was employed as the primary MR approach. Sensitivity analyses, including MR-Egger regression and the weighted median method, were conducted to assess the robustness of the findings against potential pleiotropy28.
Polygenic risk score construction and evaluation
Polygenic Risk Scores (PRS) were constructed to evaluate the collective predictive capacity of common variants identified through the Genomic SEM for the HCF29. The PRS-CS algorithm, a Bayesian regression framework incorporating continuous shrinkage priors, was utilized for this purpose. This algorithm provides a means of integrating the Genomic SEM summary statistics with an external LD reference panel (1000 Genomes EUR) to compute posterior SNP effect sizes. The resulting shrunken effect estimates are suitable for calculating PRS, the performance of which could potentially be evaluated in independent target cohorts.
Results
Structural equation model fitting
Based on LDSC analysis of the seven GWAS summary statistics comprising the genomic SEM for the HCF, three traits (salivary secretion disorders, chronic periodontitis, and GERD) exhibited heritability Z-scores exceeding 1.96. The remaining four traits (dental caries, CRS, Helicobacter pylori infection, and porphyromonas genus abundance) exhibited Z-scores below this threshold (Table S2). Although four traits showed non-significant heritability Z-scores, they were retained in the model. This decision was based on two key considerations: first, Genomic SEM can effectively leverage the genetic covariance between traits, which can be estimated with greater power than individual heritabilities, even when the heritability of some indicators is low or non-significant. Second, these traits are clinically and biologically integral to the multifaceted etiology of halitosis, and their exclusion would have resulted in a less comprehensive and biologically valid common factor. The point estimates for their heritability were non-zero and their standard errors were promising, suggesting they still contribute valuable information to the model. These findings suggest statistically significant heritable components for a subset of the traits, whereas others demonstrated weaker or non-significant heritability signals. The genetic covariances between each pair of traits are presented in Table S3 and Fig. 2. A one-common-factor model fitted to the genetic covariance matrix (S) was evaluated. The model fit indices were mixed, with a perfect CFI (1.000) suggesting excellent fit, but an elevated SRMR (0.221) indicating some residual error (Table S4). This pattern can occur in Genomic SEM, particularly with a limited number of indicators, where CFI may be insensitive. Despite the elevated SRMR, which suggests potential model misspecification, we proceeded with the one-factor model given the strong theoretical rationale and the exploratory nature of this study. Standardized factor loadings of the latent variable onto each observed trait, alongside estimates of residual variances for each trait, are detailed in Table S5. Collectively, these results provide evidence supporting a shared genetic factor underlying the selected halitosis-related traits within the genomic SEM. The final genomic SEM analysis generated an indirectly measured GWAS, based on 6,918,772 SNPs, to investigate the genetic architecture of the HCF.
Stability assessment of the genomic SEM via LDSC
To assess the stability and potential confounding influences within the GWAS summary statistics derived from the genomic SEM for the HCF, we employed LDSC. Following quality control procedures specific to this LDSC analysis, 1,083,268 SNPs were excluded, retaining 892,405 SNPs for the regression model. The LDSC regression applied to the common factor summary statistics yielded a mean χ² statistic of 0.5736 across the retained SNPs. The genomic inflation factor (λGC) was 1.1144, and the LDSC intercept was 0.4662 (SE = 0.0029). The total observed-scale heritability (h²) was estimated at 0.0009 (SE = 3.4333e-05). The observed λGC is modest and, in the context of our sample overlap correction via multivariate LDSC, is interpreted as reflecting true polygenicity rather than residual confounding. The mean χ² of 0.5736, while lower than typically observed in single-trait GWAS, is not indicative of data quality issues or overcorrection in this context. Instead, it reflects the nature of a common factor GWAS, where the effect of any single SNP on the latent factor is an aggregation of its effects across seven traits, leading to an attenuated average signal strength across the genome. The true signal is captured by the genome-wide significant loci that emerge from this attenuated background, representing variants with pleiotropic effects consistent with the common factor model.
