Abstract
Crotonoside (CTS) is a primary bioactive component found in Croton. current research has mainly focused on leukemia, with few reports in other cancers. Considering the traditional use of Croton, this study aims to evaluate the anti-colorectal cancer (CRC) activity of CTS and reveal its potential mechanisms. First, results in vitro show that CTS can markedly suppress CRC cell proliferation, invasion and migration, and EMT pathway (P < 0.01). Then, RNA-seq analysis was employed to identify the core target and potential mechanism of CTS against CRC, and the results suggested KIF20A is a core target. Bioinformatic analysis showed that KIF20A is overexpressed in CRC and associated with a worse prognosis (P < 0.01). KEGG and GO enrichment analyses indicated that anti-CRC activity of CTS is linked to the cell cycle. Next, Results confirmed that CTS can promote the expression of CDK1 and Cyclin B1 (P < 0.01), and induces G2/M phase arrest to exert anti-CRC effects (P < 0.01). Then, a subcutaneous tumor model was established in vivo to evaluate the anti-CRC activity of CTS; the results showed that CTS significantly inhibited CRC tumor growth and reduced both tumor weight and volume (P < 0.01). CTS can suppress the expression of Ki67, E-cadherin, vimentin, and KIF20A (P < 0.05), and promote the expression of Cyclin B1 and CDK1 (P < 0.01). In addition, molecular docking analysis revealed that the binding energy of CTS to KIF20A was − 7.9 kcal/mol, and CETSA assay showed that CTS treatment attenuated the thermal degradation of KIF20A protein. These results showed CTS can bind KIF20A tightly. Finally, overexpression of KIF20A reverses the anti-CRC effect of CTS. In summary, this study confirms that CTS can target and inhibit KIF20A, thereby inducing cell G2/M phase arrest and exerting anti-CRC effects.
Similar content being viewed by others
Introduction
Recent statistics from the International Agency for Research on Cancer, World Health Organization, indicate an estimated 20 million new cancer cases and nearly 9.7 million cancer-related deaths globally. Colorectal cancer (CRC) ranks as the third most common cancer, with around 1.93 million new cases and 900,000 deaths, accounting for 9.3% of all cancer deaths, indicating high incidence and mortality rates1,2. Studies suggest that by 2040, the number of new CRC cases is expected to exceed 3 million annually, with 1.6 million deaths3; CRC imposes a heavy burden on global public health and socioeconomics4. In addition to conventional surgical treatments, chemotherapy remains the primary treatment for CRC, with 5-fluorouracil, oxaliplatin, irinotecan, and regorafenib as the main chemotherapeutic agents. They form the basis of common CRC chemotherapy regimens such as FOLFOX and XELOX5,6. Moreover, targeted therapies and immunotherapies are widely used in CRC treatment. However, the clinical benefits of these therapies are still limited7,8; Consequently, increasing numbers of patients are turning to traditional Chinese medicine for CRC management, and there is an urgent need to explore more safe and effective anti-CRC drugs from traditional Chinese medicine.
Crotonoside (CTS), also known as isoguanosine, is an indole alkaloid; and is the main bioactive alkaloid in Crotonis fructus, which is a traditional Chinese medicine9. It can treat acute myeloid leukemia by inhibiting APOBEC3G and FLT3, showing antitumor potential10,11,12. Despite concerns about the toxicity and bioavailability of Croton, studies have shown that the maximum tolerated dose of CTS in mice is 100 mg/kg. Both in vivo and in vitro results demonstrated that CTS exhibits robust antitumor efficacy within the maximum tolerated dose. Tissue distribution studies have shown that CTS mainly accumulates in the liver, and is also detected in the spleen and lungs. Thus, these results indicate the potential of CTS as an antitumor drug within a safe dosage13. However, apart from acute myeloid leukemia, CTS has not been reported in other tumors. Croton are commonly used in gastrointestinal disease treatment14,15. Notably, recent studies have shown that silver nanoparticles containing croton extract exhibit promising therapeutic effects on colon cancer in rats16. Based on this, the present study first evaluated the effects of CTS on CRC cell proliferation, invasion, and migration in vitro. Transcriptomic analysis was employed to uncover the underlying anti-CRC mechanisms of CTS, which were comprehensively validated in vitro and in vivo by Western blotting, flow cytometry, and other assays. The results demonstrate that CTS markedly inhibits CRC-cell proliferation, invasion, and migration, and a significant suppression of CRC progression was also observed in vivo. Mechanistically, CTS exerts anti-CRC effects by targeting KIF20A to regulate the cell cycle, and our findings offer a promising therapeutic agent for CRC treatment.
