Abstract
The spectrum of insecticidal activity of the Cry1B.3, Cry1Da_7, and Vip3Cb1 proteins targeting lepidopteran insect pests produced by MON 89151 cotton was characterized with microbially-produced protein-incorporated diet bioassays against selected insect species from four orders and nine families. Cry1B.3 and Cry1Da_7 are Bacillus thuringiensis (Bt) insecticidal proteins, whereas the Vip3Cb1 protein is derived from Paenibacillus spp. in the P. popilliae-containing clade. Notably, Cry1B.3 has the same functional region as the Cry1B.2 protein in MON 94637 soybean, and Cry1Da_7 is identical to the protein expressed in MON 95379 maize; therefore, existing spectrum of activity data for Cry1B.2 and Cry1Da_7 were utilized to inform the characterization of Cry1B.3 and Cry1Da_7 in MON 89151 cotton. Overall, our results demonstrate that Cry1B.3, Cry1Da_7, and Vip3Cb1 exhibit a narrow spectrum of activity limited to the lepidopteran pests tested. Thus, these insecticidal proteins are valuable additions to the cotton trait portfolio to manage lepidopteran pests with a high degree of specificity.
Similar content being viewed by others
Introduction
Cotton, Gossypium hirsutum L., is a globally important crop that supplies essential raw materials to various industries. However, cotton production faces significant yield losses due to insect pests, particularly from the orders Hemiptera, Thysanoptera, and Lepidoptera1,2,3. Notably, damage by lepidopteran pests to various cotton tissues across the growing season can be particularly devastating4. Historically, growers have relied on foliar applications of broad-spectrum chemical insecticides for pest management5. However, the increasing costs of application and the development of resistance to some insecticides have resulted in the need to further develop alternative pest management technologies for cotton that are consistent with integrated pest management strategies6. In 1996, Monsanto® (now Bayer CropScience LP) introduced the first genetically modified (GM) cotton crop, trademarked as Bollgard® (MON 531). This trait produced the insecticidal crystal (Cry) Cry1Ac protein from Bacillus thuringiensis (Bt) for the control of economically important lepidopteran pests including tobacco budworm (TBW), Heliothis virescens (Fabricius) (Lepidoptera: Noctuidae), corn earworm (CEW), Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae), and pink bollworm, Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae)7.
The introduction of Bollgard® transformed and improved cotton protection against lepidopteran pests, providing environmental and economic benefits through reduced insecticide applications and decreased yield losses5,8,9. The value added by insecticidal traits promoted a rapid adoption of GM cotton worldwide10. However, the main threat to existing cotton traits is the emergence of insect resistance11,12. To address this challenge, various practices are implemented in cotton cultivation to delay the development of resistance in target pests13, thereby extending the durability of the deployed traits in the field. One of those practices is the development of insect-protected events with new modes of action, which are then commercialized as a single trait product or combined in a pyramid strategy with multiple traits14,15. The Bayer CropScience LP cotton trait portfolio has been expanded over time with the addition of Bollgard® 2 cotton (MON 15985) expressing Cry1Ac and Cry2Ab2 proteins in a pyramid7, and Bollgard® 3 (MON 15985 × COT102) expressing a pyramid of Cry1Ac, Cry2Ab2, and the vegetative (Vip) Vip3Aa19 Bt proteins16. Recently, Bayer CropScience LP has developed Bollgard® 4 (MON 89151 × MON 15947), the fourth generation GM cotton product, that provides effective protection against feeding from lepidopteran pests such as TBW, CEW, and fall armyworm (FAW), Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). MON 15947 is a segregant line of MON 15985 cotton developed through a natural breeding selection process that produces only the Cry2Ab2 protein, while MON 89151 expresses two Bt insecticidal proteins, Cry1B.3 (GenBank accession number PP436398), and Cry1Da_7 (GenBank accession number MN045176), as well as the Vip3Cb1 (GenBank accession number PP436397) protein derived from Paenibacillus spp. in the P. popilliae-containing clade.
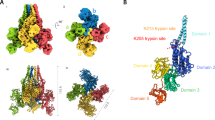
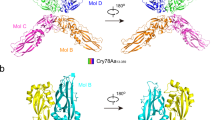
The Cry1B.3 protein, expressed in MON 89151, shares identical sequences in domains I, II, and III with the Cry1B.2 (GenBank accession number UWS13095) protein expressed in MON 94637 soybean17. The only difference between these two proteins lies in their protoxin sequences (Fig. 1). Since domains I–III are responsible for insecticidal activity18,19 and the protoxin domain is cleaved and removed before the protein becomes active20, both Cry1B.3 and Cry1B.2 are expected to exhibit similar activity spectrum profiles. The Cry1Da_7 protein is also expressed in MON 95379 maize21, sharing 100% sequence identity to Cry1Da_7 in MON 89151 cotton. As members of the Cry1 protein family, Cry1B.2, Cry1B.3, and Cry1Da_7 have a similar mode of action to previously commercialized Cry proteins, providing protection against lepidopteran pests17,22. Upon ingestion, Bt proteins undergo solubilization and proteolytic activation into a three-domain active form (tryptic core), followed by interactions with cell surface receptors, oligomerization, pore formation, and midgut cell lysis23,24. In contrast, the Vip3Cb1 protein exhibits sequence and structural similarities to other members of the Vip3C protein family. The mode of action of Vip3Cb1 resembles that of commercially available Vip3Aa proteins25,26, which have been utilized in various GM crops for their effectiveness against lepidopteran pests27,28. Vip3 proteins exert their insecticidal toxicity in a sequence of events similar to Cry proteins, but evidence suggests Vip3A may target different midgut binding sites from Cry proteins29,30,31. Expressing Cry1B.3, Cry1Da_7, and Vip3Cb1 proteins as part of event MON 89151 in Bollgard® 4 is designed to provide value to growers by diversifying their toolbox for lepidopteran pest management. However, prior to registration and introduction of the product into the market, a detailed characterization of each protein’s spectrum of activity is warranted to help inform selection of species for subsequent testing with non-target organisms for ecological risk assessment32.