Risk genetic loci
In the GWAS derived from the genomic SEM for the HCF, we identified 23 genetic variants that surpassed the genome-wide significance threshold (P < 5 × 10⁻⁸) (Fig. 3, Table S6). Functional annotation performed using the FUMA platform revealed that the majority of these associated loci reside within intergenic (55.1%) and intronic (36.9%) regions. Smaller proportions were located in downstream (0.6%), 3’ untranslated regions (UTR3, 0.2%), non-coding RNA exonic (0.2%), ncRNA intronic (6.7%), and upstream (0.2%) regions. Notably, no variants were annotated to coding exonic regions (0%). Subsequent analysis identified 23 independent lead SNPs (r² < 0.1) (Table S7). Based on our analysis, all 23 lead SNPs constitute novel risk loci for the HCF, as these variants were not previously detected at genome-wide significance within the GWAS summary statistics of the individual input traits (Table S8). We queried the GWAS Catalog database to ascertain prior associations of these lead SNPs with other phenotypes (Table S9). For example, the lead SNP rs10001274 has been previously associated with Supramarginal gyrus volume. Lead SNP rs12271161 demonstrated associations with multiple traits, including Non-accommodative esotropia, Medication use-thyroid preparations, Subjective well-being, Hypothyroidism, Proprotein convertase subtilisin/kexin type 7 levels, and Serum albumin levels. Lead SNP rs12904518 is associated with Angina pectoris, Insomnia, and Coronary artery disease. Lead SNP rs2060196 is associated with Type 2 diabetes - age of onset. Lead SNP rs4882411 shows associations with Major depressive disorder, Insomnia, Depression, Depressive symptoms, Lifetime smoking index, and Depressive symptoms. Lead SNP rs7752901 is associated with Educational attainment, and lead SNP rs9882338 is associated with Health literacy. Gene-based association analysis, conducted using MAGMA, aimed to identify specific genes implicated by the SNP-level associations with the HCF. This analysis highlighted 19 potentially associated genes (P < 0.05) (Table S10, Fig. 4). Among these, RBM5 exhibited the strongest association signal (Z = 2.3955, P = 0.0083), followed by CTD-2330K9.3 (Z = 2.2413, P = 0.0125) and MON1A (Z = 2.2384, P = 0.0126). Other genes surpassing the significance threshold (P < 0.05) included CDHR4, RBM6, MST1, SEMA3F, GPX1, APEH, TRAIP, MST1R, CAMKV, BSN, TCTA, NICN1, HIST1H3B, RNF123, GNAT1, and IP6K1.
Fine-mapping
To pinpoint potential causal variants within the identified loci, we performed statistical fine-mapping. This analysis was conducted on the 23 identified genomic risk loci. From these loci, we identified 30 distinct SNPs that were part of a 95% credible set and demonstrated a high PP (PP ≥ 0.95) of being the causal variant. Among these SNPs, we highlight three exemplary signals. The lead SNP rs12271161 (GWAS P = 4.09 × 10⁻⁶⁹), located in the AP000936.4 region, presented a highly compelling signal with a t-statistic of -17.57 and a mean PP of 1.0. Similarly, rs9625070 (GWAS P = 1 × 10⁻²⁰⁰) within the CTA-211A9.5 region (t-statistic = 42.16) and rs2439335 (GWAS P = 1 × 10⁻²⁰⁰) within the KCNB2 region (t-statistic = -45.31) both exhibited exceptionally strong associations and achieved the maximum possible PP (mean PP = 1.0) (Table 1; Fig. 5). These findings strongly implicate the respective lead SNPs at these loci as the likely causal variants driving the observed association signals.
Gene-level identification of susceptibility
We conducted a TWAS leveraging summary-data-based sCCA to identify genes whose genetically regulated expression levels are associated with the HCF. This analysis identified only one gene, APOC3, exhibiting a statistically significant association. Subsequently, we employed the FOCUS methodology to fine-map the gene-level association signals derived from the genomic SEM data. This identified three genes with a PIP exceeding 0.8, suggesting they may represent credible causal genes within their respective loci. To further solidify high-confidence gene-level associations, we integrated the TWAS and FOCUS findings. Based on the unique TWAS-significant gene and the FOCUS outputs, APOC3, located on chromosome 11, was robustly identified, satisfying both criteria with a TWAS Z-score of 17.57 (TWAS P = 4.09 × 10⁻⁶⁹) and a FOCUS PIP of 1 (Table S11, Fig. 6).
Pathway enrichment analysis
Pathway enrichment analysis indicated that genes associated with the HCF were significantly overrepresented in two Reactome pathways (FDR = 0.014) and two BioCarta pathways (FDR = 0.0038), both pertinent to the MSP-RON signaling pathway (Table S12). Furthermore, interrogation of GWAS Catalog-defined gene sets revealed highly significant enrichment (FDR < 0.05) for numerous sets related to cognitive function, behavior, health metrics, and neurological traits. Prominent examples include Extremely high intelligence (FDR = 8.98e-38), Sleep duration (FDR = 5.31e-34), Regular attendance at a gym or sports club (FDR = 2.70e-31), Subcortical volume (FDR = 5.93e-27), Regular attendance at a religious group (FDR = 1.40e-26), Brain morphology (FDR = 2.85e-26), Cortical surface area (FDR = 3.32e-25), and Subcortical volume (FDR = 2.77e-23) (Table S12).