Results
CTS suppresses the proliferation, invasion, and migration of CRC
To evaluate the possible effect of CTS on the progression of CRC, we assessed the effects of CTS on the CRC cell lines HCT-116 and SW480 in vitro. First, the CCK-8 assay was used to evaluate the effects of CTS on the proliferation of the two CRC cells, HCT-116 and SW480. The results showed that CTS could significantly suppress the proliferation of HCT-116 and SW480, with HCT-116 cells being more sensitive (Fig. 1A-B). Therefore, HCT-116 cells were chosen as the research subject for the subsequent studies, and the low, medium, and high doses were set at 5, 10, and 20 µM based on the results. Then, results showed that CTS can significantly downregulate N-cadherin and vimentin, while upregulating E-cadherin expression, indicating that CTS can inhibit the epithelial-mesenchymal transition (EMT) pathway activation and has potential anti-metastatic effects in CRC cells (Fig. 1C-D). In addition, the clonogenic assay results showed that CTS could significantly suppress the clonogenic activity of HCT-116 cells (Fig. 1E-F), and the Transwell results further confirmed that CTS could inhibit HCT-116 cell invasion and migration (Fig. 1G-H). Finally, the EdU staining results showed that CTS could significantly inhibit HCT-116 cell proliferation (Fig. 1I-J). In summary, CTS exhibits notable anti-CRC activity in vitro and represents a potential therapeutic agent for colorectal cancer.
The effect of CTS anti-CRC in vitro. (A, B) CCK-8 assay was employed to evaluate the proliferation of HCT-116 and SW480 cells (n = 3); (C, D) The effect of CTS on the EMT pathway of HCT-116 (n = 3); (E, F) The effect of CTS on the cloning ability of HCT-116 was detected by colony-formation assay (n = 3); (G, H) The Transwell assay was uesd to assess the invasion and migration abilities of HCT-116 treatment with CTS (n = 3); (I, J) The effect of CTS on HCT-116 proliferation by EdU assay (n = 3).
RNA-seq analysis of CTS’ anti-CRC effects
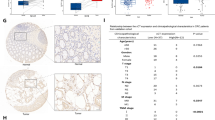
RNA-seq analysis was used to uncover the molecular mechanisms of CTS against CRC, with differential analysis conducted via the Limma package. Compared to the control group, the CTS treatment group had 2118 up-regulated and 2212 down-regulated genes (Fig. 2A). The changes in expression of the top five upregulated and five downregulated genes were shown, with KIF20A, ERCC6L, and PKA6 exhibiting the greatest expression changes (Fig. 2B). A heatmap of the TOP25 genes indicated a marked decrease in KIF20A following CTS treatment (Fig. 2C). The PPI network identified KIF20A, CDC20, and KIF11 as key targets (Fig. 2D-E). Thus, KIF20A was considered a core target of CTS against CRC. KIF20A expression profiles from the TCGA and GTEx databases revealed high KIF20A expression in CRC (Fig. 2F). OS analysis showed that high KIF20A expression significantly shortened the survival of CRC patients (Fig. 2G), aligning with previous studies and highlighting KIF20A as a therapeutic target for CRC. Western Blot results demonstrated that CTS could markedly suppress KIF20A expression, suggesting that it exerts anti-CRC effects by inhibiting KIF20A (Fig. 2H-I). Then, the mRNA levels of KIF20A, CDK1, and CCNB1 were assessed through PCR assay, results show that CTS markedly suppressed KIF20A expression while up-regulating both CDK1 and CCNB1 (Fig. S1).
RNA-seq analysis of HCT-116 treatment with CTS. (A) Differential expression genes (DEGs) screening; (B) TOP5 genes with up and down expression in HCT-116; (C) Cluster heatmap of TOP25 genes with up and down; (D, E) The core targets of CTS anti-CRC identified, the colors represent the degree values, ranked from high to low; (F) Expression of KIF20A in CRC using TCGA and GTEx databases; (G) The Impact of KIF20A Expression on the Overall Survival of CRC Patients; (H, I) CTS inhibits KIF20A expression of HCT-116 (n = 3).
