Abstract
Liver cancer has a high incidence and mortality rate globally, particularly in patients requiring intensive care unit (ICU) admission. Early prediction of in-hospital mortality for these patients is crucial, yet lacking reliable tools. This study aims to develop and evaluate machine learning (ML) models for predicting in-hospital mortality in critically ill liver cancer patients admitted to the ICU. This retrospective study used data from the MIMIC-III and MIMIC-IV databases, including 862 patients from MIMIC-III (training cohort) and 692 patients from MIMIC-IV (validation cohort). The study focused on patients diagnosed with liver cancer, identified by specific ICD codes. Four ML algorithms, namely logistic regression, random forest, XGBoost, and LightGBM, were used to predict in-hospital mortality based on clinical characteristics, laboratory results, and severity scores. Performance was evaluated using accuracy, AUROC, AUPRC, F1 score, and Kaplan-Meier curves. A total of 1,554 patients were included. The random forest model demonstrated the best performance, with an AUROC of 0.911 (95% CI: 0.855–0.956) and an AUPRC of 0.823 (95% CI: 0.718–0.905) in the internal test set, and an AUROC of 0.857 (95% CI: 0.826–0.889) and an accuracy of 0.828 (95% CI: 0.802–0.857) in the external validation set. Kaplan-Meier curves showed that all four models effectively stratified high-risk and low-risk groups. Key features influencing the prediction included APSIII, SAPSII, LODS, OASIS, and vital signs such as heart rate, temperature, and oxygen saturation. Feature importance analysis revealed that clinical severity scores played a major role in predicting mortality. This study demonstrates the potential of machine learning algorithms, particularly random forest, in predicting in-hospital mortality for critically ill liver cancer patients in the ICU. The identified clinical features provide valuable insights for clinicians in assessing patient risk and making timely interventions.
Similar content being viewed by others
Introduction
Liver cancer is one of the most common and fatal cancers worldwide. It is a leading cause of cancer death in both men and women, accounting for a significant proportion of global cancer-related deaths1. In 2020, over 830,000 people died from liver cancer, making it the third leading cause of cancer-related death worldwide2. Although treatment methods for liver cancer have improved in recent years, the prognosis remains poor, with low five-year survival rates across all stages, especially for patients with advanced liver cancer3,4,5,6. Many liver cancer patients require admission to the intensive care unit (ICU) due to complications such as acute liver failure, hepatic encephalopathy, gastrointestinal bleeding, and infections7,8. Among these, liver failure is the primary reason for ICU admission in liver cancer patients. Despite advancements in treatment, the mortality rate for ICU-admitted liver cancer patients remains extremely high, with studies showing a mortality rate exceeding 50%, and even higher for advanced-stage patients9,10. At present, the lack of early prediction and risk stratification of in-hospital mortality remains a major challenge for ICU clinicians11,12,13. Deciding which high-risk and poor-prognosis liver cancer patients should be admitted to the ICU requires considering a range of complex factors, including underlying diseases, liver function reserves, treatment plans, and the preferences of patients and their families14. These critically ill liver cancer patients generally have low long-term survival rates, and the cost of healthcare during hospitalization is high. Therefore, it is necessary to explore risk prediction models to identify high-risk groups among ICU-admitted liver cancer patients. The development of artificial intelligence has significantly improved the predictive models for estimating cancer patient mortality risk. Machine learning (ML), a novel form of artificial intelligence, can leverage large datasets and the rapid development of deep learning to transform measurement results into relevant predictive models, especially for cancer15,16. In recent years, ML has been proven effective in predicting liver cancer susceptibility, recurrence, and patient survival17,18,19. However, there is still limited data on the use of ML models to predict in-hospital mortality risk for liver cancer patients in ICU settings. Therefore, this study aims to establish four ML models, logistic regression, random forest, gradient boosting machine (GBM), and extreme gradient boosting (XGBoost) to predict the in-hospital mortality rate for ICU-admitted liver cancer patients. These models will help develop personalized prevention strategies for critically ill liver cancer patients and assist clinicians in making treatment decisions. Additionally, we aim to compare these four ML models to determine the best model for predicting in-hospital mortality in ICU-admitted liver cancer patients.
