Abstract
To investigate the expression characteristics, functional roles, and regulatory mechanisms of circPSMA4 via miR-767-3p/HIF1A in bladder cancer. The expression and in vitro functionality of circPSMA4 in T24 bladder cancer cell lines were analyzed using RNase R treatment, actinomycin D stability assay, nucleocytoplasmic separation, real-time quantitative PCR, Western blot, dual-luciferase reporter assay, cellular energy metabolism analysis, scratch assay, Transwell migration and invasion assay, CCK-8 proliferation assay, lactate and glucose uptake detection, and ATP production measurement. Further, the in vivo roles of circPSMA4 were validated through the construction of stable cell lines and tumor xenograft assays in nude mice. CircPSMA4 was identified as a circular RNA highly expressed in bladder cancer cells, directly interacting with miR-767-3p. CircPSMA4 functions as a miR-767-3p sponge, regulating miR-767-3p to influence the expression of HIF1A, thereby promoting the proliferation, migration, invasion, and metabolic activities of bladder cancer cells, as well as tumor formation. CircPSMA4 facilitates the proliferation, migration, invasion, and metabolism of bladder cancer cells through the miR-767-3p/HIF1A pathway.
Similar content being viewed by others
Introduction
Bladder cancer is a type of cancer that originates from the cells lining the inside of the bladder, with the most common type being urothelial carcinoma (also known as transitional cell carcinoma)1. The incidence of bladder cancer is relatively high in many countries and regions, especially among men. The causes of bladder cancer are complex, often resulting from smoking, occupational exposure, genetic factors, chronic bladder inflammation, environmental and lifestyle factors. Most patients present with symptoms such as hematuria, bladder irritation, and difficulty urinating. The five-year survival rate is only 78%, affecting the prognosis and quality of life of patients. The pathogenesis of bladder cancer is not yet fully understood, hence there is an urgent need to delve into its pathogenic mechanisms to improve diagnostic levels2,3,4.
In bladder cancer, circular RNAs are distinguished by their unique closed-loop structures, rendering them resistant to degradation by RNA exonucleases, thus ensuring more stable expression. This stability allows circular RNAs to exert sustained and effective regulatory functions within cells. Rich in multiple miRNA binding sites, circular RNAs can function as miRNA sponges, adsorbing and modulating the activity of specific miRNAs. Through this mechanism, circular RNAs can influence miRNA-mediated regulation of their target genes, subsequently affecting cellular biological processes. This mechanism is referred to as the competitive endogenous RNA (ceRNA) mechanism5,6,7.
MicroRNAs (miRNAs) are a class of small RNA molecules approximately 20–24 nucleotides in length that play multiple important regulatory roles within cells. miRNAs negatively regulate gene expression at the transcriptional level by binding to complementary sequences on target mRNAs, leading to the suppression of mRNA translation or degradation. Despite their small size, miRNAs play a significant role in the development of eukaryotes and gene expression, participating in various processes such as animal development, cell proliferation and death, and cell differentiation. The biogenesis of miRNAs begins with the transcription of DNA in the cell nucleus, forming primary miRNA (pri-miRNA), which is then processed into precursor miRNA (pre-miRNA) with a specific structure, and further cleaved into smaller miRNA molecules in the cytoplasm. miRNAs do not directly participate in protein synthesis but affect gene expression by regulating mRNA8,9.
As a novel circular RNA expressed in bladder cancer, the specific roles and mechanisms of circPSMA4 remain unclear. Through database searches and bioinformatics analyses, we identified miR-767-3p and HIF1A as downstream targets, suggesting that circPSMA4 may promote the proliferation, migration, and invasion of bladder cancer cells. Given that HIF1A is a gene highly expressed in cancer, it may contribute to the warburg effect.
Methods
Cell Culture
Bladder cancer cell lines BIU-87, T24, RT112, and HT-1376 were purchased from Saibain Bio Technology Co., Ltd. The cells were cultured in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin solution. The cells were incubated in a humidified incubator with 5% CO2 at 37 °C.
RNase R treatment experiment
Total RNA extracted from cells was verified for quality and concentration using a UV–Vis spectrophotometer. The amount of RNase R required was calculated based on the RNA concentration, followed by digestion at 37 °C for 10 to 30 min. RNA samples were divided into two groups: an untreated Control group and a treated group receiving RNase R treatment. The treated samples were subjected to reverse transcription at designated time points (0, 5, 15, 30, 60 min). RNase R was inactivated at 70 °C for 10 min, succeeded by immediate reverse transcription.
Actinomycin D treatment
Each experimental group was treated with Actinomycin D, which inhibits new RNA synthesis, causing the accumulation of pre-existing RNA within the cell. Different treatment durations of 8 h, 16 h, and 24 h were set to observe RNA stability changes over time. Corresponding negative Controls were established at each time point to exclude non-specific factors. Contaminating genomic DNA in the extracted total RNA was removed using specific enzymes or reagents to ensure the accuracy of subsequent experiments. The extracted RNA was reverse-transcribed into cDNA. The molecular levels of target RNA and reference RNA were detected using qPCR technology. By comparing the variations in RNA levels at different time points or experimental conditions, RNA stability was assessed, and the half-life of the target RNA calculated based on qPCR results.
Nuclear and cytoplasmic RNA isolation
Cell samples were collected and washed multiple times with PBS buffer to remove impurities from the culture medium. The samples were added to centrifuge tubes containing appropriate cell lysis solution and gently agitated to ensure complete cell lysis. The tubes were placed on ice, and cells were disrupted using a sonicator or homogenizer to release cytoplasmic and nuclear contents. The lysate was subjected to low-speed centrifugation to remove unbroken cells and large debris. The supernatant was transferred to a new centrifuge tube, and an adequate amount of proteinase K was added to digest proteins within the supernatant. The tube was placed in a constant-temperature water bath, allowing proteinase K to act effectively. Post digestion, high-speed centrifugation was performed to remove precipitated proteins and impurities. The supernatant was transferred for RNA precipitation, which was achieved by adding an equal volume of isopropanol or ethanol and gently mixing. The tube was then placed on ice to facilitate RNA precipitation, followed by low-speed centrifugation to pellet the RNA. After discarding the supernatant, the RNA pellet was washed with 75% ethanol to remove residual salts and impurities. Subsequent to ethanol removal, the RNA pellet was air-dried and dissolved in an appropriate amount of TE buffer. To isolate RNA from the nucleus, cells were subjected to hypotonic treatment post cytoplasmic RNA separation, allowing nuclear swelling and subsequent isolation. The nuclear fraction was purified using nucleic acid extraction kits or phenol–chloroform extraction. The isolated nuclei were lysed using an appropriate lysis solution, and the disrupted nuclear suspension was centrifuged to separate the supernatant, which underwent RNA precipitation, washing, and dissolution to yield pure nuclear RNA.