Domain architecture and sequence identity of the Cry1B.2 and Cry1B.3 proteins. The core insecticidal domains (I–III) of Cry1B.2 and Cry1B.3 are 100% identical to each other. These domains are derived from and share 100% sequence identity with Domains I and II of Cry1Be (GenBank: AAC32850) and Domain III of Cry1Ka2 (GenBank: AEH31431). The protoxin regions, however, differ. The protoxin of Cry1B.2 is derived from Cry1Ab (GenBank: AAA22561), whereas the protoxin of Cry1B.3 is derived from Cry1Be (GenBank: AAC32850). As a result, while each protoxin is identical to its respective donor, the protoxin regions of Cry1B.2 and Cry1B.3 are only 74% identical to each other.
The typical approach to characterizing the spectrum of activity of insecticidal traits produced by GM crops includes testing target insect pests and insect species that are taxonomically related (both closely and distantly) to these pests32,33,34. To characterize the spectrum of activity for the microbially-produced Cry1B.3, Cry1Da_7, and Vip3Cb1 proteins, selected insect species from four orders and nine families were evaluated using diet incorporation bioassays. Given that Cry1B.3 and Cry1B.2 have the same functional region (i.e., domain I–III), and that Cry1Da_7 has an identical sequence between crops (i.e., cotton and maize), existing activity spectrum data from Cry1B.2 (surrogate for Cry1B.3) and Cry1Da_7 expressed in soybean and maize, respectively, were leveraged for the characterization of Cry1B.3 and Cry1Da_7 in MON 89151. Overall, our results demonstrate that the three proteins in MON 89151 exhibit a narrow spectrum of activity, primarily targeting lepidopteran pests.
Results
Bioassays with Cry1B.2 protein
The Cry1B.2 protein showed insecticidal activity after 7-day dietary exposure bioassays against the tested target lepidopteran species. The EC50 value (i.e., the effective concentration of the test substance required to produce a 50% reduction in body mass of the larvae relative to the control after seven days) was estimated to be 0.029 µg Cry1B.2/mL diet in velvetbean caterpillar (VBC), Anticarsia gemmatalis Hübner (Lepidoptera: Erebidae), 6.2 µg Cry1B.2/mL diet in soybean looper (SBL), Chrysodeixis includens (Walker) (Lepidoptera: Noctuidae), 4.0 µg Cry1B.2/mL diet in FAW, 1.8 µg Cry1B.2/mL diet in CEW, and 0.47 µg Cry1B.2/mL diet in European corn borer (ECB), Ostrinia nubilalis (Hübner) (Lepidoptera: Crambidae) (Table 1).
For all non-lepidopteran species bioassays, the nominal (i.e., target) concentration of 700 µg Cry1B.2/mL diet at the beginning of the bioassay (Day 0) was confirmed, except for the Mexican bean beetle (MBB), Epilachna varivestis Mulsant (Coleoptera: Coccinellidae) bioassay. The nominal concentration of Cry1B.2 in diet at bioassay completion (Day End), or Day 7 for common fruit fly (CFF), Drosophila melanogaster Meigen (Diptera: Drosophilidae), was confirmed for all species except in the western corn rootworm (WCR), Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) bioassay. For MBB and WCR, the Cry1B.2 concentration was adjusted at Day 0 and at Day End, respectively, based on the activity observed when SBL, the sensitive testing species, was exposed to a 2-fold geometric dose series of the test diets, and the reference protein standard. The adjusted Cry1B.2 concentrations were estimated to be 313.79 and 483.64 µg Cry1B.2/mL diet for MBB and WCR, respectively (Table S1). No buffer effect in survival or mean body mass was observed in all non-lepidopteran bioassays. Mean body mass of MBB in the untreated control was statistically lower than the buffer control (p = 0.010; Table S2), indicating no buffer effect. There were no effects on survival or mean body mass when insects were exposed to the Cry1B.2 protein across all species (Table 1). There was no statistical difference in survival or mean body mass between Cry1B.2 treatments and the buffer control for all species (p > 0.05; Table S2), except WCR. WCR showed significantly higher mean body mass in the 350 µg Cry1B.2/mL diet treatment than in the buffer control (p = 0.015; Table S2), but not in the 700 µg Cry1B.2/mL diet treatment, the highest concentration tested.
Bioassays with Cry1B.3 protein
The Cry1B.3 protein showed insecticidal activity after 7-day dietary exposure bioassays against the tested lepidopteran species (Table 1). The EC50 value was estimated to be 5.5 µg Cry1B.3/mL diet in FAW, 1.6 µg Cry1B.3/mL diet in CEW, and 0.26 µg Cry1B.3/mL diet in ECB (Table 1).
Bioassays with Cry1Da_7 protein
The Cry1Da_7 protein showed insecticidal activity after 7-day dietary exposure bioassays against the tested target lepidopteran species. The EC50 value was estimated to be 0.096 µg Cry1Da_7/mL diet in FAW, 0.042 µg Cry1Da_7/mL diet in CEW, and 11 µg Cry1Da_7/mL diet in ECB (Table 2).