Additionally, enrichment analyses were performed to identify the signaling pathways involved in CTS’s anti-CRC action. GO result revealed that DEGs were primarily enriched in biological processes like mitosis and the cell cycle (Fig. 3A). The top 3 pathways in each GO category were selected to create a chord diagram, which showed high enrichment frequency of KIF20A in these pathways, further indicating its key role in CTS’s anti-CRC effects (Fig. 3B). KEGG enrichment analysis indicated that CTS primarily exerts anti-CRC effects through the cell cycle pathway, consistent with GO results (Fig. 3C). Western Blot analysis showed that CTS could largely boost the expression of cell-cycle associated regulatory proteins CDK1 and Cyclin B1 (Fig. 3D-E). Flow cytometry results confirmed that CTS could markedly arrest HCT-116 cells in the G2/M phase, reducing cell proliferation (Fig. 3F-G). These findings suggest that CTS inhibits KIF20A to block the G2/M cell cycle, thereby exerting anti-CRC effects. Moreover, we evaluated the proliferation of HCT116 cells after knocking down KIF20A (Fig. S2A). The results showed that silencing KIF20A significantly upregulated Cyclin B1 expression (Fig. S2B). Colony formation assays revealed that KIF20A knockdown markedly inhibited HCT116 cell proliferation, producing an effect similar to that of CTS (Fig. S2C).
CTS suppresses HCT-116 cells proliferation in vivo
To evaluate the anti-CRC efficacy of CTS in vivo, a subcutaneous tumor mouse model was established by inoculating HCT-116 cells. Results showed that CTS significantly inhibited tumor growth, reducing tumor weight and volume (Fig. 4A-C). Furthermore, we assessed the safety of CTS treatment. Organ index results indicated that CTS had no drug toxicity (Fig. S3). IHC staining showed that CTS suppressed tumor KI67 expression, indicating reduced tumor activity in vivo (Fig. 4D). Also, CTS inhibited the activation of the tumor EMT pathway in vivo (Fig. 4E-F). Western Blotting confirmed that CTS significantly suppressed the tumor EMT pathway (Fig. 4G-H). Moreover, CDK1 and Cyclin B1 were upregulated in the CTS group, consistent with in vitro findings (Fig. 4I). Lastly, IHC and Western Blotting showed that CTS markedly reduced KIF20A expression in tumor tissue (Fig. 4J-L).
CTS inhibits tumor growth in subcutaneous tumor mice. (A) Tumor image in bright field (n = 4); (B, C) Tumor volume and weight (n = 4); (D–F) The expression of KI67, E-cadherin, and Vimentin in tumor tissues (n = 3); (G–I) The effects of CTS on the EMT pathway and cell cycle in tumor tissue (n = 3); (J–L) CTS inhibits expression of KIF20A in tumor tissue (n = 3).
CTS blocks the cell cycle to exert anti-CRC effects by targeting and binding to KIF20A
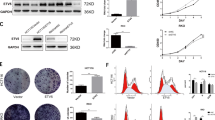
The above results indicate that CTS can suppress CRC by inhibiting KIF20A. Molecular docking shows that CTS has a binding energy of -7.9 kcal/mol with KIF20A, and forms hydrogen bonds with ASN376, SER164, and SER378 of KIF20A (Fig. 5A). CETSA results show that CTS increases the thermal stability of KIF20A protein, with the CETSA curve shifting right (Fig. 5B-C), indicating that CTS binds to and inhibits KIF20A. After constructing a plasmid to overexpress KIF20A (OE-KIF20A) (Fig. 5D), the inhibitory effects of CTS on HCT-116 cell proliferation, invasion, and migration were significantly reversed (Fig. 5E-H). In summary, CTS exerts anti-CRC effects by targeting and inhibiting KIF20A to block the cell cycle.
CTS targets and inhibits KIF20A to suppresses CRC progression by inducing G2/M phase arrest. (A) Molecular docking of CTS with KIF20A; (B, C) CETSA assay evaluates the binding of CTS to KIF20A (n = 3); (D) Verification of KIF20A overexpression efficiency (n = 3); (E–H) Overexpressing KIF20A reverses the anti-CRC effects of CTS (n = 3).
Discussion
Colorectal cancer (CRC) ranks as the third most common cancer globally. Its incidence and mortality rates are on the rise, making prevention efforts urgent. Although current therapeutic strategies like surgery, chemotherapy, and immunotherapy have significantly extended the CRC patient survival period, the clinical efficacy of these treatments are restricted due to factors such as drug resistance, metastasis, immune evasion, and severe adverse reactions17,18. Thus, there is an urgent need to further explore the molecular mechanisms underlying CRC progression and to develop more effective therapeutic agents for CRC.