Lactate assay
In the presence of NAD, lactate dehydrogenase (LDH) catalyzed the conversion of lactate to pyruvate, simultaneously generating NADH. Under alkaline conditions, the chromogen reacted with pyruvate to form a complex that shifted the equilibrium towards lactate oxidation, completing the reaction. NADH and lactate equimolarly correlated. The absorbance at 340 nm was measured using a spectrophotometer, and lactate levels were determined via colorimetric analysis.
Glucose uptake assay
5 µl of the standard or sample was added to a PCR tube, with the option to use 20 µl when necessary. If sample volume was minimal with an appropriate concentration of glucose, volumes of 1–2 µl or even smaller were used. 185 µl of Glucose Assay Reagent was added to reach a final volume of 190 µl. Following vortex mixing and centrifugation at 5000×g to settle the liquid, the PCR tubes were heated at 95 °C for 8 min and then cooled to 4 °C. Post-cooling, 180 µl was extracted from each tube to a clean 96-well plate, ensuring no bubbles formed. The absorbance at 630 nm (alternatively 620–650 nm) was measured. Absorbance readings were completed within 30 min post-reaction to obtain optimal results, and glucose concentration was calculated based on a standard curve.
Seahorse energy metabolism analysis
Extracellular acidification (ECAR) and oxygen consumption rates (OCR) were measured using the Seahorse XF96 analyzer to reflect real-time glycolytic activity in hippocampal cells. Cells were seeded in a 96-well XF-PS plate at a density of 30,000 cells per well and cultured in Seahorse XF RPMI medium supplemented with 2 mM sodium pyruvate, 10 mM glucose, and 2 mM L-glutamine. Cell glycolytic rates were evaluated using the Seahorse XF Glycolysis Stress Test Kit according to manufacturer’s instructions.
Western blot
Protein extraction commenced, processing samples with RIPA lysis buffer and protease inhibitors, and protein concentration was determined using a BCA protein assay kit. Subsequently, proteins were separated by SDS-PAGE electrophoresis and transferred from the gel to a PVDF membrane via blotting. Post-transfer, the membrane was blocked with a 5% skim milk or 5% BSA solution to prevent nonspecific binding. Then, the membrane was incubated overnight at 4 °C with primary antibodies HIF1A, HK2, GLUT1, and LDHA. The membrane was washed, then incubated with secondary antibodies at room temperature for 2 h. Finally, ECL chemiluminescence reagents were used for substrate luminescence reaction and the imaging was captured with an exposure machine to obtain and analyze the optical density values of the protein bands.
qRT-PCR
Total RNA was extracted from tissues and cells using Trizol reagent, following the manufacturer’s instructions. RNA concentration and quality were determined for each sample using a microplate reader at a 260/280 nm ratio. RNA was converted to cDNA using ReverTra AceTM qPCR RT Kit, adhering to the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was performed using AceQ R qPCR SYBR Green Master Mix and a real-time PCR amplification system. Each qRT-PCR reaction was performed in triplicate, with GAPDH used as an internal Control.
PSMA4: F: 5′-AGT GTG GCA GGC ATA ACT TCT-3′; R: 5′-TCA CAA GGT ATT GGC TCC TGA-3′. miR-767-3p: F: 5′-ATC GTG GCC AGT GCT GAA CGT-3′; R: 5′-TGC CAA CGT GCC AAA CAC GTG C-3′. HIF1A: F: 5′-TCA CCA GAC AGA GCA GGA AA-3′; R: 5′-TCA CCA GAC AGA GCA GGA AA-3′. GAPDH: F: 5′-TTG CAA TGC AAC TTC AGA TG-3′; R: 5′-TGC ATG ATA TAC GTG ATT G-3′.
Scratch test
A marker pen was used to draw horizontal lines on the back of a 6-well plate, ensuring each line intersected at least five times per well at intervals of 0.5–1 cm. Cells in the logarithmic growth phase were seeded into the 6-well plates at a density of 60,000 cells per well, then incubated at 37 °C in a 5% CO2 incubator for 24 h. The following day, scratches were created at the marked lines using a 200 µL pipette tip. The cells were washed with PBS, serum-free medium was added, and the scratches were photographed under a 4× objective lens, ensuring centered, vertical scratches and consistent background. The test samples and concentration gradients were added, and samples were taken and photographed at 0 and 24-h time points. The results were analyzed using Image J software by selecting 6 to 8 random horizontal lines and calculating the mean cell distance or scratch area.
CCK-8 assay
100 µL of cell suspension was seeded into each well of a 96-well plate and pre-incubated under 37 °C with 5% CO2. 10 µL of CCK solution was added per well, avoiding bubble formation, and the plate was incubated for 1–4 h. The absorbance at 450 nm was measured using a microplate reader at 24, 48, and 72-h time points.
Transwell assay
The upper surface of the Transwell chamber’s lower membrane was coated with or without Matrigel dilution and air-dried at 4 °C to hydrate the basement membrane. The basement membrane was hydrated using serum-free medium with BSA. The serum-starved cells were digested, resuspended in serum-free medium at an appropriate density, and the cell suspension was seeded into the Transwell chambers. Medium containing FBS or chemokines was added to the lower chamber of a 24-well plate, avoiding air bubbles. After 12–48 h of incubation, the chambers were removed, cells from the upper surface were wiped away with a cotton swab, and PBS washes, neutral formalin fixation, PBS washes, crystal violet staining, and final PBS washes were performed. Images were captured and cells were counted using a microscope for analysis.