For all non-lepidopteran species bioassays, the nominal concentration at Day 0, Day 3 and Day End of 50 µg Cry1Da_7/mL diet was only confirmed for western tarnished plant bug (WTP), Lygus hesperus Knight (Hemiptera: Miridae) and the Neotropical brown stink bug (NBS), Euschistus heros (Fabricius) (Hemiptera: Pentatomidae) bioassays (Table S3). Bioassays with WCR, southern corn rootworm (SCR), Diabrotica undecimpunctata howardi Barber (Coleoptera: Chrysomelidae), Colorado potato beetle (CPB), Leptinotarsa decemlineata (Say) (Coleoptera: Chrysomelidae), and MBB were also initially prepared at a nominal concentration of 50 µg Cry1Da_7/mL diet (named assay 1 in Table S3). However, due to low Cry1Da_7 stability in these diets, the Cry1Da_7 nominal concentration was increased to 500 µg Cry1Da_7/mL diet for all consecutive bioassays (named assay 2 in Table S3). The nominal concentration at Day 0, Day 3 and Day End for all consecutive bioassays using 500 µg Cry1Da_7/mL diet was not confirmed. The Cry1Da_7 concentration was adjusted based on the activity observed when FAW, the sensitive testing species, was exposed to a 2-fold geometric dose series of the test diets and the reference protein standard. The adjusted Cry1Da_7 concentrations were estimated to be 106 µg Cry1Da_7/mL diet for WCR, 65 µg Cry1Da_7/mL diet for SCR, 58 µg Cry1Da_7/mL diet for CPB, and to 65 µg Cry1Da_7/mL diet for MBB (Table S3). No buffer effect in survival was observed in all non-lepidopteran bioassays except MBB assay 2. In this bioassay, MBB survival in the buffer control was significantly lower than the untreated control (p = 0.024; Table S4), indicating a buffer effect. In general, there were no effects on survival when non-lepidopteran insects were exposed to the Cry1Da_7 protein (Table 2). There was one significant difference in MBB in assay 2, but the survival observed at 500 µg Cry1Da_7/mL diet was higher than in the buffer control (p = 0.024; Table S4).
Bioassays with Vip3Cb1 protein
The Vip3Cb1 protein showed insecticidal activity after 7-day dietary exposure bioassays against the tested target lepidopteran species. The EC50 value was estimated to be 0.054 µg Vip3Cb1/mL diet in FAW, 0.20 µg Vip3Cb1/mL diet in CEW, 0.28 µg Vip3Cb1/mL diet in ECB, and 0.14 µg Vip3Cb1/mL diet in TBW (Table 3).
For all non-lepidopteran species bioassays, the nominal concentration of 250 µg Vip3Cb1/mL diet at Day 0 was confirmed. The nominal concentration of Vip3Cb1 in diet at Day End, or Day 6 for CFF, was also confirmed for all bioassays except for SCR and CPB. The Vip3Cb1 concentration at Day End, for SCR and CPB, was adjusted based on the activity observed when FAW, the sensitive testing species, was exposed to a 2-fold geometric dose series of the test diets and the reference protein standard, and was calculated using a time-weighted mean35. The adjusted Vip3Cb1 concentrations were estimated to be 141.7 and 170.5 µg Vip3Cb1/mL diet for SCR and CPB, respectively (Table S5). No buffer effect in survival or mean body mass was observed in all non-lepidopteran bioassays, except CPB. For CPB, the mean body mass in the buffer control was significantly lower than the untreated control (p = 0.023; Table S6), indicating a buffer effect. In general, there were no effects when non-lepidopteran insects were exposed to the Vip3Cb1 protein, but an effect was observed in CFF and MBB (Table 3). There was no statistical difference in survival between Vip3Cb1 treatments and the buffer control for all species (p > 0.05; Table S6), except CFF. In CFF, survival was significantly lower when exposed to 250 µg Vip3Cb1/mL (p = 0.037; Table S6), but no effect on survival was observed at 50 or 100 µg Vip3Cb1/mL (p > 0.05; Table S6). Furthermore, there was no statistical difference in mean body mass or development time to pupa for CFF between treatments and the buffer control for all species (p > 0.05; Table S6), except MBB. Mean body mass of MBB was significantly lower when exposed to 250 µg Vip3Cb1/mL, the highest concentration tested (p = 0.021; Table S6), but no effect on mean body mass was observed at 100 µg Vip3Cb1/mL (p > 0.05; Table S6).
Discussion
Characterizing the spectrum of activity of insecticidal substances produced by GM crops is essential to inform subsequent testing with non-target organisms for ecological risk assessment. The current approach for spectrum of activity testing aims to include a range of target and taxonomically diverse additional insect taxa32,33,34 which are readily available with established and reliable laboratory bioassays. The utilization of spectrum of activity data from Cry1B.2 in MON 94637 soybean for Cry1B.3 was feasible due to the high degree of similarity between the two proteins. The tryptic core of Cry1B.2, comprising domains I–III is identical to that of Cry1B.3, and these proteins differ only in the protoxin domain (Fig. 1). Since the tryptic core is widely recognized as the active component of three-domain Cry proteins, and protoxins are not known to contribute to protein activity18,19,36, Cry1B.2 should serve as a suitable surrogate for the Cry1B.3 protein. This expectation is further supported by the similar activity levels (i.e., EC50 values within 2-fold of each other, which is within expected bioassay variation) observed for the Cry1B.2 and Cry1B.3 proteins against key lepidopteran pests, including CEW, FAW, and ECB (Table 1). Similarly, spectrum of activity data generated for MON 95379 maize was leveraged for Cry1Da_7, since the Cry1Da_7 produced by MON 95379 maize shares 100% sequence identity with that found in MON 89151 cotton.
Target and other insect species across four orders and nine families were evaluated against Cry1B.2 in MON 94637 soybean (surrogate for Cry1B.3 in MON 89151 cotton), Cry1Da_7 in MON 95379 maize, and Vip3Cb1 proteins. All three proteins were tested against the target lepidopterans FAW, CEW, and ECB. Cry1B.2 was additionally tested against VBC and SBL, while Vip3Cb1 was also tested against TBW (Table 4). The inclusion of VBC and SBL reflects the target pests listed for MON 94637 soybean, whereas TBW is listed as a target of MON 89151 cotton.
