Abstract
Mitochondrial permeability transition (MPT)-driven necrosis is associated with kidney renal clear cell carcinoma (KIRC), but its role in prognosis remains unclear. This study develops a prognostic model for KIRC outcomes using MPT-driven necrosis-related genes (MPTDNRGs). Differentially expressed genes (DEGs) from TCGA-KIRC samples were analyzed and categorized based on MPTDNRGs scores. Three key genes—IL2RA, CD7, and CXCL13—were identified as significant prognostic markers and used to construct a risk model, validated through public datasets and reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The risk score and age were the independent prognostic factors. A nomogram incorporating these factors demonstrated good clinical utility. The high-risk group was enriched in immune-related pathways, such as systemic lupus erythematosus, while the low-risk group showed enrichment in metabolism-related pathways, including butanoate metabolism. Significant differences in 25 immune cells were observed between the risk groups, with the high-risk group exhibiting higher TIDE scores, suggesting a greater likelihood of immune escape. Additionally, a ceRNA network revealed complex interactions, such as CXCL13-hsa-miR-670-5p-AL121985.1, and predicted 25 transcription factors for key MPTDNRGs. This study presents a novel prognostic model for KIRC based on three MPTDNRGs, offering valuable insights into KIRC prognosis and potential therapeutic targets.
Similar content being viewed by others
Introduction
The incidence of renal cell carcinoma (RCC) is increasing worldwide and ranks first among cancers of the upper urinary tract system1. Kidney renal clear cell carcinoma (KIRC) is the most common type of RCC, accounting for almost 75–80% of RCC2. The study found that KIRC is both the most invasive subtype of kidney cancer and one of the solid tumor subtypes that are most resistant to conventional chemotherapy protocols3. KIRC is not sensitive to radiotherapy and chemotherapy4, so surgical resection is still the main treatment method for KIRC patients. However, after radical treatment of local RCC, up to 30% of patients experience tumor recurrence5. Therefore, targeted therapy has become one of the hot research directions of KIRC therapy6. In addition, early KIRC is often asymptomatic, which makes it difficult to detect, adding challenges for subsequent treatment. When patients are diagnosed with advanced metastasis, 30% of patients have a 5-year survival rate of less than 15%7. Therefore, exploring key genes is necessary for early diagnosis and treatment, and has positive significance for improving the prognosis of KIRC.
Necrosis triggered by mitochondrial permeability transition (MPT) is a type of cell demise that occurs outside of the apoptotic pathway, caused by oxidative stress and an excessive accumulation of calcium ions in the cell membrane. It mainly participated in the release of mitochondrial materials during cell death. Recent studies have shown that mitochondria have a role in the governance of mechanisms controlling cell death8, and this regulation is related to the transformation of membrane permeability. Once the transformation of mitochondrial membrane permeability occurs, cells will eventually die due to apoptosis or necrosis9,10. Scientific research has confirmed that MPT-driven necrosis is related to a multitude of diseases, based on the alterations in the molecular mechanisms that oversee cell death, such as brain injury, amyotrophic lateral sclerosis, fulminant death receptor-induced hepatitis11, and liver cancer predispositions12. In lung adenocarcinoma, the activity of mitochondrial pathways has been demonstrated to significantly influence the tumor immune microenvironment and the response to immunotherapy13. This study posits that a particular mitochondria-associated mechanism, namely MPT-driven necrosis, may also play a crucial role in the formation of an immunosuppressive microenvironment in kidney renal clear cell carcinoma (KIRC). This hypothesis, if validated, would enhance our comprehension of the involvement of mitochondria in cancer immunity. However, few studies have been conducted on necrotic and clear cell renal carcinoma driven by MPT. Therefore by identifying the genes that drive MPT, this might contribute to the formulation of new predictive strategies for the clinical course of KIRC patients.
The study leveraged transcriptome data to identify the key genes involved in MPT-mediated cell death in KIRC, followed by a validation process using clinical samples from patients with KIRC. Ultimately, a predictive model was formulated utilizing pivotal genes, laying a theoretical foundation for investigating the influence of MPT-associated necrosis genes in KIRC. This also holds substantial importance for the prediction of outcomes and the treatment strategies for patients.
Methods
Data source
The Cancer Genome Atlas-KIRC (TCGA-KIRC) transcriptomic and clinical information were obtained from TCGA database (http://cancergenome.nih.gov/). Totally 598 samples were acquired, comprising 526 KIRC tumor tissue samples (only 522 samples with survival information) and 72 paracancerous samples as training cohort. The 101 KIRC samples in transcriptom dataset (E-MTAB-1980) were gained from ArrayExpres database (https://www.ebi.ac.uk/biostudies/arrayexpress) as validation cohort14. The KIRC transcriptom dataset (GSE40435) was acquired from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds), including 101 KIRC tumor tissue samples and 101 paracancerous samples as validation cohort15. Moreover, 39 MPT driven necrosis-related genes (MPTDNRGs) were procured from M17902, M3873 and M16257 in Molecular signatures database (MSigDB) database (https://www.gsea-msigdb.org/gsea/msigdb).
Identification of candidate MPTDNRGs
In TCGA-KIRC dataset, differentially expressed genes (DEGs) were screened out from KIRC and paracancerous samples (|log2fold change (FC)| > 1 and adj.P (Benjamini-Hochberg) < 0.05) by DESeq2 package (v 1.36.0)16, denoted as DEGs-KIRC. The scores of MPTDNRGs were calculated for both KIRC and paracancerous samples by single sample gene set enrichment analysis (ssGSEA) algorithm of gene set variation analysis (GSVA) package (v 1.48.3)17. Then, differences in MPTDNRGs scores between KIRC and paracancerous samples were compared (P < 0.05). The KIRC samples were divided into high and low score groups according to MPTDNRGs score’s median value. Afterwards, DEGs were screened out from different score groups, denoted as DEGs-MPT (|log2FC| > 1 and adj.P (Benjamini-Hochberg) < 0.05). Volcano plot and heatmap of KIRC-DEGs and MPT-DEGs were plotted using ggplot2 package (v 3.3.6)18 and circlize package (v 0.4.15)19, respectively. Thereafter, candidate MPTDNRGs were acquired by overlapping KIRC-DEGs and MPT-DEGs using eulerr package (v 7.0.0)20. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of candidate MPTDNRGs were analyzed by ClusterProfiler package (v 4.8.2) (adj.P (Benjamini-Hochberg) < 0.05)21. GO and KEGG results were presented via ggpubr package (v 0.6.0)22 and treemap package (v 2.4-4)23, respectively24,25,26. Moreover, a protein-protein interaction (PPI) network was constructed according to candidate MPTDNRGs using STRING database (confidence score > 0.4). And the MCODE in Cytoscape software (v 3.9.1)27 was utilized to identify and visualize significant gene clusters within PPI network.