Cell type annotation and enrichment analysis
Utilizing CELLECT for cell type enrichment analysis based on the Tabula Muris dataset, we explored the partitioning of heritability for the HCF across various cell types. Among those tested, Brain_Non-Myeloid_neuron exhibited the lowest P-value (P = 0.024), suggesting a potential enrichment of HCF heritability within this cell type. This was followed by Trachea_blood_cell (P = 0.038) (Table S13).
Heritability enrichment across genomic functional and regulatory regions
Analysis of heritability enrichment across genomic functional categories, performed using S-LDSC (Table S14), revealed significant patterns (FDR < 0.05) in multiple annotation classes. Specifically, significant positive or negative heritability enrichment was detected for Conserved_LindbladToh (conserved elements) (Enrichment = 16.62, P = 1.93e-11), DHS_Trynka.Extend.500 (500 bp-extended DNase I hypersensitive sites) (Enrichment = 1.99, P = 3.24e-07), regions marked by H3K4me1 histone modification (H3K4me1_peaks_Trynka, H3K4me1_Trynka, H3K4me1_Trynka.Extend.500; Enrichments = 3.18, 1.80, 1.41; P = 2.54e-03, 9.84e-03, 3.22e-04, respectively), regions marked by H3K9ac histone modification (H3K9ac_peaks_Trynka, H3K9ac_Trynka.Extend.500; Enrichments = 5.70, 2.40, 1.80; P = 2.47e-02, 4.28e-02, 9.84e-03, respectively), Intron_UCSC.Extend.500 (500 bp-extended intronic regions) (Enrichment = 1.24, P = 1.28e-03), Repressed_Hoffman.Extend.500 (500 bp-extended repressed chromatin regions) (Enrichment = 0.84, P = 1.28e-03), and WeakEnhancer_Hoffman.Extend.500 (500 bp-extended weak enhancer regions) (Enrichment = 2.71, P = 3.20e-02). Furthermore, significant enrichment was noted for categories including FetalDHS_Trynka and FetalDHS_Trynka.Extend.500 (fetal DHS sites), H3K27ac (PGC2), and H3K4me3 (Trynka.Extend.500) (Table S14). These findings underscore the significant contribution of specific genomic regulatory elements, such as conserved regions, DHS sites, and regions characterized by particular histone modifications, to the overall genetic architecture of the HCF.
Identification of potential causal risk factors for the halitosis common factor via Mendelian randomization
To systematically interrogate exposures potentially causally associated with the HCF, we conducted an extensive two-sample MR analysis leveraging exposure GWAS data from the IEU OpenGWAS database. Employing the IVW method as the primary analysis, we identified approximately 95 exposures demonstrating potential causal associations with the HCF (Table S15). Several factors exhibited potential causal effects indicative of an increased risk. These encompassed anthropometric traits, including Body mass index (OR = 1.040, 95% CI = 1.028–1.053, P < 0.001), Waist circumference (OR = 1.033, 95% CI = 1.018–1.048, P < 0.001), Hip circumference (OR = 1.017, 95% CI = 1.005–1.028, P = 0.004), and various adiposity measures like Leg fat percentage (right) (OR = 1.065, 95% CI = 1.044–1.087, P < 0.001) and Arm fat mass (right) (OR = 1.035, 95% CI = 1.023–1.046, P < 0.001). Indicators reflecting negative affect and psychological distress were also associated with increased risk, such as Neuroticism score (OR = 1.008, 95% CI = 1.002–1.014, P = 0.006), Depressed affect (OR = 1.049, 95% CI = 1.018–1.082, P = 0.002), Feelings of being ‘fed-up’ (OR = 1.211, 95% CI = 1.073–1.365, P = 0.002), Feeling miserable (OR = 1.043, 95% CI = 1.005–1.083, P = 0.025), Experiencing mood swings (OR = 1.067, 95% CI = 1.028–1.107, P = 0.001), and Major depression (OR = 1.056, 95% CI = 1.030–1.081, P < 0.001). Additionally, poorer self-rated health (Overall health rating: OR = 1.202, 95% CI = 1.145–1.263, P < 0.001), a higher Number of self-reported non-cancer illnesses (OR = 1.113, 95% CI = 1.038–1.194, P = 0.003), GERD (OR = 1.086, 95% CI = 1.065–1.107, P < 0.001), and Ulcerative colitis (OR = 1.003, 95% CI = 1.000–1.005, P = 0.039) showed potential risk associations. Regarding lifestyle, Smoking status: Current (OR = 1.204, 95% CI = 1.019–1.422, P = 0.029) and more Time spent watching television (TV) (OR = 1.111, 95% CI = 1.054–1.170, P < 0.001) were linked to increased risk. Conversely, the MR analysis identified factors potentially associated with a decreased risk. These included markers of higher cognitive function and educational attainment, such as Intelligence (OR = 0.972, 95% CI = 0.962–0.983, P < 0.001), Cognitive performance (OR = 0.973, 95% CI = 0.961–0.985, P < 0.001), Qualifications: College or University degree (OR = 0.884, 95% CI = 0.855–0.915, P < 0.001), and Years of schooling (OR = 0.970, 95% CI = 0.944–0.997, P = 0.027). In terms of lifestyle and social factors, engagement in Strenuous sports