Our results demonstrate that Cry1B.2, Cry1Da_7, and Vip3Cb1 proteins exhibit high activity against target lepidopteran pest species. FAW, CEW, and ECB showed EC50 values ranging from 0.04 to 11 µg/mL for all three proteins. Cry1B.2 activity was further evaluated against VBC and SBL, resulting in EC50 values of 0.029 and 6.2 µg/mL, respectively. Vip3Cb1 activity was also assessed against TBW, with an EC50 value of 0.14 µg/mL. In addition to these results, Cry1Da_7 has previously demonstrated efficacy against FAW strains resistant to Cry1F and Vip3A23. Cry1B.2 has also been shown to be active against SBL, VBC, southern armyworm, Spodoptera eridania (Stoll), and black armyworm, Spodoptera cosmioides (Walker) (both Lepidoptera: Noctuidae)17. Furthermore, Vip3Cb1 has shown in-planta activity against CEW, FAW, ECB, and the southwestern corn borer, Diatraea grandiosella Dyar (Lepidoptera: Crambidae) when expressed in maize as a fusion protein with a chloroplast-targeting peptide (CTP) at its N-terminus26. The fusion protein is expected to be processed with the CTP cleaved off during translocation into the chloroplast. Small variations at the N-termini of chloroplast-targeted proteins are often observed due to incomplete processing of the CTP, but such variations are not expected to alter protein activity, as reported previously37,38.
Bioassays with Cry1B.2, Cry1Da_7, and Vip3Cb1 for non-lepidopteran species were performed at concentrations (Tables 1, 2 and 3) comparable to or above the estimated environmental concentration39 in MON 89151 cotton. Protein stability was assessed in all non-lepidopteran artificial diets. At Day End (or Day 7 and 6 for CFF in Cry1B.2 and Vip3Cb1, respectively), Cry1B.2 and Vip3Cb1 were shown to be stable in all diets except WCR and MBB diets for Cry1B.2, and SCR and CPB diets for Vip3Cb1. Despite these proteins being less stable in these diets, the species were still tested at concentrations close to or above estimated environmental Cry1B.3 and Vip3Cb1 concentration in MON 89151 cotton39. On the other hand, Cry1Da_7 was found to be stable only in WTP and NBS diets, with high levels of degradation observed in all of the other three diets (Cry1Da_7 stability in MBB diet was estimated using the adjusted protein concentration for SCR, since these two species use the same diet; Table S3). To compensate for the degradation, WCR, SCR, CPB, and MBB bioassays were repeated with an increased Cry1Da_7 concentration, from 50 to 500 µg Cry1Da_7/mL diet (Table S4). This approach ensured that sufficient Cry1Da_7 protein was present at the end of these studies to achieve concentrations close to or above estimated environmental Cry1Da_7 concentration in MON 89151 cotton. In addition to confirming sufficient exposure levels through protein stability testing, no activity on survival or development was observed across all non-lepidopteran species with Cry1B.2, Cry1Da_7, and Vip3Cb1 (except MBB and CFF), (Tables 1, 2 and 3). The minor decrease in mean body mass of MBB and survival of CFF was restricted to the highest concentration tested (i.e., 250 µg Vip3Cb1/mL diet, Table S6) and well above estimated environmental concentration, suggesting that these effects may not be biologically relevant.
Considering the high insecticidal activity observed against tested lepidopteran pests (i.e., FAW, CEW, TBW, ECB), along with the absence of activity on all other species at exposure levels close to or above estimated environmental concentration, it is evident that Cry1Da_7, Cry1B.3, and Vip3Cb1 in MON 89151 exhibit a narrow spectrum of activity primarily within Lepidoptera (see also Cry1Da_7 maize EPA registration decision40 that includes other non-lepidopteran species tested). Consequently, these insecticidal proteins represent valuable additions to the cotton trait portfolio, providing growers with essential tools to protect their crops from lepidopteran damage.
Materials and methods
Test substances used in bioassays
The coding sequence for Vip3Cb1 was cloned into the pET SUMO (Thermo Fisher Scientific, Walthman, MA) vector and expressed in Rosetta 2 (DE3) E. coli (Sigma-Aldrich, St. Louis, MO). Fermentation was conducted following the SUMO tag protein method described by Wang et al.37, but at a temperature of 20 °C and without the addition of iron. DNA sequencing of the Vip3Cb1 gene was confirmed both before and after fermentation. The recombinant Vip3Cb1 protein was purified from ~ 9 kg E. coli cell paste. Briefly, the thawed cell paste was suspended in lysis buffer containing 50 mM Tris, 300 mM NaCl, 2 mM MgCl2, 10 mM imidazole, 0.5 mM Dithiothreitol (DTT), 50 U/mL Benzonase, 0.2 g/L lysozyme, 2 mM benzamidine HCl, 0.5 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), pH 8.0. The homogenized E. coli cells were lysed by passing through a Microfluidics (IDEX Material Processing Technologies, Westwood, MA) microfluidizer at ~ 20,000 pounds per square inch (psi). The resulting extract was clarified by centrifugation. The clarified extract was diluted and loaded to Ni-NTA (QIAGEN, Germantown, MD) affinity column and step-eluted with 40-, 200-, and 300-mM imidazole in 25 mM sodium carbonate, 300 mM NaCl and 0.5 mM DTT, pH 9.0. The 6xHis-SUMO Vip3Cb1-containing fractions were diluted and subjected to proteolysis by 6xHis-tagged SUMO protease (Bayer CropScience LP, Chesterfield, MO) to remove the 6xHis-SUMO tag. Then the protein solution was passed through the Ni-NTA column to capture the cleaved 6xHis-SUMO tag and the His-tagged SUMO protease. The tag-free Vip3Cb1 retained in the flowthrough was collected and then concentrated by ultrafiltration, and buffer exchanged by diafiltration into 25 mM sodium carbonate/bicarbonate, 25 mM NaCl, pH 10 and stored at -80 °C until use.