Dual-luciferase reporter assay
Dual-luciferase reporter plasmids were constructed with wild-type hsa_circ_PSMA4 or mutant hsa_circ_PSMA4 inserted upstream of the luciferase gene. Cells were transfected with the respective wild-type or mutant hsa_circ_PSMA4 plasmids. After 48 h, cells were collected and lysed to release luciferase, luciferase substrates were added, and luciferase activity was measured using a detection system. Luciferase activity was compared between wild-type and mutant hsa_circ_PSMA4 plasmids to determine the binding ability of hsa_circ_PSMA4 with hsa-miR-767-3p. Similarly, plasmids with wild-type or mutant hsa-miR-767-3p were constructed and transfected to evaluate hsa-miR-767-3p binding capacity with HIF1A mRNA, comparing luciferase activity.
Establishment of stable cell lines
The overexpression plasmids for circPSMA4, microRNA-767-3p (F: 5′-UGA GGU AGU AGG UUG UGU GGU U-3′; 5′-CCA CAC AAC CUA CUA CCU CAU U-3′), and HIF1A shRNA (F: 5′-CCG GGC CAG AAG AAA GAA GAC CAA CTC GAG TTG GTC TTC TTT CTT CTG GCT TTT TG-3′; R: 5′-AAT TCA AAA AGC CAG AAG AAA GAA GAC CAA CTC GAG TTG GTC TTC TTT CTT CTG G-3′) and HIF1A overexpression were downloaded from NCBI. A tripartite packaging system was utilized, including transfer plasmids, packaging plasmid pSPAX2, and envelope plasmid pMD2.G. 293 T cells were seeded in 6 cm dishes, allowed to reach 70–80% confluence, and the medium was changed. Solution A was prepared by adding 2.5 µg of pCDH plasmid, 1.5 µg of pSPAX2, and 1.5 µg of pMD2.G in opti-MEM to form a 100 µL solution, and solution B was prepared by adding 30 µL PolyJet DNA transfection reagent in opti-MEM. Solutions A and B were mixed gently, added to the 293 T cell culture supernatant, the medium was changed after 12 h, and the supernatant was collected after 48 h, centrifuged at 2000 rpm for 10 min, cell debris was discarded, and the supernatant was filtered using a 0.45 µm membrane. Logarithmic-phase cells were digested and resuspended, seeded into a 6-well plate, and upon reaching 80–90% confluence, DMEM medium containing varying concentrations (0–10 µg/mL, 6 gradients) of puromycin was added. After 24 h, cell status was observed and the minimum concentration (2 µg/mL) causing complete cell death was determined for puromycin selection of stable strains. Logarithmic-phase cells were infected by mixing virus supernatant and complete medium at a 3:1 ratio, infected for 48 h, and replaced with fresh medium containing 2 µg/mL puromycin, continuing the culture.
Measurement of ATP
The level of ATP in the cells was measured using an ATP assay kit. Briefly, 2 × 106 cells per well were seeded in 60 mm dishes. After overnight culture, the cells were lysed with ATP lysis buffer. The supernatants were collected after centrifugation at 12,000 rpm for 10 min at 4 °C, and the signal was detected using a luminometer.
Xenograft tumor model in nude mice
Thirty-six male 6-week-old nude mice were purchased from Henan Scripps Biotechnology Co. The nude mice were housed in an SPF environment with a 12-h cycle of light and dark. Adequate food and water were provided. 1 × 106 T24 tumor cells were prepared per grafting site, mixed with an equal volume of matrix gel, and drawn into a 1 mL syringe. Wearing sterile gloves, the mouse was held by pinching the neck skin with left thumb and forefinger, the tail was fixed with the left ring and little fingers, and the axilla or groin was sterilized with 75% ethanol thrice. The syringe containing the tumor cell–matrix gel mixture was inserted at a 45-degree angle at the selected site, taking care not to pierce the peritoneum, and injected subcutaneously near a horizontal position. 1 × 106 tumor cells in the gel mixture were injected subcutaneously, the needle was promptly removed, and gentle finger pressure was applied to the injection site for around one minute before placing the mouse back in the cage, positioning it on the bedding to prevent aspiration in case of vomiting. Mouse recovery was checked after 2–3 h. Tumor dimensions were measured with a caliper twice weekly and tumor volume was calculated. The formula for calculating tumor volume: V = 0.52 × L × W2. The nude mice were euthanized when they showed loss of appetite, depression or when the diameter of the tumor reached 1.5 cm. Nude mice were euthanized using CO2. The flow rate was 30% of the volume of the container per minute, and the euthanasia was performed gently to minimize the anxiety of the nude mice. Nude mice were confirmed dead after 20 min of respiratory arrest. This study was approved by the Laboratory Animal Welfare Ethics Review Committee of Hebei University, all methods were performed in accordance with the relevant guidelines and regulations and in accordance with the relevant ARRIVE guidelines and the 3Rs principles.
We conducted a search in the circBANK database using “PSMA4” as the keyword and obtained the molecular structure of circRNA PSMA4. By intersecting the downstream target genes of circRNA PSMA4 using the Venn method through the circBANK, Interactome, and CSCD databases, we identified miRNA-767-3p as the sole downstream target gene of circRNA PSMA4. Furthermore, we predicted the downstream target genes of miRNA-767-3p using the miRDB, miRNAminer, and TargetScan databases and found HIF1A to be the only target gene downstream of miRNA-767-3p through the intersection of Venn diagrams.
Clinical sample collection
A total of 40 patients with bladder cancer (BCa) admitted to the Affiliated Hospital of Hebei University were selected. During surgery, BCa tissue and adjacent tissue (3 cm from the tumor margin, pathologically confirmed to be free of cancer infiltration) were collected. Inclusion criteria: (1) Pathological diagnosis of BCa, including non-muscle-invasive BCa and muscle-invasive BCa; (2) Complete clinical and pathological records; (3) Patients had not received any neoadjuvant treatments such as chemotherapy, radiotherapy, or immunotherapy prior to pathological tissue sampling; (4) Patients or their families fully understood the purpose and process of the study, voluntarily participated, and signed written informed consent forms; (5) Patients were able to cooperate with regular postoperative follow-ups to assess prognosis. Exclusion criteria: (1) Concurrent malignant tumors; (2) Concurrent systemic diseases that may affect patient survival or treatment outcomes; (3) Pregnancy or lactation; (4) Patients with mental disorders or those unable to cooperate with treatment. This study was approved by the Ethics Committee of the Affiliated Hospital of Hebei University. Ethics approval number: HDFY-LL-2022-087. All research involving human subjects was conducted in accordance with the Declaration of Helsinki.