or other exercises (OR = 0.715, 95% CI = 0.554–0.923, P = 0.01) and Walking for pleasure (OR = 0.849, 95% CI = 0.749–0.963, P = 0.011) were associated with lower risk. Lower likelihood of Past tobacco smoking (OR = 0.972, 95% CI = 0.949–0.995, P = 0.019) and higher Cereal intake (OR = 0.956, 95% CI = 0.919–0.993, P = 0.022) suggested potential protective effects. Furthermore, later Age first had sexual intercourse (OR = 0.938, 95% CI = 0.917–0.959, P < 0.001) and later Age at first live birth (OR = 0.907, 95% CI = 0.869–0.946, P < 0.001) were also associated with reduced risk. Sensitivity analyses were performed for these primary findings to evaluate potential biases such as horizontal pleiotropy and heterogeneity. For the majority of the reported significant associations, Cochran’s Q test P-values exceeded 0.05, indicating no significant heterogeneity was detected. Similarly, MR-Egger regression intercept P-values were greater than 0.05, providing no evidence of significant directional horizontal pleiotropy in these analyses, thus bolstering confidence in the robustness of the IVW estimates.
Polygenic risk score construction from summary data
We constructed PRS for the HCF using the PRS-CS algorithm applied to the genomic SEM-derived GWAS summary statistics. An examination of the summed contributions from per-chromosome aggregated PRS (Table S16) revealed considerable variation in genetic contributions to susceptibility across different chromosomes. Considering all SNPs included in the PRS, chromosome 2 (PRS Score Sum = 18.85) and chromosome 1 (PRS Score Sum = 17.68) exhibited the highest cumulative PRS scores. This suggests that common variants residing on these chromosomes contribute most substantially, in aggregate, to the polygenic risk for the HCF.
Discussion
This study represents the first application of Genomic SEM to elucidate the shared genetic architecture underlying seven key halitosis-related phenotypes, offering novel genetic insights into the biological basis of halitosis. The core achievement was the successful identification and validation of a latent HCF, followed by an mvGWAS that pinpointed 23 genome-wide significant and novel associated loci. Fine-mapping provided high-confidence evidence for 30 putative causal SNPs within these loci. Furthermore, an integrated suite of post-GWAS analyses—including TWAS, MAGMA, GSEA, cell-type enrichment, S-LDSC heritability partitioning, and MR—collectively provided multi-faceted evidence aimed at characterizing the HCF’s biological functions, relevant cellular contexts, and potential causal relationships with other traits. A central contribution of this work is the initial characterization of halitosi’s shared genetic structure, moving beyond previous research focused predominantly on single related disorders or halitosis in isolation. By uncovering the common genetic underpinnings of these frequently co-occurring conditions, this study lays a foundation for a more comprehensive understanding of the integrated mechanisms driving halitosis.
To elucidate the genetic architecture underlying halitosis, LDSC was performed on seven clinically relevant phenotypes, revealing a significant network of shared genetic correlations. Notably, a strong positive genetic correlation was identified between GERD and dental caries, suggesting substantial overlap in genetic susceptibility pathways. This finding is consistent with proposed pathophysiological mechanisms wherein GERD-induced alterations, such as reduced oral pH via acid reflux and potential salivary dysfunction, may create an environment conducive to dental caries formation and shifts in the oral microbiome favoring VSC-producing anaerobes. Furthermore, dental caries can serve as reservoirs for bacterial retention. These processes are collectively implicated in halitosis pathogenesis30,31. Significant positive genetic correlations were also observed between GERD and chronic periodontitis, Helicobacter pylori infection, CRS, and salivary secretion disorders. These intercorrelations further underscore the multifactorial nature of halitosis predisposition, potentially involving shared systemic factors. For instance, GERD-associated upper airway effects potentially predisposing to CRS32, with subsequent VSC production from post-nasal drip decomposition contributing to halitosis7. Additionally, GERD is a potential risk factor for sicca symptoms33, which could impair salivary clearance and buffering capacity, thereby promoting halitosis. Understanding this shared genetic architecture is pivotal for dissecting the etiology of halitosis and motivated the subsequent mvGWAS designed to identify specific genetic loci associated with this HCF.