The recombinant Cry1Da_7 protein was purified from Bt cell paste containing a plasmid expressing the Cry1Da_7 protein. The cell paste was washed with 10 mM Tris, 1% Triton X-100, 0.5 M NaCl, pH 8 then extracted by 200 mM sodium carbonate, 10 mM DTT, 1 mM Ethylenediaminetetraacetic Acid (EDTA), 2 mM benzamidine, 2 mM Phenylmethylsulfonyl Fluoride (PMSF), plus benzonase. The clarified extract was buffer exchanged into 10 mM 3-(cyclohexylamino)-1-propane-sulfonic acid (CAPS), 1 mM DTT, 1 mM benzamidine HCl by diafiltration then sequentially passed through Q Sepharose Fast Flow resin to remove the impurities and Diethylaminoethyl (DEAE) Sepharose Fast Flow resin (Cytiva, Marlborough, MA) following a step elution with 10 mM CAPS, 1 mM DTT, 1 mM benzamidine, 250 mM NaCl. The Cry1Da_7 containing solution was buffer exchanged into 25 mM sodium carbonate, pH 10.5 by diafiltration and further concentrated by ultrafiltration. The resulting Cry1Da_7 protein solution was stored at -80 °C until use.
The coding sequence for Cry1B.2 was cloned into a Bt protein expression vector and expressed in the Bt strain EG10650 according to the method described by Chen et al.17. DNA sequence of Cry1B.2 gene was confirmed before and after fermentation. The recombinant Cry1B.2 protein was purified from ~ 7 kg Bt cell paste. The cell paste was washed four times sequentially with 50 mM Tris, 5 mM MgCl2, 1 mM Benzamidine, pH 8.0, 50 mM Tris, 5 mM MgCl2, 1 mM Benzamidine, 2% Triton X-100, pH 8.0, 50 mM Tris, 1.0% Sodium Deoxycholate, pH 8.0 and 50 mM sodium carbonate/bicarbonate, 250 mM NaCl, pH 10.0. The washed Bt pellet was extracted in 50 mM CAPS, 10 mM DTT, 1 mM PMSF, 1 mM Benzamidine, 100 µM tosyl phenylalanyl chloromethyl ketone (TPCK), pH 11.5. The clarified extract was fractionated using ammonium sulfate precipitation. The resulting pellet was redissolved in 20 mM CAPS, 10 mM DTT, 1 mM PMSF, 1 mM Benzamidine, 100 µM TPCK, pH 11.5, diluted and then passed through Diethylaminoethyl (DEAE) Sepharose fast flow resin. The Cry1B.2 containing fractions were eluted with 20 mM CAPS, 10 mM DTT, 1 mM Benzamidine, 250 mM NaCl, pH 11.5, concentrated by ultrafiltration, and buffer exchanged by diafiltration into 20 mM CAPS, pH 11.2 then stored at -80 °C until use. A similar production and purification process was applied to generate Cry1B.3 used against the three lepidopterans described in Table 1.
Selection of insect species and bioassay procedures
The insects selected for bioassays were either the intended target insect pest species for MON 95379 maize, MON 94637 soybean, MON 89151 cotton, or non-lepidopteran insect pest species representing relevant taxa found in the agro-ecosystem. Bioassay methodologies allowed ad libitum feeding with protein-incorporated artificial species-specific diet under laboratory conditions in order to evaluate survival, body mass, and development time (Table 4). All insects were exposed orally to proteins in diet, which is the most ecologically relevant route of potential exposure to plant-incorporated protectants in the environment33,34,41. Bioassay methodologies followed established regulatory guidelines under Good Laboratory Practice requirements (40 CFR Part 160)42, and published methodologies in peer-reviewed literature43,44,45. In total, 14 insect species from four orders and nine families were evaluated against Cry1B.2 (12 species), Cry1B.3 (3 species), Cry1Da_7 (9 species) and Vip3Cb1 (11 species) (Table 4).
Lepidopteran species
Test concentrations for bioassays consisted of a 2-fold geometric series of protein dilutions, a buffer control, and an untreated control (only for Cry1B.2 and Cry1Da_7), which elicited a dose-response from each species and estimation of an EC50 value, the effective concentration of the test substance required to produce a 50% reduction in body mass of the larvae relative to the control after seven days. The buffer control and all protein treatments contained the same volume of buffer, whereas the untreated control contained only purified Milli-Q® grade water. The buffer control and untreated control treatments each had three in-bioassay replicates. Test concentrations were prepared by diluting each protein with buffer and mixing the dilution into an agar-based, multiple species diet. All diets were prepared, incorporated with protein, water and/or buffer and used on the day of bioassay initiation. The treatment diet was dispensed into 128-well trays with a volume of 0.5 or 1 mL per well, and a total of 16 or 24 wells per treatment, respectively. Each well was infested with a single lepidopteran larva. Larvae were fed ad libitum for seven days in an environmental chamber programmed at 27 °C, 60% relative humidity, and a lighting regime of 14:10 h light:dark (Table 4). A treatment was excluded from concentration-response modeling if there was > 20% contamination of infested wells, or < 3 survived larvae per treatment was observed. Statistical analysis was conducted using SAS46,47 software or a verified R/Shiny application48 for each bioassay using the endpoints shown in Table 4. Estimation of EC50 values was performed using a three-parameter logistic regression model.