Methods
Data source
This retrospective study used data from the Medical Information Mart for Intensive Care (MIMIC-III and MIMIC-IV) databases. MIMIC-III contains data from over 60,000 ICU patients treated at the Beth Israel Deaconess Medical Center (BIDMC), Harvard University, between 2001 and 201220. MIMIC-IV (version 2.0) extends the data range to include ICU patients from 2008 to 201921. Both databases include rich clinical characteristics, laboratory results, vital signs, and medical records, providing high-quality data for this study. Since the data used in this study are from public databases, patient consent and institutional review board (IRB) approval were not required. All research procedures strictly followed the ethical standards set by the 1964 Declaration of Helsinki and its subsequent amendments. After completing an online training course and the Protecting Human Research Participants exam, we were granted permission to extract data from the MIMIC-III and MIMIC-IV databases. Methodologically, the study adheres to the guidelines of the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) statement22.
Cohort selection
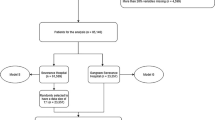
Patients were excluded from the study if they met any of the following criteria: (1) age under 18 years at the time of first ICU admission; (2) multiple ICU admissions. We selected patients who met the diagnostic criteria from the MIMIC-III and MIMIC-IV databases. Patients with ICD-9 codes starting with 155 or 197.7 and ICD-10 codes starting with C22 or C78 were included in the study. We randomly selected the MIMIC-III database as the training cohort and the MIMIC-IV database as the validation cohort. To prevent data leakage, we only selected samples from MIMIC-IV where the admission occurred after 2014. In the end, 862 patients from the MIMIC-III database were included in the training cohort, and 692 patients from the MIMIC-IV database were included in the validation cohort. The specific process is shown in Fig. 1.
Features and outcome
Baseline characteristics and admission information included age and gender. Comorbidities were collected based on the ICD codes recorded in both databases, including hypertension, diabetes, chronic kidney disease, myocardial infarction, congestive heart failure, atrial fibrillation, valvular disease, chronic obstructive pulmonary disease, stroke, hyperlipidemia, and liver disease. The Charlson Comorbidity Index (CCI)23,24 was also included. Additionally, severity scores were collected, including the Sequential Organ Failure Assessment (SOFA) score25, Oxford Acute Severity of Illness Score (OASIS)26, Acute Physiology Score III (APSII)27, Systemic Inflammatory Response Syndrome (SIRS) score, Logistic Organ Dysfunction Score (LODS)28, and the Model for End-Stage Liver Disease (MELD) score29. Furthermore, the initial vital signs and laboratory results from the 24 h before ICU admission were measured. Detailed laboratory tests and vital signs are shown in Table S1. Since these features were routinely measured at the time of patient admission and are easy to obtain in practice, no further feature selection was performed. The missing data rate for all variables included in the study was kept below 30%. To reduce the impact of missing data on the classification process, we used the Predictive Mean Matching (PMM) method for imputation. PMM predicts the mean of missing values and finds the closest observed values from the dataset to fill in the missing data30,31. This method has performed well in handling missing values and effectively estimates the missing data while reducing potential bias. We calculated the mean and standard deviation for each feature in the training set and performed Z-standardization on the features. The primary outcome of the study was in-hospital mortality, defined by the patient’s status at discharge (whether alive or dead).
Statistical analysis
For normally distributed continuous data, the mean and standard deviation were used for description. For between-group comparisons of normally distributed continuous data, a t-test was used. For data that did not follow a normal distribution, the Wilcoxon Mann-Whitney U test was used. Categorical data were described as frequencies and percentages, and between-group differences were compared using the chi-square test or Fisher’s exact test. A p-value of less than 0.05 was considered statistically significant. Baseline characteristics were reported for the training and validation cohorts. All statistical analyses were performed using R (version 4.0.1).
Model development and evaluation
In this study, we used random stratified sampling to split the MIMIC-III dataset into a training set (80%) and an internal test set (20%). The internal test set was created to compare its performance with that of the external test set (MIMIC-IV) and to prevent overfitting. To address the class imbalance in the training data, we applied the Synthetic Minority Over-sampling Technique (SMOTE) within the training set to balance the number of classes. The internal test set and external test set were kept unchanged to ensure unbiased model evaluation. When testing the model on the external test set, all samples from the MIMIC-III dataset were used as the training set. The study focused on applying 4 popular machine learning algorithms (logistic regression, random forest (RF), extreme gradient boosting (XGBoost), and LightGBM) to predict in-hospital mortality in liver cancer ICU patients.