Statistical analysis
The data were analyzed using GraphPad Prism 9.0, representing results as mean ± standard deviation. Student’s t-test was used for comparison between two groups and one-way ANOVA for multiple groups. A P value of P < 0.05 denoted statistical significance. All experiments were conducted independently three times.
Results
CircRNA PSMA4 was a novel circular RNA highly expressed in bladder cancer
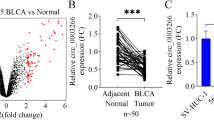
Through the collection of databases such as circBANK, it was discovered that circRNA PSMA4 was a circular RNA constructed by a head-to-tail junction from position 78836531 to position 78839040 on chromosome 15 (Fig. 1A). When treated with RNase R+, which digested most mRNA samples but had no digesting effect on circRNA, qPCR amplification of RNA treated with RNase R + showed no significant decrease in the levels of both mRNA and circRNA of PSMA4 in the untreated group. However, the levels of PSMA4 mRNA significantly decreased in the RNase R-treated group, while the circRNA levels remained unchanged, indicating that circPSMA4 was a circular RNA (Fig. 1B). Upon treating the same RNA samples with actinomycin D and observing them for 8 h, 16 h, and 24 h, it was found that the degradation rate of circPSMA4 over time was much slower than that of PSMA4 mRNA (Fig. 1C). By isolating RNA from the cell nucleus and cytoplasm and using β-ACTIN and U6 as internal Controls, it was observed that the expression level of circPSMA4 in the cytoplasm was significantly higher than in the nucleus. On the contrary, the expression level of PSMA4 mRNA was significantly higher in the nucleus than in the cytoplasm (Fig. 1D). Using qRT-PCR to detect the expression levels of circPSMA4 in bladder cancer cell lines BIU-87, T24, RT112, and HT-1376 cells, it was found that T24 cells exhibited the highest circPSMA4 expression (Fig. 1E).
Identification and expression analysis of circRNA PSMA4. (A) Schematic diagram of circRNA PSMA4 structure; (B) qPCR analysis of circPSMA4 and PSMA4 mRNA after RNase R treatment; (C) Degradation curves of circPSMA4 and PSMA4 mRNA post-actinomycin D treatment; (D) Expression levels of circPSMA4 and PSMA4 mRNA in the nucleus and cytoplasm; (E) Expression levels of circPSMA4 in different bladder cancer cell lines. N = 3; **P < 0.01.
CircPSMA4 as a novel circRNA promoted the proliferation, invasion, and metastasis of bladder cancer in terms of cell behavior
After constructing plasmids of OE-NC, OE-circPSMA4, si-NC, and si-circPSMA4 and transfecting them into T24 cells for 48 h, cell behavior assays were conducted. qRT-PCR detected the transfection efficiency. Compared with the Control group, the expression level of circPSMA4 circRNA in the OE-NC group did not show a significant difference. However, the OE-circPSMA4 group exhibited a particularly marked increase in circRNA expression compared to the OE-NC group. Conversely, the si-circPSMA4 group showed a significant reduction in circPSMA4 expression compared to the si-NC group (Fig. 2A). CCK-8 assays demonstrated that the OD value at 450 nm did not change significantly in the OE-NC group compared to the Control group. In contrast, the OD value at 450 nm in the OE-circPSMA4 group increased significantly compared to the OE-NC group. Similarly, the OD value at 450 nm in the si-circPSMA4 group decreased significantly compared to the si-NC group (Fig. 2B). Colony formation assays revealed that the number of colonies at 450 nm showed no significant change in the OE-NC group compared to the Control group. However, the OE-circPSMA4 group exhibited a notable increase in colony numbers compared to the OE-NC group, whereas the si-circPSMA4 group showed a marked decrease compared to the si-NC group (Fig. 2C). Scratch assays indicated that the scratch width did not significantly change in the OE-NC group compared to the Control group. However, the scratch width significantly narrowed in the OE-circPSMA4 group compared to the OE-NC group, while it significantly widened in the si-circPSMA4 group compared to the si-NC group (Fig. 2D). Transwell assays demonstrated that the number of migratory and invasive cells did not significantly change in the OE-NC group compared to the Control group. However, the OE-circPSMA4 group showed a significant increase in the number of migratory and invasive cells compared to the OE-NC group, whereas the si-circPSMA4 group displayed a marked reduction compared to the si-NC group (Fig. 2E). We then examined ATP levels in each group. The results showed that there was no significant difference in ATP concentration between the Control, OE-NC and SI-NC groups. The ATP concentration was significantly higher in the OE-circPSMA4 group relative to the OE-NC group. The ATP concentration was significantly lower in the si-circPSMA4 group relative to the si-NC group (Fig. 2F).
Effects of circPSMA4 on proliferation, colony formation, migration, and invasion of bladder cancer cells. (A) qRT-PCR detecting changes in circPSMA4 expression levels post-overexpression and interference; (B) CCK-8 assay analyzing the effect of circPSMA4 on BCa cell proliferation; (C) Colony formation assay evaluating circPSMA4’s effect on cell cloning capabilities; (D) Scratch assay detecting the impact of circPSMA4 on cell migration; (E) Transwell assay assessing circPSMA4’s effect on cell invasion capabilities; (F) Evaluate the effect of circPSMA4 on ATP concentration in cells. N = 3; **P < 0.01; *P < 0.05; nsP > 0.05.
miRNA-767-3p was the sole downstream miRNA of circPSMA4
By intersecting datasets from circBANK, Interactome, and CSCD databases, miRNA-767-3p emerged as the unique miRNA (Fig. 3A). Dual luciferase experiments revealed no significant changes in luciferase intensity between circPSMA4-MUT with miR-NC or miRNA-767-3p; however, circPSMA4-WT exhibited distinct binding with miRNA-767-3p, resulting in a marked decrease in luciferase intensity (Fig. 3B). qRT-PCR assays in bladder cancer cell lines BIU-87, T24, RT112, and HT-1376 revealed the expression levels of miR-767-3p, with T24 cells displaying the lowest levels (Fig. 3C). Additionally, transfection efficiency detected by qRT-PCR indicated that compared to the Control group, miR-767-3p expression in the OE-NC group did not show significant differences; however, miR-767-3p expression in the OE-circPSMA4 group drastically decreased compared to the OE-NC group, and increased significantly in the si-circPSMA4 group compared to the si-NC group (Fig. 3D).