Coleopteran, hemipteran and dipteran species
Each protein was incorporated into species-specific diet at two to four nominal concentrations (see Tables S2, S4, and S6) based on 1–2 fold of the 95th percentile protein concentration value, i.e., estimated environmental concentration, found in MON 95379 maize (for Cry1Da_7), MON 94637 soybean (for Cry1B.2), and MON 89151 cotton (for Vip3Cb1)39. Nominal concentration is defined as the amount of the protein added that results in a target protein concentration in the test diet, before any potential losses due to degradation, or other factors that could affect the actual concentration that insects may be exposed to throughout the duration of the bioassay. In addition to protein treatments, all bioassays had an untreated control (except in Cry1Da_7 MBB assay 1), a buffer control, and a positive control (except Cry1Da_7 bioassays when prepared at 500 µg/mL diet) (Table S4). The untreated control treatment was a species-specific diet mixed with purified Milli-Q® grade water. The buffer control treatment was a species-specific diet mixed with a volume of buffer equivalent to the volume of buffer in the highest protein level. The dietary toxin, potassium arsenate was used as the positive control treatment, and its concentration ranged from 200–500 µg/mL diet, based on the insect species. Each treatment diet included three or more in-bioassay replicates, except for the Cry1B.2 buffer control of CFF and the 20 µg/mL diet treatment of Cry1Da_7 WCR, which each had two in-assay replicates. Treatment diets were volumetrically dispersed into plates, trays, or encapsulated in parafilm diet domes as described in Tan et al.45, according to the species tested (see Table 4). All artificial diets were prepared, incorporated with protein, water, and/or buffer, and used on the day of bioassay initiation. One neonate (or one second instar for NBS) was infested per well or diet dome. Insects fed ad libitum for a specific period of time in an environmental chamber at a defined temperature, relative humidity, and light:dark photoperiod regime (Table 4). The survival observed in the untreated control showed whether the buffer affected insect survival, body mass or development, whereas the low survival observed in the positive control demonstrated that the insects could detect toxic effects attributed to the ingestion of a known gut toxin, via protein-incorporated diet bioassays. The statistical software SAS46,47 was used to calculate the means and standard errors of the endpoints shown in Table 4. Pairwise comparisons between the protein and the buffer control treatments and between the untreated control and the buffer control treatments were conducted within a linear (mixed, if needed) or generalized linear model and tested using t-tests or conducted using Fisher’s exact tests where appropriate at a 5% level of significance. All tests were two-sided pair-wise comparisons.
In addition, to assess protein stability in the diet throughout the duration of the bioassay, diet samples from the untreated control, buffer control, and protein-incorporated treatments were collected at the beginning (Day 0), periodically (Day 3 only for Cry1Da_7 test diets), and bioassay completion (Day End; for CFF Day End refers to Day 7 (Cry1B.2) and Day 6 (Vip3Cb1) collections rather than day of bioassay completion); and stored at -80 °C. Stock solutions were prepared by diluting the diet samples with additional buffer control diet and buffer to contain an equivalent amount of diet matrix and buffer as incorporated into the highest protein concentration treatment. A reference standard was created using the same lot of purified protein and containing an equivalent amount of diet matrix and buffer as the other stock solutions. All stock solutions were diluted into at least five non-zero dosing solutions in a 2-fold geometric dose series of the protein expected to elicit a response from the sensitive insect species, allowing the estimation of EC50 values with FAW for Vip3Cb1 and Cry1Da_7, and with SBL for Cry1B.2. Dosing solutions were mixed with an agar-based, multiple species diet and then dispensed in 1 mL aliquots into 16 wells per treatment level in a 128-well tray. Each well was infested with a single unfed larva. Larvae were allowed to feed ad libitum for a period of seven days in an environmental chamber at a temperature of 27 ± 5 °C, 60 ± 10% relative humidity, and a 14 h:10 h light:dark photoperiod. The number of surviving insects per treatment/concentration and the combined mass of the surviving insects per treatment/concentration was used to estimate EC50 values and their associated 95% confidence intervals (CI) with a three-parameter logistic regression model.
For Cry1B.2 and Vip3Cb1, the nominal concentration in the Day 0 test diet samples was confirmed by the overlapping 95% CI of the EC50 values for the Day 0 test diet sample and the reference standard. The nominal concentration in Day End test diet samples was confirmed by the overlapping 95% CI of the EC50 values from the Day End test diet samples and the Day 0 test diet samples. When the nominal concentration was not confirmed, a ratio of EC50 values for the test and reference diets was used to adjust for protein concentration. When necessary, a time-weighted mean calculation, accounting for the exponential decay of the test substance over the exposure period35, was applied to the concentration adjustment of Vip3Cb1.
For Cry1Da_7, the nominal concentration at bioassay completion was confirmed when the 95% CI of the reference standard, Day 0, Day 3, and Day End EC50 values overlapped with each other. When one of the 95% CI values did not overlap with the reference, a stability index was calculated by dividing the reference standard EC50 value by the largest EC50 value of the test diets. This stability index was used for protein concentration adjustment of Cry1Da_7.
Data availability
All relevant data are within this publication and its Supporting Information files.
References
Greene, J. K., Turnipseed, S. G., Sullivan, M. J. & May, O. L. Treatment thresholds for stink bugs (Hemiptera: Pentatomidae) in cotton. J. Econ. Entomol. 94, 403–409. https://doi.org/10.1603/0022-0493-94.2.403 (2001).
Smith, R. in Proceedings of the Beltwide Cotton Conference. 856–858 (National Cotton Council).
Cook, D. & Threet, M. in 2022 Beltwide Cotton Conferencesv. 145–200.
Luttrell, R. G. & Jackson, R. E. Helicoverpa zea and Bt cotton in the united States. GM Crops Food: Biotechnol. Agric. Food Chain. 3, 1–15 (2012).