To evaluate model performance, we used confusion matrix metrics, including accuracy, area under the receiver operating characteristic curve (AUROC), area under the precision-recall curve (AUPRC), accuracy, and F1 score. We also compared the performance of models trained with SMOTE to those trained without SMOTE to evaluate the impact of class balancing. In addition, we plotted calibration curves for both SMOTE and non-SMOTE models and used the Brier score to further assess the calibration and overall performance of the models. We used 1,000 bootstrap iterations to calculate the confidence intervals for all performance metrics, including AUROC, AUPRC, accuracy, and F1 score. The model with the highest AUROC in both the internal test set and external test set was considered to have the best predictive performance. The modeling work was carried out using Python (version 3.7.3).
Kaplan-Meier curve
In the external validation cohort, we used each machine learning model to calculate the predicted probability of in-hospital mortality for each patient. The optimal threshold was defined as the cutoff point that achieved the maximum sum of sensitivity and specificity in the training set. Patients were stratified into high-risk and low-risk groups based on this threshold. Kaplan-Meier (KM) survival curves were constructed using the survival package in R, with ICU admission as the starting point and either in-hospital death (event) or discharge (censored) as the endpoint. A log-rank test was performed to compare the survival differences between the two risk groups, and a P-value less than 0.05 was considered statistically significant. This time-to-event analysis was used to evaluate the models’ ability to stratify mortality risk over time and to assess their clinical utility in early prognostic evaluation of critically ill liver cancer patients.
SHAP analysis
To evaluate the importance of individual features, we used feature ranking as a method to assess their impact on the final prediction. The contribution of each input variable to the model output was determined using Shapley values, which are based on cooperative game theory. To address the opaque nature of machine learning algorithms and facilitate clinical interpretation, we applied SHAP and Local Interpretable Model-agnostic Explanations (LIME) to help explain the predictions made by the most effective models. We calculated feature SHAP values for the best-performing models from both the internal and external test sets. Then, we used beeswarm plots to show the relationship between the top 20 features’ values and their SHAP values, based on the global SHAP values from the training, internal test, and external test sets. Additionally, we presented scatter plots of the top 20 features in the training set to help visualize the dependence between feature values and their corresponding SHAP values.
Results
Baseline characteristics
A total of 1,554 patients were included in this study, with 862 patients in the training cohort extracted from the MIMIC-III database and 692 patients in the validation cohort extracted from the MIMIC-IV database. In the training cohort, 226 patients (26.2%) died during hospitalization, while in the validation cohort, 184 patients (26.6%) died. Table 1 shows the baseline characteristics of the training and validation groups. The training cohort had a higher proportion of patients with mild liver disease (26.8% vs. 15.3%), while the validation cohort had a higher proportion of patients with severe liver disease (14.0% vs. 8.7%). In terms of scores, the Charlson Comorbidity Index was significantly lower in the training cohort (2.98 ± 1.35) compared to the validation cohort (3.44 ± 1.26). Additionally, the SIRS score was lower in the validation cohort (2.58 ± 0.87 vs. 2.87 ± 0.92). Although there were no significant differences in SOFA, OASIS, and SAPS II scores between the two groups, the in-hospital mortality rate was similar (26.6% in the validation cohort vs. 26.2% in the training cohort), suggesting comparable disease severity between the two groups. See Table 1 for details.
Model performance
We used 4 models to predict in-hospital mortality using all features. As shown in Table 2, which reports the results of models trained without SMOTE, the logistic regression model had the poorest predictive performance, followed by XGBoost, LightGBM, and random forest. In the internal test set, the random forest model performed the best, with the highest AUROC of 0.911 (95% CI: 0.855–0.956), AUPRC of 0.823 (95% CI: 0.718–0.905), and accuracy of 0.861 (95% CI: 0.809–0.908), although its F1 score 0.745 (95% CI: 0.632–0.836) was slightly lower than that of logistic regression 0.722 (95% CI: 0.610–0.818). In the external test set, random forest also performed excellently, with an AUROC of 0.857 (95% CI: 0.826–0.889) and accuracy of 0.828 (95% CI: 0.802–0.857), although its F1 score decreased to 0.577 (95% CI: 0.502–0.641). LightGBM and XGBoost performed similarly across both datasets, with LightGBM showing strong performance in the internal test set with an AUPRC of 0.786 (95% CI: 0.662–0.888), but its performance slightly decreased in the external test set with an AUPRC of 0.717 (95% CI: 0.657–0.773).