Leveraging the validated HCF model, our mvGWAS successfully identified 23 independent, novel genomic risk loci associated with this shared genetic susceptibility at genome-wide significance, substantially expanding the known genetic landscape of halitosis. Our fine-mapping analyses further refined these signals, nominating 30 high-confidence putative causal SNPs. Annotation of these loci revealed potential biological links. For instance, rs17117475 and rs9625070 are near loci with currently uncharacterized function (RP11-11K13.1, CTA-211A9.5), yet their extreme statistical significance and high PIP values underscore their potential critical role in halitosis susceptibility. Moreover, rs2439335, lies near the KCNB2 gene, encoding the voltage-gated potassium channel Kv2.2, crucial for neuronal excitability34. Given the potential involvement of neural regulation in halitosis-related physiology, genetic variation influencing KCNB2 function might contribute to halitosis by affecting relevant neural circuits, a hypothesis requiring functional validation. To probe the potential biological functions of these novel loci, pleiotropy analysis using the GWAS Catalog was performed. Results showed rs10001274 is associated with schizophrenia-related brain structure35; rs12271161 links to diverse phenotypes including strabismus36, thyroid function37, and subjective well-being38; and rs775290 associates with educational attainment39. These pleiotropic associations suggest that some genetic risk factors for HCF may participate in broader biological processes extending beyond traditional oral biology. Collectively, these findings underscore the polygenic basis of HCF as a complex trait. Future functional studies are warranted to elucidate the precise biological functions of these identified loci and their mechanistic roles in halitosis development.
To elucidate the functional mechanisms underlying previously identified GWAS signals for halitosis, an integrative post-GWAS analysis strategy was employed. Initially, TWAS identified APOC3 as the sole gene exhibiting a significant association. Subsequent FOCUS provided robust support for APOC3’s causality. The APOC3 gene encodes Apolipoprotein C-III, a protein known to exert a pivotal inhibitory effect on plasma triglyceride (TG) metabolism, primarily by suppressing lipoprotein lipase (LPL) activity and hepatic uptake of triglyceride-rich lipoproteins40,41. Although direct evidence linking APOC3 to halitosis is currently lacking, a plausible mechanistic link can be hypothesized. Genetically influenced alterations in APOC3 expression can impact lipid profiles, and emerging evidence implicates both ApoC-III in activating inflammatory pathways42 and intracellular triglyceride metabolism in the regulation of macrophage inflammatory responses43. Therefore, it is biologically plausible that APOC3-mediated effects on lipid homeostasis and systemic inflammation could contribute to halitosis susceptibility, potentially by fostering a pro-inflammatory milieu that influences the oral microbiome or local tissue inflammation in the periodontium or gastrointestinal tract. Secondly, gene-based association analysis using MAGMA highlighted RBM5 as the gene most significantly associated with the previously derived HCF. RBM5 encodes an RNA-binding protein recognized as a critical regulator of alternative splicing of pre-mRNAs44 and a key participant in the control of apoptosis, suggesting a potential tumor suppressor function45. Considering the fundamental role of apoptosis in maintaining tissue homeostasis and modulating inflammation46, variations affecting RBM5 expression or function might indirectly influence halitosis development, possibly by perturbing cellular turnover balance or inflammatory signaling responses within oral tissues. In conclusion, this work successfully translates GWAS findings into biologically plausible hypotheses by nominating APOC3 and RBM5 as high-priority candidate effector genes for halitosis. The implicated biological pathways, notably lipid metabolism, RNA processing, and apoptosis, furnish critical insights into the condition’s potential genetic underpinnings and provide a panel of prioritized targets warranting subsequent functional validation to definitively establish their roles in halitosis pathogenesis.
Cell-type enrichment analysis pinpointed key cellular contexts where HCF genetic risk might operate. HCF heritability was significantly enriched in brain non-myeloid neurons and trachea blood cells. The enrichment in brain neurons aligns with the observed genetic correlations between HCF and traits like neuroticism, depression, and cognition from the MR analysis, and resonates with emerging evidence linking oral health to brain structure47,48. Enrichment in trachea blood cells suggests involvement of systemic inflammatory or immune response pathways49. This is consistent with the known capacity of oral diseases to trigger or exacerbate systemic inflammation50 and aligns with the MR findings linking HCF to inflammatory conditions like GERD and UC. In concert, these functional analyses successfully connect HCF GWAS signals to relevant cell types, providing crucial clues to the genetic basis of HCF.