Carpenter, J. E. & Gianessi, L. P. National Center for Food and Agricultural Policy, Washington, D.C., (2001).
Naranjo, S. E. Impacts of Bt Transgenic cotton on integrated pest management. J. Agric. Food Chem. 59, 5842–5851. https://doi.org/10.1021/jf102939c (2011).
Perlak, F. J. et al. Development and commercial use of Bollgard® cotton in the USA - Early promises versus today’s reality. Plant J. 27, 489–501 (2001).
Betz, F. S., Hammond, B. G. & Fuchs, R. L. Safety and advantages of Bacillus thuringiensis-protected plants to control insect pests. Regul. Toxicol. Pharmacol. 32, 156–173. https://doi.org/10.1006/rtph.2000.1426 (2000).
Brookes, G. & Barfoot, P. Global impact of biotech crops: environmental effects, 1996–2010. GM Crops Food: Biotechnol. Agric. Food Chain. 3, 129–137 (2012).
James, C. Brief 49 The International Service for the Acquisition of Agri-biotech Applications (Ithaca, 2014).
Dhurua, S. & Gujar, G. T. Field-evolved resistance to Bt toxin Cry1Ac in the Pink bollworm, Pectinophora Gossypiella (Saunders) (Lepidoptera: Gelechiidae), from India. Pest Manag. Sci. 67, 898–903. https://doi.org/10.1002/Ps.2127 (2011).
Zhang, H. N. et al. Early Warning of Cotton Bollworm Resistance Associated with Intensive Planting of Bt Cotton in China. PLoS ONE, 6 (2011).
Knight, K. M., Head, G. P. & Rogers, D. J. Successful development and implementation of a practical proactive resistance management plan for Bt cotton in Australia. Pest Manag. Sci. 77, 4262–4273. https://doi.org/10.1002/ps.6490 (2021).
Carrière, Y., Fabrick, J. A. & Tabashnik, B. E. Can pyramids and seed mixtures delay resistance to Bt crops? Trends Biotechnol. 34, 291–302. https://doi.org/10.1016/j.tibtech.2015.12.011 (2016).
Carrière, Y., Crickmore, N. & Tabashnik, B. E. Optimizing pyramided Transgenic Bt crops for sustainable pest management. Nat. Biotechnol. 33, 161 (2015).
Levine, S. L., Mueller, G. M. & Uffman, J. P. Assessing the potential for interaction between the insecticidal activity of two genetically engineered cotton events combined by conventional breeding: an example with COT102 × MON 15985. Regul. Toxicol. Pharmacol. 79, 35–41. https://doi.org/10.1016/j.yrtph.2016.05.003 (2016).
Chen, D. et al. Bacillus thuringiensis chimeric proteins Cry1A.2 and Cry1B.2 to control soybean lepidopteran pests: new domain combinations enhance insecticidal spectrum of activity and novel receptor contributions. PLoS ONE. 16, e0249150. https://doi.org/10.1371/journal.pone.0249150 (2021).
Palma, L., Munoz, D., Berry, C., Murillo, J. & Caballero, P. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins 6, 3296–3325. https://doi.org/10.3390/toxins6123296 (2014).
Höfte, H. & Whiteley, H. R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53, 242–255 (1989).
Deist, B. R., Rausch, M. A., Fernandez-Luna, M. T., Adang, M. J. & Bonning, B. C. Bt toxin modification for enhanced efficacy. Toxins (Basel). 6, 3005–3027. https://doi.org/10.3390/toxins6103005 (2014).
EPA, U. S. & Registration Decision for the New Active Ingredients: Bacillus thuringiensis Cry1B.868 protein and the genetic material (Vector PV-ZMIR522223) necessary for its production in MON 95379 maize (OECD unique identifier MON-95379-3), and Bacillus thuringiensis Cry1Da_7 protein and the genetic material (Vector PV-ZMIR522223) necessary for its production in MON 95379 maize (OECD unique identifier MON-95379- 3. (2024).
Horikoshi, R. J. et al. A new generation of Bt maize for control of fall armyworm (Spodoptera frugiperda). Pest Manage. Sci. (2021).
Wang, Y. et al. Bacillus thuringiensis Cry1Da_7 and Cry1B.868 protein interactions with novel receptors allow control of resistant fall armyworm, Spodoptera frugiperda (J.E. Smith). Appl. Environ. Microb. 85, e00579–00519 https://doi.org/10.1128/AEM.00579-19 (2019).
Pigott, C. R. & Ellar, D. J. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71 (2007).
Ferré, J., Bel, Y., Lázaro-Berenguer, M. & Hernández-Martínez, P. Vip3 insecticidal proteins: structure and mode of action. Adv. Insect Physiol. 65, 93–122. https://doi.org/10.1016/bs.aiip.2023.09.006 (2023).
Ciche, T. et al. The Vip3Cb1 protein exhibits sequence and structural similarities to other members of the Vip3C protein family. The mode of action of Vip3Cb1 resembles that of commercially available Vip3Aa
Wen, Z. et al. More than 10 years after commercialization, Vip3A-expressing MIR162 remains highly efficacious in controlling major lepidopteran maize pests: laboratory resistance selection versus field reality. Pestic. Biochem. Physiol. 192, e105385 (2023).
Yang, F., Kerns, D. L., Little, N. S., Santiago González, J. C. & Tabashnik, B. E. Early warning of resistance to Bt toxin Vip3Aa in Helicoverpa zea. Toxins 13, 618. https://doi.org/10.3390/toxins13090618 (2021).
Chakroun, M., Banyuls, N., Bel, Y., Escriche, B. & Ferré, J. Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 80, 329–350. https://doi.org/10.1128/mmbr.00060-15 (2016).
Gomis-Cebolla, J. et al. Insecticidal spectrum and mode of action of the Bacillus thuringiensis Vip3Ca insecticidal protein. J. Invertebr. Pathol. 142, 60–67 (2017).