KIF20A, a member of the kinesin superfamily, also called mitotic kinesin-like protein 2 (MKlp2), is overexpressed in most cancers, such as CRC, breast cancer, and gliomas, and serves as a diagnostic and prognostic factor19. KIFs precisely regulate mitosis to ensure orderly progression, transport intracellular membrane organelles, mRNA, and protein complexes to sustain cell activity, and transport chromosomes during mitosis, playing a key role in cell shape and proliferation. In early tumor development, upregulated KIF family proteins can cause excessive spindle separation, leading to premature formation and separation of sister chromatids, resulting in abnormal DNA aneuploidy and distribution20. Studies have shown that KIF20A accumulates in the nucleus and participates in cell division. In the interphase of the cell cycle, KIF20A and myosin II are located at the Golgi complex’s division hotspot, regulating the fission of Rab6 + vesicles. Chromosomes and KIF20A form a complex that promotes cleavage furrow formation in a Cdk1-dependent manner21. KIF20A is more critical in the late phase of cytokinesis. Mad2 is a negative regulator of KIF20A and it inhibits the loading of KIF20A onto the mitotic spindle. During the late mitosis, the spindle formation mediated by KIF20A is also regulated by Mad2 22. KIF20A is recognized as a promising therapeutic target for cancer in regulating the cell cycle.
In CRC, overexpression of KIF20A is positively correlated with tumor invasion, distant metastasis, and clinical staging. Notably, KIF20A has been shown to be associated with G2/M phase arrest of the cell cycle. For example, genistein can induce G2/M phase arrest by inhibiting KIF20A, thereby suppressing the progression of gastric cancer23; Cucurbitacin B can also induce G2/M phase arrest in melanoma cells via the FOXM1-KIF20A pathway24. KIF20A is one of the targets of FOXM1 for normal spindle formation and chromosome separation. It is related to the proliferation and drug resistance of tumor cells. The specific binding of FOXM1 to the forkhead responsive element of KIF20A significantly enhances KIF20A’s transcriptional activity. By regulating KIF20A expression, tumor chemosensitivity can be adjusted25. Studies have shown that KIF20A is highly expressed in oxaliplatin-resistant CRC cells. Knocking out KIF20A enhances CRC cell sensitivity to oxaliplatin and inhibits NUAK1 activation, a kinase associated with CRC progression and poor prognosis26,27. Another study indicates that KIF20A can enhance chemoresistance in CRC by modulating the JAK/STAT3 pathway28. In prostate cancer, high FOXM1 expression promotes docetaxel resistance by upregulating KIF20A. Knocking out FOXM1 induces G2/M arrest and apoptosis, enhancing docetaxel sensitivity29. Also, targeting KIF20A can overcome doxorubicin resistance in breast cancer30. In summary, many studies have confirmed that KIF20A is a regulator of G2/M phase arrest and is a highly promising therapeutic target for CRC.
Crotonis fructus is a traditional Chinese medicine with laxative properties, used for moistening the intestines, promoting defecation, and detoxifying accumulations. CTS, an indole alkaloid separated from it, is one of its major bioactive components with considerable antitumor activity13. Currently, CTS has been seldom reported in cancers other than acute myeloid leukemia. This study confirms that CTS can target and inhibit KIF20A both in vitro and in vivo. It exerts anti-CRC effects by inducing G2/M phase cell-cycle arrest, suppressing CRC cell proliferation, invasion, and migration. Although there are concerns about CTS potential drug-induced toxicity, our study did not observe diarrhea or other organ-specific toxicities. These findings suggest that CTS may be an effective CRC treatment strategy when administered within a safe dosage range. Nevertheless, this study has certain limitations that need to be addressed in future work. For example, while molecular docking and CETSA assays confirmed that CTS can target KIF20A, additional evidence is required to strengthen this conclusion. Furthermore, referring to previous studies, we set the CTS administration dose at 70 mg/kg/d 11. Although no significant adverse reactions were noted at this dose, the long-term safety profile of CTS administration requires further evaluation in subsequent studies; 3) Further study is needed to reveal the mechanism by which CTS targets KIF20A to suppress CRC.
In summary, our findings demonstrate that CTS suppresses CRC cell proliferation by targeting and inhibiting KIF20A, causing G2/M phase cell-cycle arrest. These results highlight that CTS is likely to be an anti-CRC agent and lays the foundation for further research on CTS.