When comparing models trained with SMOTE to those without SMOTE, we found that models trained without SMOTE consistently outperformed those trained with SMOTE across key performance metrics, particularly AUROC, AUPRC, and Brier score. For example, in the external test set, the random forest model without SMOTE achieved a Brier score of 0.1254, which was lower than the SMOTE-trained version (0.1447), indicating better calibration and overall performance. These findings suggest that applying SMOTE did not improve, and in some cases slightly degraded, the predictive performance and calibration of the models.
Additionally, we performed ROC analysis to further confirm the predictive ability of these four models for in-hospital mortality. As shown in Fig. 2A and B, logistic regression had the worst predictive performance, followed by XGBoost and LightGBM. Overall, the random forest model showed the best predictive performance in both the internal and external test sets. Furthermore, we plotted Kaplan-Meier survival curves for the high-risk and low-risk groups identified by the four models in the external test set (Fig. 3). The log-rank test showed that the high-risk and low-risk groups identified by all four models were effective (P < 0.001), indicating that all four models can be used for early mortality risk prediction in liver cancer patients entering the ICU. Figure S1 shows the predicted probability distributions for the four models in the training, internal test, and external test sets.
The detailed results for models trained with SMOTE are provided in Table S2, while a direct comparison of model performance between SMOTE and non-SMOTE training is presented in Fig. S3. The corresponding calibration curves and comprehensive Brier score analyses are shown in Fig. S4.
Feature importance analysis
To identify the important features that affect the model’s output, we performed feature importance analysis using the best-performing random forest model. We calculated the SHAP values for the features in the training set, internal test set, and external test set, and ranked them. Figure 4 shows the top 20 features for each dataset. Although the feature order slightly varies across datasets, many high-ranking features are common, indicating that these features generally have strong predictive power in the model. In the training set, APSIII, SAPSII, and LODS were among the top features, followed by OASIS, average respiratory rate, and average temperature. These features appeared consistently in all datasets, highlighting their importance in predicting in-hospital mortality risk for liver cancer ICU patients. The feature ranking in the internal test set was highly consistent with that of the training set, though certain features, like average SPO2 and minimum heart rate, had a different order. However, APSIII, SAPSII, LODS, and OASIS remained at the top, showing the stability and consistency of these clinical scores across different datasets. The feature importance ranking in the external test set was slightly different, especially with higher rankings for maximum ALB and minimum SBP, but key features like APSIII, SAPSII, and LODS still appeared in the top 20, further confirming the stability of these features in cross-dataset applications. The horizontal axis and color of the points indicate that the random forest model successfully captured the relationship between scores like APSIII, SAPSII, LODS, OASIS, and SOFA, with higher scores being associated with a higher probability of in-hospital mortality. More details on the relationship between feature values and SHAP values are shown in Fig. S2.
Summary of SHAP values for the top 20 features in the training set, internal test set, and external test set. (A), (B), and (C) represent the training set, internal test set, and external test set, respectively. The color in the plot indicates the size of each feature’s value, with the color gradient from blue to red representing an increase in variable value from low to high. The horizontal axis of the points represents the SHAP values for the corresponding features, reflecting the extent of the feature’s impact on the model’s predictions. Positive SHAP values indicate a positive effect, while negative SHAP values indicate a negative effect.