Chakroun, M. & Ferré, J. In vivo and in vitro binding of Vip3Aa to Spodoptera frugiperda midgut and characterization of binding sites by (125)I radiolabeling. Appl. Environ. Microbiol. 80, 6258–6265. https://doi.org/10.1128/Aem.01521-14 (2014).
Rose, R. I. White paper on tier-based testing for the effects of proteinaceous insecticidal plant-incorporated protectants on non-target invertebrates for regulatory risk assessment. (2007).
Raybould, A. Problem formulation and hypothesis testing for environmental risk assessments of genetically modified crops. Environ. Biosaf. Res. 5, 119–125. https://doi.org/10.1051/ebr:2007004 (2006).
Romeis, J. et al. Deriving criteria to select arthropod species for laboratory tests to assess the ecological risks from cultivating arthropod-resistant genetically engineered crops. Chemosphere 90, 901–909. https://doi.org/10.1016/j.chemosphere.2012.09.035 (2013).
OECD. Test No. 211: Daphnia magna Reproduction Test. OECD, Paris, (2012).
Yanfei, W. et al. Bacillus thuringiensis Cry1Da_7 and Cry1B.868 protein interactions with novel receptors allow control of resistant fall armyworms, Spodoptera frugiperda (J.E. Smith). Appl. Environ. Microbiol. 85, 1–15 (2019). https://doi.org/10.1128/AEM.00579-19
Wang, C. X. et al. Risk assessment of homologous variants of biotech trait proteins using a bridging approach. GM Crops Food. 15, 336–351. https://doi.org/10.1080/21645698.2024.2420412 (2024).
Wang, C. et al. Production and characterization of homologous protoporphyrinogen IX oxidase (PPO) proteins: evidence that small N-terminal amino acid changes do not impact protein function. PLoS ONE. 19, e0311049. https://doi.org/10.1371/journal.pone.0311049 (2024).
Bal, H. K. et al. Ecological risk assessment for Cry1Da_7, Cry1B.3 and Vip3Cb1 proteins expressed in MON 89151 cotton: an insect-protected cotton with targeted activity against lepidoptera. Transgenic Res. (Submitted).
EPA, U. S. (U.S. Environmental Protection Agency, Washington, D.C.). (2024).
EPA, U. S. (ed) USDA-APHIS (ed Animal and Plant Health Inspection Service and Biotechnology Regulatory Services and Biopesticides and Pollution Prevention Division) U.S. Department of Agriculture, Animal and Plant Health Inspection Service and U.S. Environmental Protection Agency, Biotechnology Regulatory Services and Biopesticides and Pollution Prevention Division, Washington, D.C., (2007).
EPA, U. S. Good Laboratory Practice Standards. (1989).
Bolognesi, R. et al. Characterizing the mechanism of action of double-stranded RNA activity against Western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE. 7, e47534. https://doi.org/10.1371/journal.pone.0047534 (2012).
Bachman, P. M. et al. Characterization of the activity spectrum of MON 88702 and the plant-incorporated protectant Cry51Aa2.834_16. PLoS ONE. 12, e0169409. https://doi.org/10.1371/journal.pone.0169409 (2017).
Tan, J. et al. Development and survival of Orius insidiosus (Say) nymphs on encapsulated bee pollen-based diet in a Tier-I toxicity assay. Environ. Entomol. 40, 1613–1621. https://doi.org/10.1603/en11060 (2011).
SAS. Copyright 2016 by SAS Institute, Inc., Cary, North Carolina, (2016).
SAS. Copyright 2002–2012 by SAS Institute, Inc., Cary, North Carolina, (2012).
R Core Team. R: A language and environment for statistical computing, (2022). https://www.R-project.org/
Acknowledgements
We thank Carly Spielberg, Lauren Konecny, David Dyck, Benjamin Chiavini, Em Rendleman, and Becky Roper for helping with the bioassays and performing quality checks on data in tables, and Peter Asiimwe for his leadership during the characterization of the spectrum of activity of Cry1B.2 and Cry1Da_7. We also thank teams working at the Union City, TN and Waterman, IL Bayer insectaries for providing the insects used in these studies. We thank Jun Zhang for his assistance with protein production work. We also thank Scott Saracco and Collin Preftakes for their thoughtful reviews of early drafts of this manuscript.
Author information
Authors and Affiliations
Contributions
CJE, FS, and CK wrote this publication. CJE, FS, JT, HKB, CRB, TX, SLL, YY, and CK contributed to the overall study strategy and design. CJE and SMR conducted dietary exposure bioassays with lepidopteran and non-lepidopteran insect species exposed to Cry1B.2. CJE conducted dietary exposure bioassays with non-lepidopteran insect species exposed to Vip3Cb1. BW conducted protein stability assessment bioassays of Cry1B.2 and Vip3Cb1. JT conducted dietary exposure bioassays with lepidopteran insect species exposed to Cry1B.3. JT, JF, and GM conducted the dietary exposure and protein stability assessment bioassays of Cry1Da_7. JT conducted the dietary exposure bioassays of Vip3Cb1 lepidopteran dietary exposure bioassays. CM led the statistical analyses. YLG and BL executed protein production, purification and characterization. YLG wrote the Materials and Methods section of test substances used in bioassays. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
All co-authors are employees of Bayer CropScience LP, a developer of seeds and traits technologies and crop protection products.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Esquivel, C.J., Stubbins, F., Bal, H.K. et al. Characterization of the spectrum of insecticidal activity of Cry1B.3, Cry1Da_7, and Vip3Cb1 proteins produced by lepidopteran-protected MON 89151 cotton. Sci Rep 15, 40200 (2025). https://doi.org/10.1038/s41598-025-23841-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-23841-y