To explore the systemic etiological network of halitosis beyond local factors and assess potential causal relationships, an extensive two-sample MR analysis was conducted. The results strongly support the hypothesis that HCF may represent an oral manifestation of broader systemic factors and dysregulation, rather than being solely a localized oral issue. The MR analysis provided evidence for potential causal associations between multiple exposures and HCF. Consistent with the GSEA findings of IBD gene set enrichment, MR showed that genetically predicted higher risk for UC was associated with increased HCF risk. Furthermore, genetically predicted higher BMI and related adiposity measures were associated with increased HCF risk, corroborating observational findings that identify higher BMI as a potential predictor of halitosis51. Similarly, genetically predicted current smoking status was linked to increased HCF risk, aligning with substantial epidemiological evidence establishing tobacco use as a recognized halitosis risk factor52. The MR analysis further unveiled complex genetic links between HCF and neuropsychiatric and cognitive traits. Genetically predicted higher neuroticism, depressive symptoms, and mood swings were associated with increased HCF risk, consistent with observational studies identifying psychological factors as risk factors for subjective halitosis53. Potential mechanisms could involve stress-induced xerostomia, altered oral hygiene habits, or even gut inflammation leading to extra-oral halitosis54. Conversely, genetically predicted higher intelligence, cognitive performance, and longer educational attainment exhibited protective effects against HCF, consistent with reports linking clinical halitosis to lower education levels55. This protection might be mediated through various pathways including better oral health knowledge/behaviors, higher health literacy, and socioeconomic advantages56. Additionally, genetically predicted higher levels of physical activity were associated with reduced HCF risk, possibly reflecting the beneficial effects of exercise on overall health, including potentially salivary function and periodontal health57. In conclusion, the MR findings, together with GSEA results, paint a multi-dimensional, systemic picture of HCF etiology. The results strongly suggest that HCF arises from a complex interplay between genetic predisposition, systemic immune/inflammatory status, metabolic factors, lifestyle exposures, and neuropsychological characteristics.
Limitations
This study, while innovative, has several limitations that warrant consideration. First, and most fundamentally, the HCF is a statistical abstraction derived from genetically correlated traits, not a direct biological measure of halitosis itself. The premise that these seven traits adequately represent the genetic risk for halitosis is a core assumption of our model. The selection of different or additional traits could alter the composition of the HCF and subsequent findings. Therefore, our results should be interpreted as identifying loci associated with the shared genetic liability of these specific conditions, which serves as a proxy for halitosis risk. Second, our one-factor model, while theoretically grounded, showed mixed fit indices, with an elevated SRMR suggesting some degree of model misspecification. Although we proceeded based on the strong a priori hypothesis and exploratory goals, more complex models (e.g., bifactor or two-factor models) might provide a more nuanced fit to the data, and future studies could explore these alternatives. Third, four of the seven input GWAS datasets had non-significant SNP-based heritability. While Genomic SEM can still leverage genetic covariance in such cases, the inclusion of traits with weak genetic signals may have introduced noise and could potentially limit the power to detect a more robust common factor. Fourth, our analyses were conducted on GWAS summary statistics from populations of predominantly European ancestry. This limits the generalizability of our findings to other populations, and further research in diverse ancestral groups is crucial to validate and extend these results. Finally, the findings from this study are statistical in nature and do not establish definitive causality. The identified loci, genes (such as APOC3), and causal risk factors from MR analysis represent high-confidence hypotheses that require extensive in vivo and in vitro functional validation to elucidate their precise biological roles in the pathogenesis of halitosis.
Conclusion
Leveraging genomic SEM, our novel mvGWAS elucidated the shared genetic architecture of halitosis via a latent common factor. Employing a suite of post-GWAS methodologies, we robustly identified 23 genome-wide significant SNP loci, all previously unreported in the context of this shared HCF. Furthermore, integrating sCCA-TWAS with FOCUS, we precisely pinpointed APOC3 as a high-confidence candidate causal gene. Genetic correlation and MR analyses further illuminated the shared genetic architecture underlying HCF and multiple traits, notably neuropsychiatric characteristics and inflammatory conditions. Moreover, through MR, we identified numerous putative causal risk factors, including BMI, smoking, depression, and cognitive function. Despite the inherent limitations of the approach, this work provides a novel and comprehensive map of the genetic landscape of halitosis-related disorders, offering a rich set of hypotheses for future functional and clinical investigation.
Data availability
The datasets analyzed during the current study are publicly available. Details of each GWAS are provided in Supplementary Table S1.
References
Silva, M. F. et al. Estimated prevalence of halitosis: A systematic review and meta-regression analysis. Clin. Oral Investig. 22, 47–55 (2018).
Wu, J. et al. Prevalence, risk factors, sources, measurement and treatment - a review of the literature. Aust Dent. J. 65, 4–11 (2020).
Ortiz, V., Filippi, A. & Halitosis Monogr. Oral Sci. 29, 195–200 (2021).
Bollen, C. M. L. & Beikler, T. Halitosis: the multidisciplinary approach. Int. J. Oral Sci. 4, 55–63 (2012).
Mei, H. et al. hTAS2R38 polymorphisms modulate oral microbiota and influence the prevalence and treatment outcome of halitosis. Microbiome 13, 85 (2025).
Li, Z. et al. Halitosis: Etiology, prevention, and the role of microbiota. Clin. Oral Investig. 27, 6383–6393 (2023).