Interaction between circPSMA4 and miR-767-3p. (A) Analysis through databases indicates that the target gene of circPSMA4 is miR-767-3p; (B) Dual-luciferase reporter assay confirming direct interaction between circPSMA4 and miR-767-3p; (C) Expression levels of miR-767-3p in bladder cancer cell lines; (D) Effect of circPSMA4 on miR-767-3p expression levels. N = 3; **P < 0.01; *P < 0.05; nsP > 0.05.
CircPSMA4 enhanced the Warburg effect and promoted migration, invasion, and metastasis in bladder cancer via miRNA-767-3p
By constructing Control and overexpression plasmids for circPSMA4 and miR-767-3p NC and mimics plasmids, T24 cells were co-transfected and divided into groups: OE-NC, OE-circPSMA4, OE-circPSMA4 + mimics-NC, and OE-circPSMA4 + miR-767-3p mimics. In the CCK-8 assay, the OD value at 450 nm significantly increased in the OE-circPSMA4 group compared to the OE-NC group, but decreased notably in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4A). Clonal formation assays revealed a considerable increase in colony numbers in the OE-circPSMA4 group compared to the OE-NC group, while the OE-circPSMA4 + miR-767-3p mimics group showed a pronounced decrease in colonies compared to the OE-circPSMA4 + mimics-NC group (Fig. 4B). Transwell assays demonstrated a notable rise in the number of migrating and invading cells in the OE-circPSMA4 group compared to the OE-NC group; conversely, the number dropped significantly in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4C). Scratch wound assays showed that the scratch width significantly increased in the OE-circPSMA4 group in comparison to the OE-NC group, yet it narrowed conspicuously in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4D). Lactate production assays indicated a substantial increase in lactate levels in the OE-circPSMA4 group compared to the OE-NC group, but a significant reduction was observed in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4E). Glucose uptake assays indicated a marked increase in glucose uptake in the OE-circPSMA4 group compared to the OE-NC group, but a notable decrease was observed in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4F). Seahorse energy metabolism assays indicated a significant elevation in Glycolysis (baseline glycolysis), Glycolysis Capacity (glycolytic capacity), and Glycolysis Reserve (glycolytic potential) in the OE-circPSMA4 group compared to the OE-NC group, with the levels significantly decreasing in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4G). The OCR results showed a notable increase in Basal Respiration, ATP production, Maximal Respiration, and Spare Respiratory Capacity in the OE-circPSMA4 group compared to the OE-NC group, which markedly decreased in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4H). Western blot results showed an increase in the expression levels of HK2, GLUT1, and LDHA proteins in the OE-circPSMA4 group compared to the OE-NC group, with a subsequent decrease detected in the OE-circPSMA4 + miR-767-3p mimics group compared to the OE-circPSMA4 + mimics-NC group (Fig. 4I). Finally, we measured the ATP content in each group. The results showed that the ATP concentration was significantly higher in the OE-circPSMA4 group compared to the OE-NC group. Compared to the OE-circPSMA4 + mimics-NC group, the ATP concentration was significantly lower in the OE-circPSMA4 + miR-767-3p mimics group (Fig. 4J).
CircPSMA4 promotes the Warburg effect and migration/invasion of bladder cancer cells through miR-767-3p. (A) The effects of circPSMA4 overexpression and miR-767-3p interference on prostate cancer cell activity were analyzed through CCK-8 experiments; (B) The effects of circPSMA4 overexpression and miR-767-3p interference on prostate cancer cell proliferation were analyzed through a colony formation assay; (C) The effects of circPSMA4 overexpression and miR-767-3p interference on the migration and invasion abilities of prostate cancer cells were analyzed through Transwell experiments; (D) The effects of circPSMA4 overexpression and miR-767-3p interference on the migration ability of prostate cancer cells were analyzed through scratch assay; (E,F) The effects of circPSMA4 overexpression and miR-767-3p interference on prostate cancer cell behavior were analyzed through lactic acid detection and glucose uptake experiments; (G,H) Seahorse energy metabolism analysis showing circPSMA4’s effect on glycolysis and mitochondrial respiration of BCa cells. (I) Western blot was used to detect the protein expression levels of HK2, GLUT1, and LDHA in the OE-NC group, OE-circPSMA4 group, OE-circPSMA4 + miR-NC mimics group, and OE-circPSMA4 + miR-767-3p mimics group; (J) Evaluation of the effects of circPSMA4 and miR-767-3p on cellular ATP concentration. N = 3;**P < 0.01;*P < 0.05.
miR-767-3p modulated bladder cancer by affecting HIF1a
Using Venn diagrams from miRDB, miRNAminer, and TargetScan databases, HIF1A was identified as a downstream research target (Fig. 5A). Dual-luciferase assays indicated no significant luciferase intensity changes between HIF1A-MUT and miR-NC, miRNA-767-3p; however, micRNA-767-3p and HIF1A-WT demonstrated evident binding, significantly decreasing luciferase intensity (Fig. 5B). qRT-PCR and Western blot assays collectively showed the expression levels of HIF1A in bladder cancer cell lines BIU-87, T24, RT112, and HT-1376, revealing that T24 cells exhibited the highest mRNA and protein expression of HIF1A (Fig. 5C). Western blot assays determined the transfection efficiency; compared to the Control group, the OE-NC group showed minimal variation in HIF1A expression, whereas the OE-circPSMA4 group exhibited a notable increase, and the si-circPSMA4 group showed a significant decrease in HIF1A expression compared to the si-NC group (Fig. 5D).