Discussion
In this retrospective study, we developed and validated machine learning algorithms based on clinical features using the public databases MIMIC-III and MIMIC-IV to predict in-hospital mortality in critically ill liver cancer patients. The MIMIC-III and MIMIC-IV datasets represent data from different time periods. We used the MIMIC-III dataset as the training set and the MIMIC-IV dataset as the external test set. This time-based data division strategy, where earlier data is used for model development and later data for validation, helps effectively reduce the risk of data leakage and overfitting. This design makes the model’s development and validation more externally valid because it avoids using future information for training, making it more aligned with real-world clinical settings. The random forest model showed the best overall performance, with an AUROC of 0.844 in the external test set. Using advanced machine learning techniques, we identified several important clinical features related to in-hospital mortality, including scores like APSIII, SAPSII, LODS, OASIS, as well as vital signs such as average respiratory rate, temperature, heart rate, oxygen saturation, and laboratory indicators like minimum lactate, maximum creatinine, minimum WBC, and INR. These results provide insights that need further consideration. In solid tumors, liver cancer patients have a high ICU in-hospital mortality rate. Studies show that the mortality rate due to liver cancer rupture can range from 25% to 75% during the acute phase32,33. ICU admission-related annual medical costs in the U.S. amount to $3 billion, with an average cost of $116,200 per admission34. Advanced cirrhosis patients are likely to be admitted to the ICU due to critical conditions such as sepsis, kidney failure, or respiratory failure. Although some studies report improved prognosis for cirrhosis patients admitted to the ICU, the prognosis remains poor, with mortality rates as high as 45% or even higher. These findings highlight the importance of predicting and intervening in in-hospital mortality for critically ill liver cancer patients. However, clinicians still face challenges in identifying high-risk ICU patients. Therefore, it is crucial to develop and promote reliable predictive models to identify these patients early and provide effective interventions to improve their prognosis.
Given the increasing applicability and effectiveness of supervised machine learning algorithms in disease prediction modeling, the scope of research is expanding. Well-known supervised learning classifiers, such as XGBoost, random forest, LightGBM, and neural networks, have gradually been applied in clinical practice15,35. With the help of machine learning classification, decision support models assisted by machine learning have proven to be more advantageous than traditional linear models. In this study, random forest, LightGBM, and XGBoost demonstrated their ability to capture nonlinear relationships, and their performance surpassed that of logistic regression, which is consistent with previous reports36. As an ensemble learning method, random forest has several significant advantages. By building multiple decision trees and making voting decisions, it effectively avoids overfitting that may occur with a single decision tree, thereby improving the model’s robustness and generalization ability. Additionally, random forest can handle high-dimensional feature spaces and maintain high prediction accuracy, especially when the number of features far exceeds the number of samples. These advantages make random forest the most effective model in this study. We used Kaplan-Meier survival curves to assess the effectiveness of different models in predicting in-hospital mortality risk for liver cancer patients admitted to the ICU. By dividing patients into high-risk and low-risk groups based on each model’s predicted probabilities, we clearly observed that the survival probability of the high-risk group was significantly lower than that of the low-risk group. This indicates that the clinical data routinely collected has the ability to distinguish between high-risk and low-risk liver cancer ICU patients. In addition, we explored the effect of applying the SMOTE to address potential class imbalance. We compared the performance of models trained with SMOTE and those trained without SMOTE. The models without SMOTE consistently achieved better discrimination and calibration, so we ultimately adopted the results without SMOTE for our final analysis. These findings are consistent with previous research37,38, which has reported that resampling techniques such as SMOTE may sometimes degrade model calibration.
We used the visualization capabilities of SHAP to analyze the impact of individual feature values on the model’s output. The scores of APSIII, SAPSII, LODS, and OASIS had the greatest impact. These scoring systems are widely used in clinical practice to assess the severity of illness in critically ill patients. The results show that early in the ICU admission of liver cancer patients, these scores can effectively help predict in-hospital mortality. APSIII mainly considers clinical physiological indicators such as partial pressure of oxygen, blood pressure, temperature, and laboratory results like serum electrolyte levels in the early ICU phase of liver cancer patients. SAPSII primarily assesses acute pathophysiological status, covering a comprehensive evaluation of blood gas analysis, kidney function, liver function, and more. LODS assesses the severity of multiple organ failure and provides strong predictive support for liver cancer ICU patients. It scores based on the function of multiple organs, while the OASIS score combines physiological data and clinical features, offering high sensitivity in predicting mortality risk for critically ill patients.
However, there are several important limitations in this study. First, the retrospective and observational design may introduce unavoidable selection bias and confounding factors, which could affect the generalizability of the findings. Second, the data were exclusively derived from the publicly available MIMIC-III and MIMIC-IV databases, which are limited to a single healthcare system in the United States. Therefore, the model’s performance may not fully represent patient populations in other institutions or countries, and further external validation using multi-center datasets is warranted. Third, our dataset did not include pathological or radiological information related to liver cancer, making it impossible to determine tumor stage or incorporate it into the prediction models. The absence of this information may have introduced bias, as disease stage is an important prognostic factor. Finally, the study focused on machine learning techniques and primarily reported discrimination metrics such as AUROC and AUPRC. Future studies should also emphasize model calibration, clinical utility assessment, and prospective validation before clinical implementation.