Mokeem, S. A. & Halitosis A review of the etiologic factors and association with systemic conditions and its management. J. Contemp. Dent. Pract. 15, 806–811 (2014).
Nini, W., Chen, L., Jinmei, Z., Lufei, W. & Jingmei, Y. The association between halitosis and periodontitis: A systematic review and meta-analysis. Clin. Oral Investig. 28, 341 (2024).
Kudo, Y. et al. Changes in halitosis value before and after Helicobacter pylori eradication: a single-institutional prospective study. J. Gastroenterol. Hepatol. 37, 928–932 (2022).
Islam, W. T., Azhar, A., Ahmed, T. F. & Shaikh, A. C. Investigating the prevalence of halitosis and its associated factors amongst the general population of karachi, Pakistan. JPMA J. Pak Med. Assoc. 74, S79–S84 (2024).
Zhang, Y., Lo, K. L., Liman, A. N., Feng, X. P. & Ye, W. Tongue-coating microbial and metabolic characteristics in halitosis. J. Dent. Res. 103, 484–493 (2024).
Nakano, Y., Yoshimura, M. & Koga, T. Correlation between oral Malodor and periodontal bacteria. Microbes Infect. 4, 679–683 (2002).
Foo, L. H., Balan, P., Pang, L. M., Laine, M. L. & Seneviratne, C. J. Role of the oral microbiome, metabolic pathways, and novel diagnostic tools in intra-oral halitosis: A comprehensive update. Crit. Rev. Microbiol. 47, 359–375 (2021).
Tam, V. et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20, 467–484 (2019).
Wu, Y. et al. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other Gastrointestinal disorders and depression. Nat. Commun. 12, 1146 (2021).
Shungin, D. et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat. Commun. 10, 2773 (2019).
Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 3, 513–525 (2019).
Lee, Y. H., Shin, S. I. & Hong, J. Y. Investigation of volatile sulfur compound level and halitosis in patients with gingivitis and periodontitis. Sci. Rep. 13, 13175 (2023).
van den Broek, A. M. W. T. & Feenstra, L. Baat, C. A review of the current literature on aetiology and measurement methods of halitosis. J. Dent. 35, 627–635 (2007). de.
Nakano, Y., Yoshimura, M. & Koga, T. Methyl mercaptan production by periodontal bacteria. Int. Dent. J. 52 (Suppl 3), 217–220 (2002).
Moshkowitz, M., Horowitz, N., Leshno, M. & Halpern, Z. Halitosis and gastroesophageal reflux disease: A possible association. Oral Dis. 13, 581–585 (2007).
Adler, I. et al. Helicobacter pylori and oral pathology: relationship with the gastric infection. World J. Gastroenterol. 20, 9922–9935 (2014).
Hur, H. J. et al. Association of polygenic variants with type 2 diabetes risk and their interaction with lifestyles in Asians. Nutrients 14, 3222 (2022).
Akdeniz, B. C. et al. Finemap-MiXeR: A variational bayesian approach for genetic finemapping. PLOS Genet. 20, e1011372 (2024).
Lu, M. et al. TWAS atlas: A curated knowledgebase of transcriptome-wide association studies. Nucleic Acids Res. 51, D1179–D1187 (2023).
Timshel, P. N., Thompson, J. J. & Pers, T. H. Genetic mapping of etiologic brain cell types for obesity. Elife 9, e55851 (2020).
Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula muris. Nature 562, 367–372 (2018).
Zheng, T. et al. Inflammatory cytokines mediating the effect of oral lichen planus on oral cavity cancer risk: A univariable and multivariable Mendelian randomization study. BMC Oral Health. 24, 375 (2024).
Ge, T., Chen, C. Y., Ni, Y., Feng, Y. C. A. & Smoller, J. W. Polygenic prediction via bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Dundar, A. & Sengun, A. Dental approach to erosive tooth wear in gastroesophageal reflux disease. Afr. Health Sci. 14, 481–486 (2014).
Yoshikawa, H. et al. Oral symptoms including dental erosion in gastroesophageal reflux disease are associated with decreased salivary flow volume and swallowing function. J. Gastroenterol. 47, 412–420 (2012).
Hait, E. J. & McDonald, D. R. Impact of gastroesophageal reflux disease on mucosal immunity and atopic disorders. Clin. Rev. Allergy Immunol. 57, 213–225 (2019).
Liu, J., Li, J., Yuan, G., Cao, T. & He, X. Relationship between sjogren’s syndrome and gastroesophageal reflux: A bidirectional Mendelian randomization study. Sci. Rep. 14, 15400 (2024).
Bhat, S. et al. Mono-allelic KCNB2 variants lead to a neurodevelopmental syndrome caused by altered channel inactivation. Am. J. Hum. Genet. 111, 761–777 (2024).