Identification of key MPTDNRGs
First, univariate Cox regression analysis was implemented on candidate MPTDNRGs that were screened out from PPI network to initially identify survival-related genes (HR ≠ 1, P < 0.05). Subsequently, the glmnet package (v 4.1-6)28 was used for least absolute selection and shrinkage operator (LASSO) Cox regression analysis on the genes that passed the proportional hazards (PH) hypothesis test for further key MPTDNRGs. Finally, multivariate Cox analysis was executed to obtain key MPTDNRGs (HR ≠ 1, P < 0.05). In addition, the expression levels of key MPTDNRGs in KIRC and paracancerous samples from both TCGA-KIRC dataset and GSE40435 dataset were analyzed.
Construction and validation of risk model
The risk score for each KIRC patient (N = 522) was determined according to relative expression of key MPTDNRGs and their associated LASSO Cox coeffcient. The formula was \({\rm{risk}}\;{\rm{source}} = \sum\nolimits_{i = 1}^n {({\rm{coe}}{{\rm{f}}_i}*{{\rm{X}}_i})}\), where Xi was the relative expression level of the i-th key MPTDNRG, coefi was the LASSO Cox coefficient corresponding to the i-th key MPTDNRG. To further assess validity of risk model, receiver operating characteristic (ROC) curve was plotted to determine area under the curve (AUC) by survivalROC package(v 4.1-6)29. Thereafter, KIRC patients were dichotomized into high and low risk groups according to risk score median value. The different risk groups underwent Kaplan–Meier (K–M) survival analysis to evaluate overall survival (OS) by survminer package (v 0.4.9)30. Furthermore, risk model was validated in E-MTAB-1980 dataset.
Independent prognostic analysis
To begin with, we merged risk scores and clinical features—like age, grade, gender, and tumor stage—from KIRC patients within the TCGA-KIRC dataset for constructing a univariate Cox regression model (HR ≠ 1, P < 0.05). Afterward, we derived independent prognostic factors from a multivariate Cox regression analysis (HR ≠ 1, P < 0.05) of the variables that passed the PH hypothesis test (P > 0.05). We then leveraged the rms package (version 6.7-0) to formulate a nomogram with these independent prognostic factors for predicting survival rates31. The predictive performance of the nomogram was gauged via calibration curves. Furthermore, decision curve analysis (DCA) was applied to determine if the nomogram had a greater clinical advantage compared to the independent prognostic factors alone.
Functional and annotation analyses
To delve into the pathways related to various risk groups, we began by analyzing the differential gene expression across the groups using the DESeq2 package, ranking the genes based on their log2FC. Subsequently, using the ranked list, GSEA was carried out with the ClusterProfiler package, considering results with adj.P (Benjamini-Hochberg) < 0.05 as significant. The reference gene sets for this analysis were obtained from the MSigDB database, namely the c2.cp.kegg.v2023.1.Hs.symbols.
Immune infiltration analysis
To further understand the differences from different risk groups of immune cells, the 28 immune cell32 enrichment scores for KIRC sample were determined by ssGSEA algorithm of GSVA package and to compare differences from different risk groups (P < 0.05). And Spearman correlation analyses were performed between differential immunity cells as well as between differential immunity cells and key MPTDNRGs by psych package (v 2.3.6)33. Besides, the expression of immune checkpoints [PD-L1 (CD274), PD-1 (PDCD1), CTLA-4 (CTLA4), TIGIT, LAG-3 (LAG3), GAL9 (LGALS9), TIM-3 (HAVCR2), PD-L2 (PDCD1LG2), SIRPα (SIRPA), BTLA and Siglec-7 (SIGLEC7)]34 were compared (P < 0.05). And to further assess the effectiveness of immunotherapeutic response, tumor immune dysfunction and exclusion (TIDE) scores were compared from different risk groups (P < 0.05)35. Tumor mutational burden (TMB) indicated the extent of genetic variation within the tumor cell genome. Consequently, somatic mutations in KIRC samples were analyzed utilizing the maftools package (v 2.16.0)36.
Construction of regulatory network
The miRNAs of key MPTDNRGs were predicted in miRDB database. And the lncRNAs corresponding to miRNAs were predicted in starBase database (clipExpNum > 7). Finally, a competitive endogenous RNA (ceRNA) network was created to explore molecular regulatory mechanisms of key MPTDNRGs. Additionally, the Jaspar database was utilized to predict transcription factors (TFs) associated with key MPTDNRGs.
Expression validation of key MPTDNRGs
KIRC tumor tissue samples and paracancerous tissue samples of 5 KIRC patients from the First Hospital of Shanxi Medical University were taken as the experimental group and the control group, respectively. All samples underwent reverse transcription quantitative polymerase chain reaction (RT-qPCR). This study received approval from the Scientific Research Ethics Review Committee of Shanxi Medical University, and informed consent was obtained from all participants. To validate the expression of key MPTDNRGs, total RNA was extracted from the 10 samples using TRIzol (Ambion, Austin, USA) following the manufacturer’s instructions. Reverse transcription to cDNA was performed using the SureScript First-Strand cDNA Synthesis Kit (Servicebio, Wuhan, China) according to the provided guidelines. RT-qPCR was conducted using the 2xUniversal Blue SYBR Green qPCR Master Mix (Servicebio, Wuhan, China). Primer sequences for the PCR were listed in Additional file 1. GAPDH was used as an internal reference gene. Gene expression levels were calculated using the 2−ΔΔCt method37.
Statistical analysis
The entire analytical process was carried out in R (version 4.2.3). Group differences were assessed using the Wilcoxon statistical test. A p-value below 0.05 was considered to signify statistical relevance.
Results
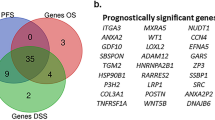
A sum of 56 candidate MPTDNRGs in PPI network were screen out
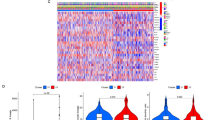
We screened out 3,314 DEGs-KIRCs, including 2,141 up-regulated and 1,173 down-regulated between KIRC and paracancerous samples from TCGA-KIRC dataset (Fig. 1a, b). Next, the scores of MPTDNRGs were calculated in KIRC and paracancerous samples, revealing a significantly higher score in KIRC samples (P < 0.0001) (Fig. 1c). Totally 408 DEGs-MPT, containing 267 up-regulated and 141 down-regulated were acquired from different scoring groups (Fig. 1d, e). Ultimately, we identified 339 candidate MPTDNRGs by overlapping DEGs-KIRC and DEGs-MPT (Fig. 1f). To uncover the biological roles and pathways linked to potential candidate MPTDNRGs, GO and KEGG enrichment analyses were performed (Fig. 2a, b). To be more specific, the potential MPTDNRGs were correlated with biological processes including T cell differentiation and the control of T cell stimulation, as indicated in the biological process (BP) entries. They were also related to cellular components such as endocytic vesicles and their membranes, as listed in the cellular component (CC) entries. Their molecular functions, noted in the molecular function (MF) entries, included cytokine receptor activity and immune receptor activity, and they were enriched in KEGG pathways including rheumatoid arthritis. Moreover, to delve into the mutual influences of candidate MPTDNRGs at the protein level, a PPI network was constructed, which contained 339 candidate MPTDNRGs and 3615 interaction pairs (Fig. 2c). And to further identify key MPTDNRGs, we analyzed significant gene clusters within PPI network, and acquired 56 candidate MPTDNRGs with 1314 interactions for subsequent analysis (Fig. 2d).