MiR-767-3p affects bladder cancer through HIF1A. (A) Databases predict HIF1A as a downstream target gene of miR-767-3p; (B) Dual-luciferase reporter assay validating miR-767-3p and HIF1A interaction; (C) Expression levels of HIF1A in various bladder cancer cell lines; (D) Effect of circPSMA4 on HIF1A expression levels. N = 3;**P < 0.01; nsP > 0.05.
miR-767-3p inhibited glycolysis in bladder cancer by suppressing HIF1a, thus impeding cellular behaviors related to cancer progression
Constructing Control and overexpression plasmids for HIF1A, alongside miR-767-3p NC and mimics plasmids, groups were transfected into T24 cells: miR-767-3p NC group, miR-767-3p mimics group, miR-767-3p mimics + OE-NC group, and miR-767-3p mimics + OE-HIF1A group. CCK-8 assays revealed that, compared to the miR-767-3p NC group, the miR-767-3p mimics group exhibited significantly reduced OD values at 450 nm; however, miR-767-3p mimics + OE-HIF1A group demonstrated increased OD values at 450 nm compared to the miR-767-3p mimics + OE-NC group (Fig. 6A). Clone formation assays showed a noticeable reduction in colonies in the miR-767-3p mimics group compared to the miR-767-3p NC group, but an increase in the miR-767-3p mimics + OE-HIF1A group compared to the miR-767-3p mimics + OE-NC group (Fig. 6B). Transwell assays indicated a significant reduction in cell migration and invasion in the miR-767-3p mimics group compared to the miR-767-3p NC group, whereas the miR-767-3p mimics + OE-HIF1A group exhibited an increase compared to the miR-767-3p mimics + OE-NC group (Fig. 6C). Scratch assays revealed a significant narrowing of scratch width in the miR-767-3p mimics group compared to the miR-767-3p NC group, but an increase in the miR-767-3p mimics + OE-HIF1A group compared to the miR-767-3p mimics + OE-NC group (Fig. 6D). Lactate production assays showed a marked decrease in lactate generation in the miR-767-3p mimics group compared to the miR-767-3p NC group, whereas the miR-767-3p mimics + OE-HIF1A group showed an increase compared to the miR-767-3p mimics + OE-NC group (Fig. 6E). Glucose uptake assays indicated a marked reduction in glucose uptake in the miR-767-3p mimics group compared to the miR-767-3p NC group, whereas the miR-767-3p mimics + OE-HIF1A group exhibited an increase compared to the miR-767-3p mimics + OE-NC group (Fig. 6F). Seahorse energy metabolism assays showed that ECAR results indicated a statistically significant decrease in glycolysis, glycolytic capacity, and glycolytic reserve in the miR-767-3p mimics group compared to the miR-767-3p NC group, whereas these parameters increased in the miR-767-3p mimics + OE-HIF1A group compared to the miR-767-3p mimics + OE-NC group (Fig. 6G). OCR results showed that basal respiration, ATP production, maximal respiration, and spare respiratory capacity all decreased in the miR-767-3p mimics group compared to the miR-767-3p NC group, but increased in the miR-767-3p mimics + OE-HIF1A group compared to the miR-767-3p mimics + OE-NC group (Fig. 6H). Western blot results revealed that the expression levels of HIF1A, HK2, GLUT1, and LDHA decreased in the miR-767-3p mimics group compared to the miR-767-3p NC group, but increased in the miR-767-3p mimics + OE-HIF1A group compared to the miR-767-3p mimics + OE-NC group (Fig. 6I). Finally, we measured the ATP content in each group. The results showed that the ATP concentration was significantly lower in the miR-767-3p mimics group compared to the miR-767-3p NC group. Compared to the miR-767-3p mimics + OE-NC group, the ATP concentration was significantly higher in the miR-767-3p mimics + OE-HIF1A group (Fig. 6J).
MiR-767-3p affects glycolysis and cellular behavior of bladder cancer cells through HIF1A. (A) The effects of miR-767-3p interference and HIF1A overexpression on prostate cancer cell activity were analyzed through CCK-8 experiments; (B) The effects of miR-767-3p interference and HIF1A overexpression on prostate cancer cell proliferation were analyzed through a colony formation assay; (C) The effects of miR-767-3p interference and HIF1A overexpression on the migration and invasion abilities of prostate cancer cells were analyzed through Transwell experiments; (D) The effects of miR-767-3p interference and HIF1A overexpression on the migration ability of prostate cancer cells were analyzed through scratch assay; (E,F) The effects of miR-767-3p interference and HIF1A overexpression on prostate cancer cell behavior were analyzed through lactic acid detection and glucose uptake experiments; (G,H) Seahorse energy metabolism analysis showing cmiR-767-3p’s effect on glycolysis and mitochondrial respiration of BCa cells; (I) Western blot was used to detect the protein expression levels of HK2, GLUT1, and LDHA in the miR-767-3p NC group, miR-767-3p mimics group, miR-767-3p mimics + OE-NC group, and miR-767-3p mimics + OE-HIF1A group; (J) Evaluation of the effects of miR-767-3p and HIF1A on cellular ATP concentration. N = 3;**P < 0.01;*P < 0.05.
CircPSMA4 facilitated tumorigenesis in bladder cancer via the miRNA-767-3p\HIF1A pathway
Lentivirus vectors were constructed for overexpressing circPSMA4 and miRNA-767-3p, and for knocking down HIF1A. Compared to the LV-OE-NC group, the LV-OE-circPSMA4 group exhibited a significant increase in tumor volume and weight. The LV-OE-circPSMA4 + miR-767-3p group showed a notable reduction in tumor volume and weight compared to the LV-OE-circPSMA4 + miR-NC group. Similarly, the LV-OE-circPSMA4 + KD-HIF1A group exhibited a marked reduction in tumor volume and weight compared to the LV-OE-circPSMA4 + KD-NC group (Fig. 7A). We also used qRT-PCR to detect the expression of circPSMA4 in cancerous and paracancerous tissues. The results showed that the expression of circPSMA4 was significantly higher in cancerous tissues than in paracancerous tissues (Fig. 7B).
CircPSMA4 promotes tumor formation in bladder cancer through the miR-767-3p/HIF1A pathway. (A) Tumor formation in nude mice showing the effects of circPSMA4 overexpression, interference with miR-767-3p, and HIF1A knockout; N = 6; (B) qRT-PCR detection of circPSMA4 expression in cancer and pericancerous tissues. N = 40;**P < 0.01.