Materials and methods
Chemicals and reagents
The Crotonoside standard (Lot: MUST-250114) (CTS) was supplied by MUST Co., Ltd (Chengdu, China). The McCoy’s 5 A Medium and Dulbecco’s Modification of Eagle’s Medium (DMEM) and were purchased from Meilun Biotech (Dalian, China). Fetal bovine serum (FBS) was purchased from Gibco (Waltham, CA, USA). The 0.25 μm PVDF membranes were obtained from Millipore (Bedford, Massachusetts). The Cell Counting Kit-8 (CCK-8), BCA assay Kit, RIPA Lysis buffer, and Fast Blocking Western reagent, and 12% SDS-PAGE gel were supplied by Beyotime CO. Ltd (Shanghai, China). The KIF20A antibody (A15377), β-actin antibody (AC026), GAPDH antibody (A19056), and HRP-conjugated AffiniPure secondary antibody (#AS014) were obtained from Abclone Biological CO. Ltd (Wuhan, China). The N-cadherin antibody (#ab76011), E-cadherin antibody (#ab314063), and Vimentin antibody (#ab92547) were procured from Abcam Biological CO. Ltd (Shanghai, China).
Cell culture
Human colorectal cancer cell lines, including HCT116 and SW480 were procured from the Shanghai Aoruishai CO. Ltd. All experiments were performed with mycoplasma-free cells. HCT116 cells were cultured in McCoy’s 5 A medium, and SW480 cells were cultured in DMEM/F12 medium. All media were supplemented with 10% FBS and 1% penicillin-streptomycin. The cells were cultured at 37 °C in an incubator with 5% CO2. Once they reached 80% confluency, they were passaged and transferred to 10 cm dishes for subsequent experiments.
CCK-8 assay
The CCK-8 assay was employed to assess cell viability in HCT116 and SW480 cells. Specifically, cells were seeded at a density of 5 × 103 cells/well in 96-well plates and cultured for 24 h. Then, they were treated with different concentrations of CTS for 24 h. After that, 10 µL of CCK-8 solution was added to each well and incubated for 2 h in a cell culture incubator. Finally, absorbance at a wavelength of 450 nm was measured using a spectrophotometer.
Transwell invasion and migration assay
Referring to previous studies31, invasion and migration experiments were conducted to evaluate the effects of CTS (1, 2, and 5µM) on HCT116 cells using transwell chambers with an 8 μm pore size. HCT116 cells were plated in the upper chamber of a 24-well plate (5 × 104 cells in 200 µL serum-free McCoy’s 5 A medium), while the lower chamber contained 700 µL of McCoy’s 5 A medium with 20% FBS. After 24 h, the 4% paraformaldehyde was used to fix cells for 30 min, and cells were then stained with a 0.1% crystal violet solution for 20 min. After washing with PBS, non-migrated cells were removed from the upper chamber with a cotton swab. The results were then visualized under a microscope and quantified utilizing ImageJ 1.38e software. Similarly, invasion assay was performed in pre-coated chambers with matrix gel.
Colony formation assay
Approximately 200 HCT-116 cells were seeded into 6-well plates for the colony formation assay and cultured with varying concentrations of CTS (5, 10, and 20µM) for 24 h. After two weeks, cells were fixed with 4% paraformaldehyde for 30 min and then stained with crystal violet. Finally, colony counting was performed using ImageJ 1.38e software.
EdU assay
Cell proliferation was detected by EdU Cell Proliferation Kit. First, HCT116 cells were treated with 5, 10, and 20 µM CTS, respectively for 24 h. After that, 10 µM EdU solution were added into the cells and incubated for 2 h. Then, the cells were washed with PBS, and were fixed and permeabilized with 0.1% Triton. The Click Additive Solution was incubated for 30 min at room temperature in the dark. Results were taken under an optical microscope.
RNA-seq
The cells were divided into two groups: Control group and CTS treatment group (10 µM). RNA-seq samples were collected using TRIzol buffer (n = 3). These samples were processed and sequenced using Illumina HiSeq (Thermo Fisher Scientific, MA, USA). Differentially expressed genes (DEGs) related to the anti-CRC effects of CTS were identified using the Limma program with the following criteria: log|FC| ≥ 1.5, P ≤ 0.05, FDR < 0.05, and FPKM value ≥ 0.1.
Bioinformatics analysis
The protein-protein interaction (PPI) network of DEGs was generated using the STRING database (https://cn.string-db.org/) with a confidence level above 0.9. Human species was selected and discrete points were removed. The PPI network parameters, such as degree and topological coefficient, were computed using Cytoscape 3.10.3 software (https://cytoscape.org/), and the Cytohubba plugin was used to identify key targets. Moreover, referring to previous studies, the expression differences of key targets in tumors and their influence on CRC patient survival were analyzed based on the TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga)32. To roughly understand the biological functions of DEGs, the CTS targets for CRC were analyzed for KEGG and GO using the ClusterProfiler and org.Hs.eg.db packages in R. Finally, results were ranked by P-value, and the terms of the highest scores were presented.