Screening of candidate MPT-driven necrosis-related genes in KIRC. a Volcano plot of DEGs-KIRC. Red upward-pointing triangles represent upregulated differentially expressed genes, blue downward-pointing triangles represent downregulated differentially expressed genes, and gray indicates non-significant genes. b Heatmap of DEGs-KIRC. Gene names are listed outside the circles. Colors indicate normalized gene expression levels, with red representing high expression and blue representing low expression. c MPTDNRGs scores were significant in control samples and KIRC samples discrepancy. ****P < 0.0001. d, e Volcano and heat maps of DEGs-MPT. f Venn map of candidate MPTDNRGs identification. KIRC kidney renal clear cell carcinoma, DEGs differentially expressed genes, MPTDNRGs MPT driven necrosis-related genes.
Further screening of candidate MPT-driven necrosis-related genes in KIRC. a, b GO and KEGG enrichment analyses for candidate MPTDNRGs. c PPI interaction network for candidate MPTDNRGs. d Using the MCODE plugin in Cytosccape to identify and visualize the PPI network of candidate MPTDNRGs yielded 56 genes. GO gene ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, PPI protein-protein interaction.
IL2RA, CD7 and CXCL13 with high expression levels in KIRC samples were identified as key MPTDNRGs
We identified 19 genes with survival-related [hazard ratio (HR) ≠ 1 and P < 0.05] (Fig. 3a), and PH hypothesis test revealed that these 19 genes satisfied the hypothesis (P > 0.05). Then, 7 genes were further screened out (Lambdamin = 0.015), namely IL2RA, CD7, CXCL13, CTLA4, CD38, IL2RG and IL10RA (Fig. 3b, c). Afterwards, IL2RA, CD7, CXCL13 and IL2RG were screened out by means of multivariate Cox analysis (HR ≠ 1 and P < 0.05) (Fig. 3d). Nevertheless, IL2RG exhibited a HR of less than 1 (HR = 0.69), contradicting the results of univariate Cox regression analysis (HR = 1.2). Accordingly, IL2RA, CD7 and CXCL13 were designated as key MPTDNRGs. And key MPTDNRGs exhibited high expression levels in KIRC samples from both TCGA-KIRC dataset and GSE40435 dataset (P < 0.0001) (Fig. 3e, f).
Screening and identification of key MPTDNRGs. a The forest maps of univariate Cox regression analysis for 56 candidate MPTDNRGs. b, c LASSO analysis of 19 genes. Left figure: The horizontal axis represents log(lambda) values, while the vertical axis denotes degrees of freedom, representing cross-validation error. Right figure: The horizontal axis represents log(lambda), while the vertical axis shows gene coefficients. d The Forest maps of multivariate Cox regression of 7 genes. Expression levels of the IL2RA, CD7 and CXCL13 in datasets (e, TCGA-KIRC dataset; f, GSE40435 dataset. ****P < 0.0001). MPTDNRGs MPT driven necrosis-related genes, LASSO Least absolute selection and shrinkage operator.
A risk model was constructed according key MPTDNRGs
Consequently, a risk model was constructed by key MPTDNRGs, with RiskScore calculated as follows: = IL2RA*0.3233 + CD7*0.3673 + CXCL13*(− 0.2197). In TCGA-KIRC dataset, the model was assessed through time-dependent ROC analysis, and AUC were 0.658, 0.614 and 0.625 at 1, 3 and 5 years, respectively (Fig. 4a). These findings indicated that our risk model had moderate and stable predictive accuracy. Figure 4b and c illustrated distribution of samples in different risk groups. Clearly and unequivocally, high risk patients had significantly worst OS than low risk group (P = 0.00014) (Fig. 4d). And we also carried out verification in E-MTAB-1980 dataset. Notably, the AUC were 0.810, 0.653 and 0.633 at 1, 3 and 5 years, respectively (Fig. 4e). And likewise, high risk patients had significantly worst OS (P = 0.036) (Fig. 4f–h). The results were consistent with training cohort.
Construction of a prognostic risk model and its prognostic value. a The ROC curves of risk model (TCGA-KIRC dataset). b Risk curve and survival time for the high- and low-risk groups (TCGA-KIRC dataset). c Heatmap of key MPTDNRG expression levels in high- and low-risk groups (TCGA-KIRC dataset). d Kaplan–Meier curves for the overall survival of patients in the high- and low-risk groups (TCGA-KIRC dataset). e The ROC curves of risk model (GSE40435 dataset). f Risk curve and survival time for the high- and low-risk groups (GSE40435 dataset). g Heatmap of key MPTDNRG expression levels in high- and low-risk groups (GSE40435 dataset). h Kaplan–Meier curves of overall survival for patients in high and low risk groups (GSE40435 dataset). ROC Receiver operating characteristic, KIRC Kidney renal clear cell carcinoma.
Only risk score and age were independent factors of prognosis
This research endeavored to determine if risk scores could act as a standalone predictor for the prognosis of patients with KIRC. As a consequence, we determined that risk scores, age, tumor grade, and stage are influential variables that affect the overall survival (OS) of KIRC patients (HR ≠ 1 and P < 0.05) (Fig. 5a). Nonetheless, the PH hypothesis test indicated that neither tumor grade nor stage fulfilled the required assumptions (P < 0.05). As a result, we proceeded with further analysis considering only risk score and age. Ultimately, we established that the risk score and age were the sole independent prognostic factors (HR ≠ 1 and P < 0.05) (Fig. 5b). Consequently, a nomogram was developed incorporating these two factors, risk score and age (Fig. 5c). The calibration plots, which closely matched the reference line, suggested that the nomogram had a favorable predictive accuracy (Fig. 5d). Furthermore, the decision curve analysis (DCA) at both 1 and 5 years showed that the nomogram had a clinical utility compared to using risk score and age in isolation (Fig. 5e–g).
Only risk score and age were independent factors of prognosis. a The result of univariate Cox regression analysis (independent prognosis). b The result of multivariate Cox regression analysis (independent prognosis). c A nomogram constructed based on independent prognostic factors. d Calibration curve for the nomogram. e–g DCA curves of 1-year, 3-year, and 5-year for nomogram model. DCA decision curve analysis.