CircPSMA4 promotes proliferation, migration, and invasion of bladder cancer cells through the HIF1A pathway
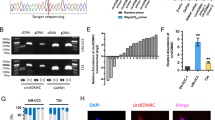
We then detected the effect of HIF1A on downstream proteins using western blot. The results showed that the relative protein expression levels of HIF1A, HK2, GLUT1, and LDHA were significantly higher in the OE-HIF1A group relative to the OE-NC group. The relative protein expression levels of HIF1A, HK2, GLUT1, and LDHA were significantly lower in the KD-HIF1A group relative to the KD-NC group (Fig. 8A). Transwell assay results showed that, compared with the OE-NC group, the migration and invasion cell counts were significantly increased in the OE-HIF1A group. Compared with the KD-NC group, the number of migrating and invading cells in the KD-HIF1A group was significantly reduced (Fig. 8B). CCK-8 assay results showed that at 48 h, the OD values in the OE-HIF1A group were significantly higher than those in the OE-NC group. Compared with the KD-NC group, the OD values in the KD-HIF1A group were significantly reduced (Fig. 8C). CircPSMA4, acting as a sponge for miRNA-767-3p, inhibited the expression of HIF1A, thereby facilitating the glycolytic pathway and the Warburg effect in bladder cancer cells. This promoted proliferation, migration, and invasion in bladder cancer (Fig. 9). The original bands generated from the Western blot experiments in this study were supplemented in Supplementary file 1. The original data from this study were supplemented in Supplementary file 2.
CircPSMA4 promotes proliferation, migration, and invasion of bladder cancer cells through the HIF1A pathway. (A) Western blot was used to detect the protein expression levels of HIF1A, HK2, GLUT1, and LDHA in the OE-NC group, HIF1A-OE group, KD-NC group, and HIF1A-KD group; (B) Transwell assay of migration and invasion ability of T24 cells in the OE-NC group, HIF1A-OE group, KD-NC group, and HIF1A-KD group; (C) CCK-8 assay to assess the proliferation capacity of T24 cells in the OE-NC group, HIF1A-OE group, KD-NC group, and HIF1A-KD group. N = 3;**P < 0.01;*P < 0.05.
Schematic of circPSMA4 promoting bladder cancer through the miR-767-3p/HIF1A pathway. Diagram depicting the molecular mechanism by which circPSMA4 sponges miR-767-3p, promoting HIF1A and subsequently enhancing glycolysis and the Warburg effect in BCa cells, leading to cell proliferation, migration, and invasion.
Discussion
Circular RNAs (circRNAs) have the capacity to interact with proteins, thereby modulating their stability or function. This mechanism is reminiscent of circRNAs acting as protein "sponges," participating in interactions between proteins, as well as between proteins and nucleic acids, thereby impacting cellular signaling and metabolic processes.
The roles and mechanisms of circRNAs in bladder cancer (BCa) are exceedingly complex and diverse. CircCASC15 promotes BCa cell proliferation by sponging miR-1224, thereby activating CREB1 expression10. CircSLC8A1 inhibits BCa cell proliferation, migration, and invasion by upregulating PTEN expression11. CircNR3C1 induces G0/G1 phase arrest by inhibiting cyclin D1 expression, thereby halting cell cycle progression12. CircCdr1as induces apoptosis in BCa cells by upregulating APAF1 expression13. Circ_0001361 promotes BCa cell invasion and metastasis by upregulating MMP9 expression14. Circ_0001429 enhances BCa cell growth and metastasis through increased expression of VEGFA15. CircCdr1as may increase BCa cell sensitivity to cisplatin chemotherapy through the circCdr1as/miR-1270/APAF1 axis13. CircPSMA4 is a circRNA highly expressed in bladder cancer cells. Rnase R treatment confirmed the circular structure of circPSMA4, as it resisted Rnase R digestion while its corresponding linear mRNA was significantly degraded. Additionally, actinomycin D treatment revealed that circPSMA4 exhibited greater stability than its linear counterpart PSMA4 mRNA. Cellular fractionation demonstrated that circPSMA4 is abundantly expressed in the cytoplasm, predominantly around the nucleus. Through the construction of overexpression and interference expression plasmids, it was observed that circPSMA4 significantly promoted BCa cell proliferation, colony formation, migration, and invasion. CCK-8, colony formation, scratch, and Transwell assays consistently indicated that overexpression of circPSMA4 enhances these malignant behaviors in BCa cells, while its interference expression markedly inhibits these behaviors. Bioinformatics analysis and dual-luciferase reporter assays revealed a direct interaction between circPSMA4 and miR-767-3p. CircPSMA4 acts as a miR-767-3p sponge, affecting miR-767-3p activity through a competing endogenous RNA (ceRNA) mechanism. This finding provides new insights into the regulatory role of circPSMA4 in BCa. Overexpression of circPSMA4 significantly enhanced glycolysis and mitochondrial respiration in BCa cells, consistent with the Warburg effect, wherein cancer cells preferentially generate energy through glycolysis rather than oxidative phosphorylation. Additionally, increased lactate production, glucose uptake, and ATP generation further corroborated the role of circPSMA4 in promoting BCa cell metabolism. CircPSMA4 likely acts as a key regulatory molecule, promoting BCa cell proliferation, migration, invasion, and metabolic reprogramming through the miR-767-3p/HIF1A axis. These discoveries provide new therapeutic targets and could aid in the development of novel diagnostic and treatment strategies for bladder cancer.
In hepatocellular carcinoma studies, hsa_circ_0000673 facilitated malignant behaviors in HCC by modulating the miR-767-3p/SET signaling pathway16. WASF3 overexpression influenced the expression of circRNA hsa-circ-0100153, which promoted breast cancer progression by sponging hsa-miR-31, hsa-miR-767-3p, and hsa-miR-93517. Knockdown of hsa_circ_0018818 inhibited tumorigenesis in non-small cell lung cancer by sponging miR-767-3p18. HIF1A promotes tumor cell proliferation and invasion by regulating related signaling pathways, such as the m6A modification of AKT1. HIF1A is a key regulatory factor in tumor angiogenesis, inducing the expression of VEGF (Vascular Endothelial Growth Factor) to promote tumor angiogenesis, thereby supplying nutrients and oxygen to tumor cells19. The expression level of HIF1A is closely related to the invasiveness, staging, and prognosis of bladder cancer. High expression of HIF1A is generally associated with high tumor grade, deep muscle layer infiltration, and poor prognosis20. HIF1A is implicated in regulating the metabolic pathways of tumor cells, maintaining energy balance and survival for tumor cells21.