RNA extraction and Real-Time quantitative PCR (RT-qPCR)
Total RNA was isolated from HCT116 cells after CTS treatment: cultures were rinsed with PBS and processed directly with the CellAmp™ Direct RNA Prep Kit. First-strand cDNA was generated with PrimeScript RT Master Mix, and transcript abundance was quantified on a SYBR® Premix Ex Taq™ platform with gene-specific primers (Sangon Biotech, Shanghai; sequences in Supplementary Table S1). Relative expression was determined by the 2^(−ΔΔCt) method, normalizing to GAPDH.
Western blot assay
Total protein was extracted from cells and tissues using RIPA lysis buffer at 4℃. The protein concentration was subsequently measured using the BCA assay, and total protein concentration in each group was normalized using RIPA lysis buffer. Then, the proteins were added with loading buffer and heated at a high temperature for 10 min. The proteins were separated by 12% SDS-PAGE before being transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). After the transfer, the membranes were blocked with 5% non-fat milk at room temperature for 1 h. Membranes were incubated with primary antibodies overnight at 4 ℃. Following primary incubation, the membrane was washed with TBST buffer, and secondary antibodies labeled with HRP were applied to the membranes and incubated at room temperature for two hours. Finally, ECL solution was evenly applied to the membranes for chemiluminescence-based signal detection via imaging. The β-actin was selected as an internal reference, and results were measured through ImageJ 1.38e software.
Flow cytometry assay
The cell cycle of the HCT-116 was assessed using flow cytometry, the HCT-116 cells were seeded in 6-well plates (5*105 per well) and cultured in an incubator for 24 h. Then, cells were treated with different concentrations of CTS (5, 10, and 20µM). Following a 24-hour treatment period, each group was treated with EDTA to detach and gather the HCT-116 cells, and the impact of CTS on the cell cycle was evaluated. Each sample contains at least 1*104 cells, and FlowJ v10 software was used to analyze results.
Animal experiments
The nude mice were subcutaneously inoculated with HCT-116 cells (1 × 106 cells in 100 µL PBS) to prepare CRC mouse model. To investigate the impact of CTS on CRC progress, mice were distributed into four groups: Control, Oxaliplatin (10 mg/kg), CTS low (35 mg/kg) and CTS high (70 mg/kg) groups. The mice have free access to water and food. Drug treatment began on day 7 after HCT-116 cells inoculation, and CTS and Oxaliplatin were administered via i.p injection once daily.Both CTS and Oxaliplatin were prepared fresh daily prior to intraperitoneal injection. The compounds were accurately weighed and dissolved in a vehicle to achieve a final composition of 10% DMSO, 40% PEG 400, 5% Tween-80, and 45% saline. The mixture was vortexed and sonicated until a clear and homogeneous solution was obtained. The control group received i.p. injection of an equivalent volume of the same vehicle. After two weeks of continuous treatment, the mice were euthanized, and the tumor sizes were measured. Then, mice were euthanized by cervical dislocation, and the tumor tissues were collected for subsequent experiments.
Approval statement
Animal experiments were approved by the Animal Ethics Committee of Shanghai Pudong New Area People’s Hospital in accordance with ethical guidelines [License number: PDYY2025015]. 4–5 weeks male nude mice (20 ± 2 g) were purchased from Shanghai Yishang Co., Ltd [License number: SCXK-2022-0011]. The study was conducted in strict accordance with ARRIVE 2.0 guidelines to ensure the ethical of animal research. All procedures were performed in accordance with the relevant guidelines and regulations.
Immunohistochemical (IHC) staining
To determine the expression level of relevant protein in tumor tissues, the following IHC protocol was utilized: the tissue was fixed in 4% paraformaldehyde for 24 h, and the fixed tissue was embedded and sectioned into 5 μm thick slices by Shanghai Ruiyu Biotechnology Co., Ltd. Then, paraffin sections were deparaffinized using xylene and rehydrated through a graded series of ethanol concentrations. Subsequently, antigen retrieval was performed in EDTA-Tris buffer under high pressure for 10 min. This was followed by incubation with hydrogen peroxide and blocked with goat serum. Next, primary antibodies along with species-specific secondary antibodies were applied to the sections. Finally, diaminobenzidine (DAB) was used for development while hematoxylin served as a counterstain.