Different risk groups-related signaling pathways
Conducting GSEA aimed to provide a more profound understanding of the associated signaling pathways and the potential biological processes that characterize the distinct risk groups. The detail results of GSEA could be found in Additional file 2 Specifically, high risk group was mainly enriched in systemic lupus erythematosus, leishmania infection etc., and low risk group was mainly enriched in oxidative phosphorylation, valine leucine and isoleicine, etc. (Fig. 6). GSEA uncovered distinct signaling pathways linked to various risk groups, thereby broadening our in-depth comprehension of KIRC.
Immune analysis of KIRC patients
The heatmap illustrated the scores of 28 immune cells (Fig. 7a). Evidently, in high risk group, except for CD56bright natural killer cells, eosinophils and neutrophils, the proportion of 25 immune cells were significantly higher (P < 0.05) (Fig. 7b). Then, we observed that the strongest correlation among differential immune cells was between T follicular helper cell and activated MDSC (r = 0.869 and P < 0.001) (Fig. 7c, d). Further, the strongest correlation was observed between CD7 and activated CD8 T cells (r = 0.856, P < 0.001), exhibiting significantly higher expression in high risk group (P < 0.0001) (Fig. 8a–c). Moving forward, we sought to determine if there existed any potential disparities in the levels of immune checkpoint expression across different risk categories. The results revealed that gene expression of 11 immune checkpoints was significantly higher in high risk group, like BTLA, CD274 and CTLA4 (P < 0.001) (Fig. 8d). TIDE score was analyzed to access the potential for tumor immune evasion. Obviously, high risk group patients exhibited significantly higher TIDE score (P < 0.05) (Fig. 8e). Additionally, the waterfall plot illustrated the top 20 mutations in tumor cells of different risk groups (Fig. 8f, g). The results indicated that VHL and PBRM1 mutations were more prevalent in different risk groups. The higher frequency mutations in high risk group were frame shift del mutation and missense mutation, while the most common mutations in low risk group were nonsense mutation, missense mutation and frame shift del mutation.
Immune analysis. a Heat map of immune cell infiltration. b Box plots of 28 types of immune cell infiltration in high and low risk groups (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns: not significant). c Correlation heat map of differential immune cells. Orange pentagrams indicate positive correlations, blue ones indicate negative correlations, with darker colors signifying stronger correlations. d Heatmap of correlation between key MPTDNRGs and differential immune cells.
Immunoassay results in high and low risk groups. a Scatterplot of correlation between CD7 and activated CD8 T-cells. b Box plot of CD7 expression in high and low risk groups. c Box plot of activated CD8 T-cells expression in high and low risk groups (****P < 0.0001). d Boxplots of immune checkpoint gene expression between high- and low-risk groups (***P < 0.001; ****P < 0.0001). e TIDE scores were different between high and low risk groups (*P < 0.05). The top 20 genes with the highest mutation frequencies in the f high-risk group and g low-risk group.
Regulatory network of key MPTDNRGs
The ceRNA network showed that 3 mRNAs of the risk model could interact with 27 miRNAs, which could in turn interact with 36 lncRNAs. The complex interaction pairs were formed, such as CXCL13-hsa-miR-670-5p-AL121985.1, IL2RA-hsa-miR-6088-AL513497.1 (Fig. 9a). A total of 25 TFs were predicted for key MPTDNRGs. Notably, IL2RA, CD7 and CXCL13 collectively predicted GATA2. CD7 and CXCL13 collectively predicted MAX and GATA3. CD7 and IL2RA collectively predicted USF2 (Fig. 9b). The results indicated the regulatory mechanism for key MPTDNRGs in KIRC.
Regulatory network and clinical validation of key MPTDNRGs. a The ceRNA network. Orange triangles represent mRNA, blue circles represent miRNA, and green circles represent lncRNA. b The TF-mRNA-miRNA network. The orange circles represent mRNA, and the green circles represent transcription factors. c–e RT-qPCR expression validation of IL2RA, CD7, CXCL13 in KIRC and control samples. MPTDNRGs MPT driven necrosis-related genes, RT-qPCR reverse transcription quantitative polymerase chain reaction, TF-mRNA-miRNA transcription factors-messenger RNA-microRNAs, KIRC Kidney renal clear cell carcinoma.
Verification of key MPTDNRGs expression
In the previous studies, we observed that IL2RA, CD7 and CXCL13 exhibited significantly higher expression levels in KIRC samples in both TCGA-KIRC and GSE40435 (P < 0.0001) (Fig. 3e, f). This prompted us to further employ RT-qPCR techniques to validate the clinical expression levels of key MPTDNRGs in patients with KIRC. Remarkably, RT-qPCR revealed that both IL2RA and CD7 showed significantly higher expression in KIRC samples (P < 0.05), while CXCL13 also showed an up-regulation trend in KIRC (P = 0.0820), consistent with our previous findings (Fig. 9c–e). Among these, no significant difference in CXCL13 expression was observed between groups. This may be attributed to insufficient statistical power due to the small sample size, or to substantial biological variability among individuals within the disease and control groups, making it difficult to achieve statistical significance. This finding warrants further validation in studies with larger sample sizes.
Discussion
The poor prognosis of KIRC has always been a big problem, and many patients still relapse after treatment. In an attempt to better prognosticate renal cancer, researchers have engaged in numerous studies. MPT-induced necrosis-related genes might play a crucial role in the processes of tumor cell proliferation, death, and dissemination38. Consequently, utilizing the clinical data and transcriptome information of KIRC available in public repositories, this study explored the necrotic genes driven by mitochondrial permeability transfer and key genes of KIRC through bioinformatics technology, and conducted clinical validation of key genes (IL2RA, CD7 and CXCL13), and constructed a new risk prognosis model, providing a new reference for clinical diagnosis and treatment of KIRC. In the TCGA training cohort, the model demonstrated an Area Under the Curve (AUC) of 0.658, 0.614, and 0.625 at 1, 3, and 5 years, respectively, indicating moderate and stable predictive accuracy. Conversely, in the external validation cohort (E-MTAB-1980), the results exhibited inconsistency: the AUC was notably high at 1 year (0.810) but significantly declined at 3 and 5 years (0.653 and 0.633, respectively). This discrepancy suggests that the genetic signature identified in this study may more accurately capture acute biological processes influencing short-term prognosis, such as early necrosis and immune responses, whereas long-term survival is likely affected by more complex clinical events, including drug resistance and late metastasis, which are not adequately addressed by current static models. Variations in patient populations and clinical management practices between databases may account for the observed fluctuations in validation outcomes. Although the model was validated using an independent cohort, its generalizability requires further confirmation through prospective, multicenter studies. As a clinical instrument, this three-gene signature holds promise for augmenting current clinical staging systems and demonstrates significant potential in the context of immunotherapy. Specifically, patients identified as high-risk may be appropriate candidates for combination immunotherapy, whereas those classified as low-risk might derive benefit from interventions such as targeted metabolic therapy. Future research endeavors will concentrate on the translation of this risk score into a viable clinical testing tool and on assessing its utility in guiding prospective clinical trials.