Conclusions
In this study, we applied four machine learning methods to predict in-hospital mortality in critically ill liver cancer patients. The results showed that the random forest model performed the best. The scores of APSIII, SAPSII, LODS, and OASIS were the most important factors influencing the model’s prediction of in-hospital mortality. This study provides clinical feature explanations that offer useful reference information for ICU clinicians in predicting patient prognosis.
Data availability
The datasets analysed during the current study are available in the Medical Information Mart for Intensive Care (MIMIC-III and MIMIC-IV) repositories at https://physionet.org/ upon registration and completion of the data use agreement.
Code availability
The preprocessing and analysis code used in this study are publicly available at https://github.com/ljwa2323/Liver_cancer_ICU.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 71 (3), 209–249 (2021).
Ferlay, J. et al. Cancer statistics for the year 2020: an overview. Int. J. Cancer. 149 (4), 778–789 (2021).
Siegel, R. L. et al. Cancer statistics, 2023. Cancer J. Clin. 73 (1), 17–48 (2023).
Balogh, J. et al. Hepatocellular carcinoma: a review. J. Hepatocellular Carcinoma 2016, 41–53 (2016).
Gosalia, A. J., Martin, P. & Jones, P. D. Advances and future directions in the treatment of hepatocellular carcinoma. Gastroenterol. Hepatol. 13 (7), 398 (2017).
Reig, M. et al. BCLC strategy for prognosis prediction and treatment recommendation: the 2022 update. J. Hepatol. 76 (3), 681–693 (2022).
Mcphail, M. J. et al. Increased survival for patients with cirrhosis and organ failure in liver intensive care and validation of the chronic liver failure–sequential organ failure scoring system. Clin. Gastroenterol. Hepatol. 13 (7), 1353–1360 (2015).
Liu, Z. et al. The trends in incidence of primary liver cancer caused by specific etiologies: results from the global burden of disease study 2016 and implications for liver cancer prevention. J. Hepatol. 70 (4), 674–683 (2019).
Weil, D. et al. Prognosis of cirrhotic patients admitted to intensive care unit: a meta-analysis. Ann. Intensiv. Care. 7, 1–14 (2017).
Van Leeuwen, D. J. et al. Acute-on-chronic liver failure 2018: a need for (urgent) liver biopsy? Expert Rev. Gastroenterol. Hepatol. 12 (6), 565–573 (2018).
Moreau, R. et al. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology 144 (7), 1426–1437 (2013). e9.
Zaccherini, G., Weiss, E. & Moreau, R. Acute-on-chronic liver failure: definitions, pathophysiology and principles of treatment. JHEP Rep. 3 (1), 100176 (2021).
Jalan, R. et al. Development and validation of a prognostic score to predict mortality in patients with acute-on-chronic liver failure. J. Hepatol. 61 (5), 1038–1047 (2014).
Hourmant, Y. et al. Impact of early ICU admission on outcome of critically ill and critically ill cancer patients: a systematic review and meta-analysis. J. Crit. Care. 61, 82–88 (2021).
Rajkomar, A. et al. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med. 1 (1), 18 (2018).
Harutyunyan, H. et al. Multitask learning and benchmarking with clinical time series data. Sci. Data. 6 (1), 1–18 (2019).
Altaf, A. et al. Artificial intelligence–based model for the recurrence of hepatocellular carcinoma after liver transplantation. Surgery 176 (5), 1500–1506 (2024).
Huang, T. et al. Machine learning for prediction of in-hospital mortality in lung cancer patients admitted to intensive care unit. Plos One. 18 (1), e0280606 (2023).
Dos Santos, H. G. et al. Machine learning to predict 30-day quality-adjusted survival in critically ill patients with cancer. J. Crit. Care. 55, 73–78 (2020).
Johnson, A. E. et al. MIMIC-III, a freely accessible critical care database. Sci. Data. 3 (1), 1–9 (2016).
Johnson, A. E. et al. MIMIC-IV, a freely accessible electronic health record dataset. Sci. Data. 10 (1), 1 (2023).