Bioinformatics analysis and experimental validation identified miR-767-3p as the sole downstream miRNA of circPSMA4, with a direct interaction between them. Sponge sequencing (circPSMA4 binding to miR-767-3p) enhanced bladder cancer cell proliferation. Transwell experiments demonstrated that decreased miR-767-3p expression increased BCa cell migration and invasion, while miR-767-3p overexpression reduced these behaviors. Downregulation of miR-767-3p was linked to increased glycolysis, lactate production, glucose uptake, and ATP generation in BCa cells, suggesting that miR-767-3p might regulate the Warburg effect in BCa cells by modulating cell metabolism. The study also discovered that miR-767-3p can directly target HIF1A mRNA, regulating its expression. HIF1A is a key transcription factor in tumor angiogenesis and metabolism. Overexpression of miR-767-3p suppressed HIF1A expression, impacting BCa cell metabolism and proliferation. Given miR-767-3p’s close association with BCa cell proliferation, invasion, migration, and metabolism, it might serve as a potential therapeutic target for bladder cancer. Modulating miR-767-3p expression levels could help Control tumor progression.
This study systematically elucidated the molecular mechanism by which circPSMA4 relieves the inhibition of HIF1A by sponge-mediated adsorption of miR-767-3p, thereby activating the glycolytic pathway and promoting the malignant phenotype of bladder cancer cells. This finding not only expands the functional understanding of circRNA in bladder cancer but also provides new insights for developing targeted therapeutic strategies based on the circPSMA4/miR-767-3p/HIF1A axis. The findings of this study suggest that circPSMA4 could serve as a novel biomarker for the diagnosis or prognosis assessment of bladder cancer; intervention strategies targeting the circPSMA4/miR-767-3p/HIF1A axis (such as circPSMA4 inhibitors or miR-767-3p mimetics) may offer new directions for bladder cancer treatment; and inhibitors of HIF1A and its downstream metabolic enzymes (such as LD HA) may enhance the efficacy of existing chemotherapy regimens. Future studies should validate the feasibility of these targets in clinical cohorts and explore their synergistic effects with immunotherapy. Although this study validated the function of circPSMA4 in vitro and in animal models, the focus was on the miR-767-3p/HIF1A axis. circPSMA4 may exert synergistic effects through other miRNAs or protein interactions, necessitating multi-omics analysis to comprehensively elucidate its regulatory network. Additionally, the animal model used subcutaneous tumor transplantation, failing to simulate the influence of the bladder cancer in situ microenvironment on tumor progression.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Fan, X. et al. Global analysis of miRNA-mRNA regulation pair in bladder cancer. World J. Surg. Oncol. 20(1), 66 (2022).
Dyrskjøt, L. et al. Bladder cancer. Nat. Rev. Disease Primers 9(1), 58 (2023).
Dobruch, J. & Oszczudłowski, M. Bladder cancer: Current challenges and future directions. Medicina (Kaunas, Lithuania) 57(8) (2021).
Richters, A., Aben, K. K. H. & Kiemeney, L. The global burden of urinary bladder cancer: An update. World J. Urol. 38(8), 1895–1904 (2020).
Yang, X. et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol. Cancer 20(1), 4 (2021).
Xie, B. et al. CircXRN2 suppresses tumor progression driven by histone lactylation through activating the Hippo pathway in human bladder cancer. Mol. Cancer 22(1), 151 (2023).
Wei, W. et al. EIF4A3-mediated biogenesis of circSTX6 promotes bladder cancer metastasis and cisplatin resistance. J. Exp. Clin. Cancer Res. CR 43(1), 2 (2024).
Li, J. et al. miRNA-143 as a potential biomarker in the detection of bladder cancer: a meta-analysis. Future Oncol. (London, England) 20(18), 1275–1287 (2024).
Gu, C. et al. UBAC2 promotes bladder cancer proliferation through BCRC-3/miRNA-182-5p/p27 axis. Cell Death Dis. 11(9), 733 (2020).
Zhuang, C. et al. Circular RNA hsa_circ_0075828 promotes bladder cancer cell proliferation through activation of CREB1. BMB Rep. 53(2), 82–87 (2020).
Lu, Q. et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer 18(1), 111 (2019).
Zheng, F. et al. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 460, 139–151 (2019).
Yuan, W. et al. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol. Oncol. 13(7), 1559–1576 (2019).
Liu, F. et al. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene 39(8), 1696–1709 (2020).
Cao, W. et al. Circ0001429 regulates progression of bladder cancer through binding miR-205-5p and promoting VEGFA expression. Cancer Biomark. Sect. A Dis. Mark. 25(1), 101–113 (2019).
Jiang, W. et al. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma malignance by decreasing miR-767-3p targeting SET. Biochem. Biophys. Res. Commun. 500(2), 211–216 (2018).
Mokhtari, M. et al. WASF3 overexpression affects the expression of circular RNA hsa-circ-0100153, which promotes breast cancer progression by sponging hsa-miR-31, hsa-miR-767–3p, and hsa-miR-935. Heliyon 9(12), e22874 (2023).
Xu, X. et al. Hsa_circ_0018818 knockdown suppresses tumorigenesis in non-small cell lung cancer by sponging miR-767-3p. Aging 12(9), 7774–7785 (2020).
Li, T. F. et al. Effects and mechanisms of N6-methyladenosine RNA methylation in environmental pollutant-induced carcinogenesis. Ecotoxicol. Environ. Saf. 277, 116372 (2024).
Lin, Q. et al. Glycoprotein α-Subunit of Glucosidase II (GIIα) is a novel prognostic biomarker correlated with unfavorable outcome of urothelial carcinoma. BMC Cancer 22(1), 817 (2022).
Loh, X. Y. et al. RNA-binding protein ZFP36L1 suppresses hypoxia and cell-cycle signaling. Can. Res. 80(2), 219–233 (2020).
Funding
This work was supported by the Hebei Medical Science Research Planning Project (No.20241097).
Author information
Authors and Affiliations
Contributions
C.G and X. Z.W study concept and design. Y. A. Y manuscript writing, Nuclear and Cytoplasmic RNA Isolation. F. Y data analysis and Glucose Uptake Assay. W.Z and F. A Seahorse Energy Metabolism Analysis and Western Blot. Y. F. S Scratch Test . All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Laboratory Animal Welfare Ethics Review Committee of Hebei University. This study has been approved by the Ethics Committee of the Affiliated Hospital of Hebei University. Ethics approval number: HDFY-LL-2022-087.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, C., Wang, X., Sun, Y. et al. CircPSMA4 as a novel circular RNA enhances the proliferation migration invasion and metabolism of bladder cancer cells through the miR-767-3p/HIF1A pathway. Sci Rep 15, 40468 (2025). https://doi.org/10.1038/s41598-025-24304-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24304-0