In this study, a nomogram incorporating risk score and age was developed, demonstrating commendable predictive accuracy and potential utility in clinical settings. A significant observation from the independent prognostic analysis was the exclusion of established clinical factors, particularly tumor stage and grade, from the final multivariate Cox model and prognostic map due to their violation of the proportional hazards (PH) assumption. Statistically, this indicates that the relationship between these factors and mortality risk varies over time, suggesting that their prognostic significance is not constant throughout the follow-up period. For instance, the substantial risk associated with an advanced stage at diagnosis may diminish for a patient who successfully navigates the initial high-risk phase. In contrast, the risk score derived from MPT-driven necrosis gene signatures (IL2RA, CD7, CXCL13) satisfied the proportional hazards assumption, suggesting that the model can offer consistent and stable predictive power for both short- and long-term patient survival. This distinction underscores the potential advantage of our genetic model: while traditional factors are essential for initial risk stratification, their variability over time may constrain long-term prognostic accuracy. Consequently, this model can serve as a complementary tool to enhance the precision of continuous risk assessment in the management of renal cancer following kidney transplantation, particularly due to its temporal stability in predicting long-term outcomes.
We identified the pivotal genes in KIRC that are linked to MPT-induced necrosis. Through the integrated analysis of the tumor microenvironment of KIRC, it was found that the expression of the immune-related gene IL2RA is related to the prognosis of clear cell carcinoma39, which is consistent with the results of our study. NK cell marker genes, including CD7 and six additional genes, can serve as a standalone biomarker for forecasting the prognosis and therapeutic responses in patients with KIRC, and are intimately associated with immunosuppression40. These studies have proved that IL-2RA and CD7 are correlated with the prognosis of patients with KIRC. However, the specific mechanism is still relatively limited and further exploration is needed. Researchers have more studies on CXCL13, and a large number of evidences show that CXCL13 is highly expressed in clear cell carcinoma41. In addition, circHIPK3 can promote the proliferation and metastasis of clear cell renal cell carcinoma (ccRCC) cells by altering miR-5083p/CXCL13 signaling42; M2 macrophages in the immune environment can secrete CXCL13, thus promoting the proliferation, migration, invasion and epithelial-mesenchymal transformation of ccRCC cells43; CXCL13 can activate the PI3K/AKT/mTOR signaling pathway by binding with CXCR5 to promote the proliferation and migration of ccRCC cells44. In conclusion, CXCL13 is closely related to KIRC, and increased CXCL13 expression is associated with poor survival outcomes in KIRC patients. This study independently confirmed the prognostic significance of several immune-related genes in kidney renal clear cell carcinoma (KIRC). While previous research has linked IL2RA and CD7 to prognosis, our study integrates these genes into a prognostic model focused on mitochondrial permeability transition (MPT)-driven necrosis, suggesting that their prognostic relevance may be partially mediated through this specific cell death pathway. Additionally, we expanded the analysis of CXCL13’s mechanism of action, revealing its association not only with proliferation and metastasis but also with immune infiltration. The high-risk microenvironment characterized by elevated CXCL13 expression exhibited dysfunctional immune cell recruitment and increased immune checkpoint expression, potentially explaining its correlation with poor survival outcomes.
In both the high- and low-risk cohorts, we observed significantly elevated TIDE scores among patients classified within the high-risk group, indicating a potential for immune escape. This observation aligns closely with the functional roles of the three pivotal genes under study. The overexpression of CXCL13, a critical chemokine involved in the formation of germinal centers and the recruitment of T follicular helper cells, signifies a highly specialized yet potentially dysfunctional T cell response that is susceptible to exhaustion45. CD7, a costimulatory molecule essential for T/NK cell activation, when persistently overexpressed, may indicate chronic and ineffective T cell activation, serving as a precursor to exhaustion46. IL2RA functions as a key marker of regulatory T cells47. Collectively, these three genes characterize a tumor microenvironment where CXCL13 and CD7 facilitate excessive immune cell recruitment and activation, while IL2RA may denote an induced immunosuppressive mechanism. In this imbalanced state of hyperactivity and suppression, the immune system is rendered incapable of effectively eradicating the tumor, ultimately resulting in immune escape. Furthermore, the current study identified a markedly increased expression of immune checkpoints in the high-risk cohort, which stands in contrast to the elevated TIDE score observed. Although immune checkpoint molecules such as PD-L1 and CTLA-4 are typically deemed essential for successful immunotherapy, within the context of this study’s high-risk characteristics, these molecules are indicative of a suboptimal response to such treatments48. This apparent paradox can be elucidated by the mechanism wherein the upregulation of immune checkpoints signifies exhaustion and dysfunction within the immune microenvironment, rather than T-cell activation49. We hypothesize that the necrotic features induced by MPTS contribute to the formation of a tumor microenvironment that is both highly inflammatory and immunosuppressive. This environment facilitates immune cell infiltration but impairs their capacity to effectively eradicate the tumor, ultimately leading to T-cell depletion. In this scenario, the overexpression of immune checkpoints serves to suppress ineffective immune responses, highlighting immune dysfunction. The inability to activate T cells with monotherapy using immune checkpoint inhibitors accounts for the elevated TIDE score50. This finding is consistent with the clinical observation that not all PD-L1-positive tumors are responsive to treatment and further supports the value of this study’s feature in identifying immunosuppressed tumors, suggesting that such tumors may require combination treatment strategies.