Molecular Docking
The human KIF20A protein structure was constructed via homology modeling using AlphaFold and Discovery Studio software. KIF20A was chosen as receptors and preprocessing of KIF20A was performed, such as adding polar hydrogens, removing water molecules, and assigning Kollman charges. Referring to previous studies33. The CASTp 3.0 web server was employed to determine the docking box, and molecular docking was performed to evaluate the binding of CTS and KIF20A using AutoDock 1.5.7 software. Finally, PyMOL software was employed to display the docking conformations of CTS with KIF20A.
The cellular thermal shift assay (CETSA)
The lysate of HCT-116 cells was prepared with RIPA lysis buffer and split into two portions. One portion was treated with 20 µM CTS for 1 h, while the other received an equal volume of DMSO. The treatment groups were subsequently divided into 6 aliquots, and were incubated at different temperatures for 10 min, starting at 35 °C with an interval of 5 °C. The samples were then centrifuged at 17,400 g for 20 min at a temperature of 4 °C. Western blot analysis was conducted on resulting supernatant liquid. Boltzmann sigmoidal function in GraphPad Prism software was used to fit the melting curve.
Plasmids construction and cell transfection
The KIF20A in HCT-116 cells was knockdown as follows: For KIF20A knockdown: The KIF20A knockdown plasmid was designed following previous research reports, synthesized and packaged into lentivirus by GenePharma Co., Ltd (Shanghai, China). The KIF20A in HCT-116 cells was overexpressed as follows: Human KIF20A cDNA was cloned into the pcDNA3.1-HA vector, the KIF20A cDNA and plasmid was constructed by GenePharma Co., Ltd (Shanghai, China).
Then, Lipofectamine 3000 transfection reagent was used to transfect HCT-116 cells. Following transfection for 24 h, the cell culture medium was replaced and the cells were cultured further. Finally, Western Blot assay was used to verify the efficiency of overexpression.
Statistical analysis
Statistical analysis was presented using GraphPad Prism software. The results were expressed as means ± SEM. The Student’s t-test were used to compare two groups. The one-way analysis of variance (ANOVA) was conducted for comparing between multiple groups, and Dunnett’s method was used to post hoc test. Statistical significance was denoted by *p < 0.05, **p < 0.01.
Data availability
The material and dataset for this study can be obtained from the corresponding author on reasonable request.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74 (3), 229–263 (2024).
Xi, Y. & Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 14 (10), 101174 (2021).
Morgan, E. et al. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut 72 (2), 338–344 (2023).
Li, D. et al. Cancer survival in Cixian of China, 2003–2013: a population-based study. Cancer Med. 7 (4), 1537–1545 (2018).
Leowattana, W., Leowattana, P. & Leowattana, T. Systemic treatment for metastatic colorectal cancer. World J. Gastroenterol. 29 (10), 1569–1588 (2023).
Bekaii-Saab, T. A decade of progress: advances in the third-line treatment of patients with metastatic colorectal cancer. Am. J. Manag Care. 30 (2), S23–s30 (2024).
Fan, A. et al. Immunotherapy in colorectal cancer: current achievements and future perspective. Int. J. Biol. Sci. 17 (14), 3837–3849 (2021).
Li, Q. et al. Signaling pathways involved in colorectal cancer: pathogenesis and targeted therapy. Signal. Transduct. Target. Ther. 9 (1), 266 (2024).
Lin, S. C. et al. Alleviation of Collagen-Induced arthritis by Crotonoside through modulation of dendritic cell differentiation and activation. Plants (Basel). 9 (11), 1535 (2020).
Ma, C. et al. The identification of APOBEC3G as a potential prognostic biomarker in acute myeloid leukemia and a possible drug target for Crotonoside. Molecules 27 (18), 5804 (2022).
Li, Y. Z. et al. Crotonoside exhibits selective post-inhibition effect in AML cells via Inhibition of FLT3 and HDAC3/6. Oncotarget 8 (61), 103087–103099 (2017).
Ma, Y. et al. Combination of diethyldithiocarbamate with 12-O-tetradecanoyl phorbol-13-acetate inhibits the growth of human myeloid leukemia HL-60 cells in vitro and in xenograft model. Biosci. Biotechnol. Biochem. 84 (10), 2069–2076 (2020).
Yan, P., Zhang, L., Peng, C. & Zhang, R. Pharmacokinetics and tissue distribution of Crotonoside. Xenobiotica 48 (1), 28–36 (2018).
Wang, X. et al. Effects of essential oil from Croton tiglium L. on intestinal transit in mice. J. Ethnopharmacol. 117 (1), 102–107 (2008).
Song, H. K. et al. Crotonis fructus extract inhibits 12-O-Tetradecanoylphorbol-13-Acetate-Induced expression of matrix Metalloproteinase-9 via the activator Protein-1 pathway in MCF-7 cells. J. Breast Cancer. 20 (3), 234–239 (2017).