Collins, G. S. et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. J. Br. Surg. 102 (3), 148–158 (2015).
Charlson, M. E. et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40 (5), 373–383 (1987).
Quan, H. et al. Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am. J. Epidemiol. 173 (6), 676–682 (2011).
Vincent, J-L. et al. The SOFA (Sepsis-related Organ Failure Assessment) Score To Describe Organ dysfunction/failure: on Behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine (see Contributors To the Project in the appendix) [Z] (Springer, 1996).
Johnson, A. E., Kramer, A. A. & Clifford, G. D. A new severity of illness scale using a subset of acute physiology and chronic health evaluation data elements shows comparable predictive accuracy. Crit. Care Med. 41 (7), 1711–1718 (2013).
Knaus, W. A. et al. The APACHE III prognostic system: risk prediction of hospital mortality for critically III hospitalized adults. Chest 100 (6), 1619–1636 (1991).
Le Gall, J-R. et al. The logistic organ dysfunction system: a new way to assess organ dysfunction in the intensive care unit. Jama 276 (10), 802–810 (1996).
Kamath, P. S. et al. A model to predict survival in patients with end–stage liver disease. Hepatology 33 (2), 464–470 (2001).
Van Buuren, S. & Groothuis-Oudshoorn, K. Mice: multivariate imputation by chained equations in R. J. Stat. Softw. 45, 1–67 (2011).
Li, P., Stuart, E. A. & Allison, D. B. Multiple imputation: a flexible tool for handling missing data. Jama 314 (18), 1966–1967 (2015).
Shimabukuro-Vornhagen, A. et al. Critical care of patients with cancer. Cancer J. Clin. 66 (6), 496–517 (2016).
Yoshida, H. et al. Spontaneous ruptured hepatocellular carcinoma. Hepatol. Res. 46 (1), 13–21 (2016).
Halpern, N. A. & Pastores, S. M. Critical care medicine in the united States 2000–2005: an analysis of bed numbers, occupancy rates, payer mix, and costs. Crit. Care Med. 38 (1), 65–71 (2010).
Hyland, S. L. et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat. Med. 26 (3), 364–373 (2020).
Purushotham, S. et al. Benchmarking deep learning models on large healthcare datasets. J. Biomed. Inform. 83, 112–134 (2018).
Van Den Goorbergh, R. et al. The harm of class imbalance corrections for risk prediction models: illustration and simulation using logistic regression. J. Am. Med. Inform. Assoc. 29 (9), 1525–1534 (2022).
Ke, J. X. C. et al. The effect of resampling techniques on the performances of machine learning clinical risk prediction models in the setting of severe class imbalance: development and internal validation in a retrospective cohort. Discover Artif. Intell. 4 (1), 91 (2024).
Acknowledgements
This research was supported by the Clinical Medicine Science and Education Innovation Group, Sichuan College of Traditional Chinese Medicine. The authors gratefully acknowledge their generous funding and support, which made this study possible.
Funding
This research was funded by the Center of Excellence-International Collaboration Initiative Grant at West China Hospital, Sichuan University (Grant Number: 139170052), the Sichuan Science and Technology Program (Grant Numbers: 2023YFS0200, 2021YFS0091), Chongqing medical scientific research project (Joint project of Chongqing Health Commission and Science and Technology Bureau) (Grant Numbers: 2025QNXM034) and the 1·3·5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (Grant Number: ZYAI24070).
Author information
Authors and Affiliations
Contributions
Z.Z. and J.X. contributed equally to this work. Z.Z. and J.X. designed the study, performed data preprocessing, and conducted statistical analyses. J.L. and X.Y. contributed to data curation and model development, while L.D. and X.Z. provided methodological guidance and critical revisions to the manuscript. Y.X. and M.L. assisted with data interpretation and visualization. S.H. supervised the project, verified the underlying data, and finalized the manuscript. All authors read and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The MIMIC-III and MIMIC-IV databases were publicly released after receiving approval from the Institutional Review Boards (IRB) of Beth Israel Deaconess Medical Center and the Massachusetts Institute of Technology in Boston, USA.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, Z., Xia, J., Luo, J. et al. Using machine learning for early prediction of in-hospital mortality during ICU admission in liver cancer patients. Sci Rep 15, 40507 (2025). https://doi.org/10.1038/s41598-025-24369-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24369-x