Evidence suggests that the presence of systemic lupus erythematosus correlates with a reduced likelihood of renal cancer, in an inverse relationship51. Through GSEA in our research, the systemic lupus erythematosus pathway was found to be enriched in the high-risk group. Our results do not contradict earlier studies, implying that the connection between systemic lupus erythematosus and KIRC is multifaceted, not merely a simple inverse association. It is possible that patients with KIRC have a poor prognosis when the systemic lupus erythematosus pathway is active. Or it may be caused by the use of immunotherapy. The relationship between leishmania infection and clear cell carcinoma of the kidney has not been reported. The oxidative phosphorylation pathway has been extensively studied in KIRC, in which oxidative phosphorylation is reported to play an important role. Deletion of chromosome 3p has also been associated with down-regulation of oxidative phosphorylation (OXPHOS)52. Clear cell cancer cells metabolize glucose primarily through glycolysis even when oxygen is plentiful. Thus, there is activation of the hypoxic response pathway in KIRC under normal oxygen conditions53. It has been demonstrated that the HIF1 signaling pathway influences mitochondrial behavior and shifts the metabolic pathway in cancer cells from oxidative phosphorylation to glycolysis54. The findings indicate a reduction in the activity of the Krebs cycle and the electron transport chain (OXPHOS) related to the Warburg effect at the protein level. Yet, it is noteworthy that the downregulation of these Krebs cycle elements and the majority of nuclear-encoded OXPHOS proteins was not evident at the mRNA level and was not detected by RNA-seq analysis alone. The researchers hypothesized that maintaining OXPHOS transcription levels similar to that of orthoxic cells, while it is beneficial for meeting the energy requirements of tumors, may provide a mechanism for rapidly inducing OXPHOS activity, which needs more and further experiments and studies to verify52. Overexpression of protein tyrosine phosphatase receptor gamma(PTPRG) can activate oxidative phosphorylation, inhibit apoptosis, inhibit epithelial-mesenchymal transformation, promote G1/S cell cycle arrest, and have anticancer effects55. The Gene Set Enrichment Analysis (GSEA) results from the current study indicate that the high-risk group is enriched in immune and inflammatory pathways, whereas the low-risk group is enriched in metabolic pathways, thereby identifying two distinct subtypes of kidney renal clear cell carcinoma (KIRC). We hypothesize that the high-risk subtype is driven by mitochondrial permeability transition (MPT) necrosis and exhibits high immunogenicity. The release of cellular contents due to MPT may provoke a robust innate immune response and chronic inflammation, ultimately leading to immune dysregulation and exhaustion, which aligns with a high Tumor Immune Dysfunction and Exclusion (TIDE) score56,57. Conversely, the low-risk subtypes may be driven by metabolic processes, maintaining normal mitochondrial function and oxidative metabolism, and displaying weaker immunogenic signals. This results in an ‘immune cold’ microenvironment and is associated with a more favorable prognosis.
The GATA transcription factor family is a zinc finger transcription factor belonging to GATA family proteins 1–6, and GATA transcription factors have been found to contribute to cell proliferation, apoptosis and tumorigenesis in a multitude of solid cancers58. The regulatory network analysis we conducted predicted the significance of GATA2 and GATA3 in the GATA family of transcription factors, highlighting their involvement in the MPT process in KIRC. The mRNA levels and protein expression levels of GATA2/3/6 in KIRC tissues were significantly decreased compared with normal tissues. Moreover, univariate analysis showed that decreased GATA2 expression level was associated with advanced tumor disease, positive distant metastasis, and lymph node metastasis status59. The presence of infiltrating immune cells showed a strong correlation with the expression patterns of GATA, and our analysis of immune cell infiltration also noted an increase in the proportion of these immune cells. Moreover, GATA2 was negatively correlated with B cells and positively correlated with CD8 + T cells, CD4 + T cells and neutrophils. Interestingly, CD8 + T cells were also the immune cells most strongly associated with a key gene, CD7. Due to homologous inhibition of phosphatase and tensin, increased GATA2 expression levels may promote the proliferation of breast cancer cells by stimulating AKT phosphorylation53. Furthermore, GATA2 was identified as the sole transcription factor binding to the promoters of all three pivotal genes, indicating its potential role as a central regulatory hub in the expression of prognostic markers. Considering the critical function of GATA factors in cell fate determination and immune response, we propose that dysregulation of GATA2 acts as a pivotal molecular switch in high-risk kidney renal clear cell carcinomas (KIRCs), leading to the concurrent overexpression of these genes. This overexpression is associated with transcriptional disruptions and aggressive phenotypes, including immune dysregulation and mitochondrial permeability transition (MPT)-driven necrosis. This hypothesis underscores the necessity of future investigations to confirm the central regulatory role of GATA2, which is crucial for elucidating the fundamental mechanisms underlying KIRC progression and may uncover novel therapeutic targets. For instance, the direct binding of GATA2 to the promoters of three genes was confirmed using ChIP-qPCR. The overexpression or knockdown of GATA2 in KIRC cell lines was employed to assess its impact on the expression of these genes and on cell phenotype. Within the ceRNA network, hub miRNAs, such as hsa-miR-670-5p, have the capacity to interact simultaneously with CXCL13 and multiple lncRNAs, potentially exerting significant regulatory functions. Future investigations can validate the interactions between miRNAs and mRNAs/lncRNAs through dual-luciferase reporter assays. These functional experiments will provide robust mechanistic evidence, thereby substantiating the bioinformatics predictions of this study.
Previous studies have shown that 3p loss and VHL mutation of chromosome almost always occur in the early stage of KIRC, and then additional aneuploidy produced by PBRM1, SETD2 or BAP1 mutations and defects and errors in DNA repair and mitosis drive tumor development. The acquisition and loss of key chromosomes, mutations in PI3K pathway elements, and other cancer-driving mutations confer the lethal potential of intra-tumor cloning and increase the likelihood of metastasis38.
Our study is subject to several limitations. Firstly, the limited sample size may compromise the statistical power and reliability of our findings. Future research should aim to increase the sample size and conduct larger clinical validation studies. Secondly, our study primarily utilized bioinformatics analysis and gene expression data, lacking functional experiments such as gene knockout or overexpression studies. These experiments will be incorporated in future research to substantiate the roles of the genes identified. Additionally, our estimates of immune infiltration may be biased; this could be addressed through more precise immunohistochemical testing in subsequent studies. Lastly, this study relies on data from public databases and lacks validation through clinical data. Future work will include clinical samples and incorporate animal experiments to enhance the robustness of our research findings.
Conclusions
In summary, by applying bioinformatics approaches, we discovered and clinically substantiated the key genes (IL2RA, CD7, and CXCL13) associated with cell necrosis driven by MPT in patients with KIRC, and subsequently formulated a fresh prognostic model. It offers clinicians a novel framework for more accurately forecasting patient outcomes and also presents a fresh target and approach for the management of KIRC. Nonetheless, additional research is required to assess the mechanisms through which MPT-driven necrosis influences tumor growth and advancement.
Data availability
The datasets generated and analysed during the current study are available in the [TCGA database, TCGA-KIRC], [http://cancergenome.nih.gov/]; [ArrayExpres database, E-MTAB-1980], [https://www.ebi.ac.uk/biostudies/arrayexpress]; [the Gene Expression Omnibus (GEO) database, GSE40435], [https://www.ncbi.nlm.nih.gov/gds]; [Molecular signatures database (MSigDB), MPTDNRGs]repository, [https://www.gsea-msigdb.org/gsea/msigdb]. Additional materials from this study are available by contacting the corresponding author at 390090960@qq.com.