Aboulthana, W. M. et al. Evaluation of the biological efficiency of silver nanoparticles biosynthesized using Croton tiglium L. Seeds extract against azoxymethane induced colon cancer in rats. Asian Pac. J. Cancer Prev. 21 (5), 1369–1389 (2020).
Morris, V. K. et al. Treatment of metastatic colorectal cancer: ASCO guideline. J. Clin. Oncol. 41 (3), 678–700 (2023).
Abedizadeh, R., Majidi, F., Khorasani, H. R., Abedi, H. & Sabour, D. Colorectal cancer: a comprehensive review of carcinogenesis, diagnosis, and novel strategies for classified treatments. Cancer Metastasis Rev. 43 (2), 729–753 (2024).
Jin, Z. et al. Expression, regulating mechanism and therapeutic target of KIF20A in multiple cancer. Heliyon 9 (2), e13195 (2023).
Wu, W. D., Yu, K. W., Zhong, N., Xiao, Y. & She, Z. Y. Roles and mechanisms of Kinesin-6 KIF20A in spindle organization during cell division. Eur. J. Cell. Biol. 98 (2), 74–80 (2019).
Hill, E., Clarke, M. & Barr, F. A. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. Embo J. 19 (21), 5711–5719 (2000).
Lee, S. H., McCormick, F. & Saya, H. Mad2 inhibits the mitotic Kinesin MKlp2. J. Cell. Biol. 191 (6), 1069–1077 (2010).
Liu, Y. L. et al. Genistein induces G2/M arrest in gastric cancer cells by increasing the tumor suppressor PTEN expression. Nutr. Cancer. 65 (7), 1034–1041 (2013).
Wei, J. et al. Cucurbitacin B-induced G2/M cell cycle arrest of conjunctival melanoma cells mediated by GRP78-FOXM1-KIF20A pathway. Acta Pharm. Sin B. 12 (10), 3861–3876 (2022).
Gartel, A. L. FOXM1 in cancer: interactions and vulnerabilities. Cancer Res. 77 (12), 3135–3139 (2017).
Port, J. et al. Colorectal tumors require NUAK1 for protection from oxidative stress. Cancer Discov. 8 (5), 632–647 (2018).
Yang, C., Zhang, Y., Lin, S., Liu, Y. & Li, W. Suppressing the KIF20A/NUAK1/Nrf2/GPX4 signaling pathway induces ferroptosis and enhances the sensitivity of colorectal cancer to oxaliplatin. Aging (Albany NY). 13 (10), 13515–13534 (2021).
Xiong, M. et al. KIF20A promotes cellular malignant behavior and enhances resistance to chemotherapy in colorectal cancer through regulation of the JAK/STAT3 signaling pathway. Aging (Albany NY). 11 (24), 11905–11921 (2019).
Yu, H. et al. FOXM1 modulates docetaxel resistance in prostate cancer by regulating KIF20A. Cancer Cell. Int. 20 (1), 545 (2020).
Rahman, K. U. et al. Mir-153-3p modulates the breast cancer cells’ chemosensitivity to doxorubicin by targeting KIF20A. Cancers (Basel). 15 (6), 1724 (2023).
Cheng, X. et al. Parishin B blocking TRIB3-AKT1 interaction inhibits breast cancer lung metastasis. Front. Pharmacol. 15, 1517708 (2024).
Sun, J. et al. Schisandrin B exerts anti-colorectal cancer effect through CXCL2/ERK/DUSP11 signaling pathway. Cancer Cell. Int. 25 (1), 97 (2025).
Wang, X. et al. Gentiopicroside targeting AKT1 activates HIF-1α/VEGF axis promoting diabetic ulcer wound healing. Front. Pharmacol. 16, 1506499 (2025).
Funding
This study was supported by Shanghai Pudong New Area People’s Hospital Qihang Plan Talent Cultivation Project (PRYQH202405).
Author information
Authors and Affiliations
Contributions
Yu Zhang: Validation, Methodology, Writing-original draft, Data curation. Jianguo Sun: Writing-review & editing, Validation, Data curation. Zexu Chen: Project administration, Writing-review & editing, Conceptualization, Funding acquisition. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Y., Sun, J. & Chen, Z. Crotonoside exerts anti-colorectal cancer effects by suppressing KIF20A to regulate the cell cycle. Sci Rep 15, 41022 (2025). https://doi.org/10.1038/s41598-025-24969-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24969-7