Alliey-Rodriguez, N. et al. NRXN1 is associated with enlargement of the Temporal Horns of the lateral ventricles in psychosis. Transl Psychiatry. 9, 230 (2019).
Shaaban, S. et al. Genome-wide association study identifies a susceptibility locus for comitant Esotropia and suggests a parent-of-origin effect. Invest. Ophthalmol. Vis. Sci. 59, 4054–4064 (2018).
Wu, Y. et al. Genome-wide association study of medication-use and associated disease in the UK biobank. Nat. Commun. 10, 1891 (2019).
Kim, S. et al. Shared genetic architectures of subjective well-being in East Asian and European ancestry populations. Nat. Hum. Behav. 6, 1014–1026 (2022).
Okbay, A. et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat. Genet. 54, 437–449 (2022).
Jong, M. C., Hofker, M. H. & Havekes, L. M. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler. Thromb. Vasc Biol. 19, 472–484 (1999).
Chebli, J., Larouche, M. & Gaudet, D. APOC3 SiRNA and ASO therapy for dyslipidemia. Curr. Opin. Endocrinol. Diabetes Obes. 31, 70–77 (2024).
D’Erasmo, L., Di Costanzo, A., Gallo, A., Bruckert, E. & Arca, M. ApoCIII: A multifaceted protein in cardiometabolic disease. Metab. Clin. Exp. 113, 154395 (2020).
van Dierendonck, X. A. M. H. et al. Triglyceride breakdown from lipid droplets regulates the inflammatory response in macrophages. Proc. Natl. Acad. Sci. U. S. A. 119, e2114739119 (2022).
Coomer, A. O., Black, F., Greystoke, A., Munkley, J. & Elliott, D. J. Alternative splicing in lung cancer. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 194388 (2019).
Mourtada-Maarabouni, M. & Williams, G. T. RBM5/LUCA-15–tumour suppression by control of apoptosis and the cell cycle? ScientificWorldJournal 2, 1885–1890 (2002).
Maarabouni, M. M. & Williams, G. T. The antiapoptotic RBM5/LUCA-15/H37 gene and its role in apoptosis and human cancer: Research update. ScientificWorldJournal 6, 1705–1712 (2006).
Cademartori, M. G., Gastal, M. T., Nascimento, G. G., Demarco, F. F. & Corrêa, M. B. Is depression associated with oral health outcomes in adults and elders? A systematic review and meta-analysis. Clin. Oral Investig. 22, 2685–2702 (2018).
Rivier, C. A. et al. Association of poor oral health with neuroimaging markers of white matter injury in middle-aged participants in the UK biobank. Neurology 102, e208010 (2024).
Hajishengallis, G. & Chavakis, T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat. Rev. Immunol. 21, 426–440 (2021).
Jia, L. et al. Porphyromonas gingivalis aggravates colitis via a gut microbiota-linoleic acid metabolism-Th17/treg cell balance axis. Nat. Commun. 15, 1617 (2024).
Rosenberg, M., Knaan, T. & Cohen, D. Association among bad breath, body mass index, and alcohol intake. J. Dent. Res. 86, 997–1000 (2007).
Ford, P. J. & Rich, A. M. Tobacco use and oral health. Addict. (abingdon Engl). 116, 3531–3540 (2021).
Vali, A. et al. Relationship between subjective halitosis and psychological factors. Int. Dent. J. 65, 120–126 (2015).
Gu, X. X., Jia, H. J. & Qian, X. X. Job stress can lead to intestinal inflammation and subsequent gut-originated extraoral halitosis. Oral Dis. https://doi.org/10.1111/odi.15174 (2024).
Pham, T. A. V., Ueno, M., Shinada, K. & Kawaguchi, Y. Comparison between self-perceived and clinical oral Malodor. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 113, 70–80 (2012).
Kanli, A., Kanbur, N. O., Dural, S. & Derman, O. Effects of oral health behaviors and socioeconomic factors on a group of Turkish adolescents. Quintessence Int. (berl Ger. : 1985). 39, e26–32 (2008).
Samnieng, P. et al. The relationship between seven health practices and oral health status in community-dwelling elderly Thai. Gerodontology 30, 254–261 (2013).
Acknowledgements
We gratefully acknowledge the original authors for providing the datasets used in this research.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
YH conceived and designed the study. YH, ZZ, and BL conducted the analysis and wrote the paper. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study used GWAS data from prior research. Ethical approvals and consents were obtained in the original studies.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, Y., Zhao, Z. & Lian, B. Genomic structural equation modeling identifies shared genetic architecture and novel loci for halitosis. Sci Rep 15, 36405 (2025). https://doi.org/10.1038/s41598-025-20316-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-20316-y