Abbreviations
- MPT:
-
Mitochondrial permeability transition
- KIRC:
-
kidney renal clear cell carcinoma
- MPTDNRGs:
-
MPT driven necrosis-related genes
- DEGs:
-
Differentially expressed genes
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- RCC:
-
Renal cell carcinoma
- GEO:
-
Gene expression omnibus
- MSigDB:
-
Molecular signatures database
- ssGSEA:
-
single sample gene set enrichment analysis
- GSVA:
-
Gene set variation analysis
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PPI:
-
Protein-protein interaction
- LASSO:
-
Least absolute selection and shrinkage operator
- PH:
-
Proportional hazards
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- OS:
-
Overall survival
- DCA:
-
Decision curve analysis
- TIDE:
-
Tumor immune dysfunction and exclusion
- TMB:
-
Tumor mutational burden
- ceRNA:
-
competitive endogenous RNA
- TFs:
-
Transcription factors
- CC:
-
Cellular component
- MF:
-
Molecular function
- OXPHOS:
-
Oxidative phosphorylation
- PTPRG:
-
Protein tyrosine phosphatase receptor gamma
References
Capitanio, U. et al. Epidemiology of renal cell carcinoma. Eur. Urol. 75, 74–84. https://doi.org/10.1016/j.eururo.2018.08.036 (2019).
Nabi, S., Kessler, E. R., Bernard, B., Flaig, T. W. & Lam, E. T. Renal cell carcinoma: a review of biology and pathophysiology. F1000Research 7, 307. https://doi.org/10.12688/f1000research.13179.1 (2018).
Vuong, L., Kotecha, R. R., Voss, M. H. & Hakimi, A. A. Tumor microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov. 9, 1349–1357. https://doi.org/10.1158/2159-8290.Cd-19-0499 (2019).
Wang, Q. et al. Immune-Associated gene signatures serve as a promising biomarker of immunotherapeutic prognosis for renal clear cell carcinoma. Front. Immunol. 13, 890150. https://doi.org/10.3389/fimmu.2022.890150 (2022).
Klatte, T., Rossi, S. H. & Stewart, G. D. Prognostic factors and prognostic models for renal cell carcinoma: a literature review. World J. Urol. 36, 1943–1952. https://doi.org/10.1007/s00345-018-2309-4 (2018).
Courtney, K. D. et al. Phase I dose-escalation trial of PT2385, a first-in-class hypoxia-inducible factor-2α antagonist in patients with previously treated advanced clear cell renal cell carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 36, 867–874. https://doi.org/10.1200/jco.2017.74.2627 (2018).
Ochocki, J. D. et al. Arginase 2 suppresses renal carcinoma progression via biosynthetic cofactor pyridoxal phosphate depletion and increased polyamine toxicity. Cell Metabol. 27, 1263–1280e1266. https://doi.org/10.1016/j.cmet.2018.04.009 (2018).
Gogvadze, V., Orrenius, S. & Zhivotovsky, B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol. 18, 165–173. https://doi.org/10.1016/j.tcb.2008.01.006 (2008).
Armstrong, J. S. The role of the mitochondrial permeability transition in cell death. Mitochondrion 6, 225–234. https://doi.org/10.1016/j.mito.2006.07.006 (2006).
Crompton, M. Mitochondria and aging: a role for the permeability transition? Aging Cell 3, 3–6. https://doi.org/10.1046/j.1474-9728.2003.00073.x (2004).
Kupsch, K., Parvez, S., Siemen, D. & Wolf, G. Modulation of the permeability transition pore by Inhibition of the mitochondrial K(ATP) channel in liver vs. brain mitochondria. J. Membr. Biol. 215, 69–74. https://doi.org/10.1007/s00232-007-9006-3 (2007).
Rasola, A. & Bernardi, P. The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis Int. J. Program Cell Death 12, 815–833. https://doi.org/10.1007/s10495-007-0723-y (2007).
Zhang, P. et al. Mitochondrial pathway signature (MitoPS) predicts immunotherapy response and reveals NDUFB10 as a key immune regulator in lung adenocarcinoma. J. Immunother. Cancer 13 https://doi.org/10.1136/jitc-2025-012069 (2025).
Sato, Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867. https://doi.org/10.1038/ng.2699 (2013).
Wozniak, M. B. et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the united States. PLoS One. 8, e57886. https://doi.org/10.1371/journal.pone.0057886 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14 https://doi.org/10.1186/1471-2105-14-7 (2013).
Gustavsson, E. K., Zhang, D., Reynolds, R. H., Garcia-Ruiz, S. & Ryten, M. Ggtranscript: an R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics (Oxford England) 38, 3844–3846. https://doi.org/10.1093/bioinformatics/btac409 (2022).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. Circlize implements and enhances circular visualization. Bioinformatics (Oxford England) 30, 2811–2812. https://doi.org/10.1093/bioinformatics/btu393 (2014).
Berker, Y., Muti, I. H. & Cheng, L. L. Visualizing metabolomics data with R. NMR Biomed. 36, e4865. https://doi.org/10.1002/nbm.4865 (2023).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Cheng, Q., Chen, X., Wu, H. & Du, Y. Three hematologic/immune system-specific expressed genes are considered as the potential biomarkers for the diagnosis of early rheumatoid arthritis through bioinformatics analysis. J. Transl. Med. 19 https://doi.org/10.1186/s12967-020-02689-y (2021).
Gortler, J., Schulz, C., Weiskopf, D. & Deussen, O. Bubble treemaps for uncertainty visualization. IEEE Trans. Vis. Comput. Graph. 24, 719–728. https://doi.org/10.1109/tvcg.2017.2743959 (2018).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. Publ. Protein Soc. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Liu, P., Xu, H., Shi, Y., Deng, L. & Chen, X. Potential molecular mechanisms of plantain in the treatment of gout and hyperuricemia based on network pharmacology. Evid. Based Complement. Altern. Med. eCAM 2020 (3023127). https://doi.org/10.1155/2020/3023127 (2020).
Li, Y., Lu, F. & Yin, Y. Applying logistic LASSO regression for the diagnosis of atypical crohn’s disease. Sci. Rep. 12, 11340. https://doi.org/10.1038/s41598-022-15609-5 (2022).
Heagerty, P. J., Lumley, T. & Pepe, M. S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56, 337–344. https://doi.org/10.1111/j.0006-341x.2000.00337.x (2000).
Ramsay, I. S. et al. Model selection and prediction of outcomes in recent onset schizophrenia patients who undergo cognitive training. Schizophrenia Res. Cogn. 11, 1–5. https://doi.org/10.1016/j.scog.2017.10.001 (2018).
Sachs, M. C. plotROC: a tool for plotting ROC curves. J. Stat. Softw. 79 https://doi.org/10.18637/jss.v079.c02 (2017).
Charoentong, P. et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint Blockade. Cell. Rep. 18, 248–262. https://doi.org/10.1016/j.celrep.2016.12.019 (2017).
Orifjon, S. et al. Translation and adaptation of the adult developmental coordination Disorder/Dyspraxia checklist (ADC) into Asian Uzbekistan. Sports (Basel Switzerland) 11 https://doi.org/10.3390/sports11070135 (2023).
Shibru, B. et al. Detection of immune checkpoint receptors—a current challenge in clinical flow cytometry. Front. Immunol. 12 https://doi.org/10.3389/fimmu.2021.694055 (2021).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558. https://doi.org/10.1038/s41591-018-0136-1 (2018).
Mayakonda, A., Lin, D. C., Assenov, Y., Plass, C. & Koeffler, H. P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756. https://doi.org/10.1101/gr.239244.118 (2018).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods (San Diego Calif). 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Jonasch, E., Walker, C. L. & Rathmell, W. K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 17, 245–261. https://doi.org/10.1038/s41581-020-00359-2 (2021).
Zheng, B. et al. Integrative analysis of immune-related genes in the tumor microenvironment of renal clear cell carcinoma and renal papillary cell carcinoma. Front. Mol. Biosci. 8, 760031. https://doi.org/10.3389/fmolb.2021.760031 (2021).
Wang, K. et al. Integrated analysis of single-cell and bulk transcriptome identifies a signature based on NK cell marker genes to predict prognosis and therapeutic response in clear cell renal cell carcinoma. Transl. Cancer Res. 12, 1270–1289. https://doi.org/10.21037/tcr-22-2782 (2023).
Shen, J. et al. Prognostic significance and mechanisms of CXCL genes in clear cell renal cell carcinoma. Aging 15, 7974–7996. https://doi.org/10.18632/aging.204922 (2023).
Han, B., Shaolong, E., Luan, L., Li, N. & Liu, X. CircHIPK3 promotes clear cell renal cell carcinoma (ccRCC) cells proliferation and metastasis via altering of miR-508-3p/CXCL13 signal. OncoTargets Ther. 13, 6051–6062. https://doi.org/10.2147/ott.S251436 (2020).
Xie, Y. et al. M2 macrophages secrete CXCL13 to promote renal cell carcinoma migration, invasion, and EMT. Cancer Cell Int. 21 https://doi.org/10.1186/s12935-021-02381-1 (2021).
Zheng, Z. et al. CXCL13/CXCR5 axis predicts poor prognosis and promotes progression through PI3K/AKT/mTOR pathway in clear cell renal cell carcinoma. Front. Oncol. 8, 682. https://doi.org/10.3389/fonc.2018.00682 (2018).
Harrer, C. et al. The CXCL13/CXCR5 immune axis in health and disease-implications for intrathecal B cell activities in neuroinflammation. Cells 11. https://doi.org/10.3390/cells11172649 (2022).
Hyslop, S. et al. CD7 regulates the persistence of terminally exhausted CD8 + T cells during chronic infection. Cell. Rep. 44, 116316. https://doi.org/10.1016/j.celrep.2025.116316 (2025).
Garg, G. et al. Type 1 diabetes-associated IL2RA variation lowers IL-2 signaling and contributes to diminished CD4 + CD25 + regulatory T cell function. J. Immunol. (Baltimore, Md. 1950) 188, 4644–4653. https://doi.org/10.4049/jimmunol.1100272 (2012).
Liao, L., Xu, H., Zhao, Y. & Zheng, X. Metabolic interventions combined with CTLA-4 and PD-1/PD-L1 Blockade for the treatment of tumors: mechanisms and strategies. Front. Med. 17, 805–822. https://doi.org/10.1007/s11684-023-1025-7 (2023).
Chan, Y. T. et al. Immune checkpoint molecules and glucose metabolism in HIV-Induced T cell exhaustion. Biomedicines 10 https://doi.org/10.3390/biomedicines10112809 (2022).
Chen, J. et al. Tumor extracellular vesicles mediate anti-PD-L1 therapy resistance by decoying anti-PD-L1. Cell Mol. Immunol. 19, 1290–1301. https://doi.org/10.1038/s41423-022-00926-6 (2022).
Liu, P. et al. Causal relationship between several autoimmune diseases and renal malignancies: a two-sample Mendelian randomization study. PLoS One. 19, e0297861. https://doi.org/10.1371/journal.pone.0297861 (2024).
Clark, D. J. et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 179, 964–983e931. https://doi.org/10.1016/j.cell.2019.10.007 (2019).
Akhtar, M., Al-Bozom, I. A. & Hussain, T. A. Molecular and metabolic basis of clear cell carcinoma of the kidney. Adv. Anat. Pathol. 25, 189–196. https://doi.org/10.1097/pap.0000000000000185 (2018).
Simon, M. C. Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell Metabol. 3, 150–151. https://doi.org/10.1016/j.cmet.2006.02.007 (2006).
Huang, L., Xie, Y., Han, W., Jiang, S. & Zeng, L. Oxidative phosphorylation-related signature participates in cancer development, and PTPRG overexpression suppresses the cancer progression in clear cell renal cell carcinoma. J. Immunol. Res. 2022, 8300187. https://doi.org/10.1155/2022/8300187 (2022).
Wei, K. et al. Mitochondrial DNA release via the mitochondrial permeability transition pore activates the cGAS-STING pathway, exacerbating inflammation in acute Kawasaki disease. Cell. Commun. Signaling CCS 22, 328. https://doi.org/10.1186/s12964-024-01677-9 (2024).
Giordano, L., Ware, S. A., Lagranha, C. J. & Kaufman, B. A. Mitochondrial DNA signals driving immune responses: Why, How, where? Cell. Commun. Signaling CCS 23, 192. https://doi.org/10.1186/s12964-025-02042-0 (2025).
Peters, I. et al. Decreased mRNA expression of GATA1 and GATA2 is associated with tumor aggressiveness and poor outcome in clear cell renal cell carcinoma. Target. Oncol. 10, 267–275. https://doi.org/10.1007/s11523-014-0335-8 (2015).
Yang, X. et al. Expression profile and prognostic values of GATA family members in kidney renal clear cell carcinoma. Aging 15, 2170–2188. https://doi.org/10.18632/aging.204607 (2023).
Acknowledgements
We thank the entire team for their support and assistance with this study.
Funding
This research received no specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was conducted in accordance with the principles of the Helsinki Declaration and approved by the Scientific Research Ethics Review Committee of Shanxi Medical University (Ethics Review No: KYLL-2024-028). Informed consent was obtained from all participants.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y., Lv, D., Zhang, W. et al. Explore the key genes and prognosis related to mitochondrial permeability transition driving necrosis gene in kidney renal clear cell carcinoma. Sci Rep 15, 40875 (2025). https://doi.org/10.1038/s41598-025-24779-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24779-x