Abstract
The PEST-containing nuclear protein (PCNP) has been reported to play paradoxical roles in tumorigenesis. While PCNP functions as a tumor suppressor in neuroblastoma and lung adenocarcinoma, it has been identified as a tumor promoter in ovarian cancer. However, the underlying mechanisms driving these contrasting effects remain unclear. Gene Expression Profiling Interactive Analysis (GEPIA) was employed to analyze relevance of PCNP expression with prognosis in HCC. To evaluate the role of PCNP in hepatocellular carcinoma (HCC), we analyzed the expression of PCNP in 87 HCC tissues and 80 adjacent normal tissues. Correlation between PCNP and ubiquitin-like with PHD and ring finger domains 2 (UHRF2) was transcriptionally detected, and then their effects on malignant behaviors, including proliferation, invasion, tumor formation in vitro and in vivo were evaluated. According to tissues array results, it is found that PCNP was significantly upregulated in HCC tissues, which was further confirmed in HCC cell lines by western blotting. Importantly, both the cytoplasmic and nuclear PCNP fractions were correlated with pathological grade, T staging, clinical staging and recurrence, and patients with a high level of PCNP had lower overall survival and disease-free survival and a higher recurrence rate than those with a low PCNP level, indicating that PCNP might be an independent prognostic factor for HCC patients. Functional investigations revealed that PCNP knockdown inhibited malignant behaviors, including proliferation, invasion, colony formation and tumor formation, in vitro and in vivo. Pearson correlation analysis indicated a positive association between PCNP and UHRF2, with evidence that PCNP transcriptionally upregulates UHRF2. Furthermore, ectopic overexpression of PCNP activated the ErbB3/Ras/Raf1 signaling pathway through upregulation of UHRF2. Our findings suggest that PCNP promotes HCC progression by activating the ErbB3/Ras/Raf signaling pathway through upregulation of UHRF2. Consequently, PCNP holds potential as both a diagnostic biomarker and a therapeutic target for hepatocellular carcinoma.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC), the predominant histological subtype of liver cancer, represents a significant contributor to the global cancer burden1. Over recent decades, the incidence of HCC has risen in many countries2, and it accounts for the majority of liver cancer-related diagnoses and mortality worldwide3. Despite significant improvements in the clinical diagnosis and treatment of early-stage liver cancer, the overall prognosis of HCC remains dismal4. The limited understanding of the cellular and molecular mechanisms driving HCC pathogenesis underscores the urgent need to explore its underlying biology and identify novel diagnostic biomarkers and therapeutic targets.
PEST-containing nuclear protein (PCNP) is a nuclear protein that has recently attracted attention due to its roles in tumorigenesis and cell cycle regulation5. Emerging evidence suggests that PCNP interacts with cell cycle regulators, influencing tissue fate by modulating apoptosis and proliferation through both tumor suppressors (e.g., p53, pRB) and oncogenic promoters (e.g., cyclin E, cyclin D)6. Clinically, aberrant PCNP expression has linked to the initiation and progression of multiple cancers, such as neuroblastoma, lung adenocarcinoma, and ovarian cancer. Mechanistically, PCNP disrupts key signaling pathways including PI3K/AKT/mTOR and MAPK, facilitating tumorigenesis7. Notably, PCNP has been shown to promote ovarian cancer progression via activation of the Wnt/β-catenin pathway and induction of epithelial-mesenchymal transition (EMT)8. However, the biological role and clinical significance of PCNP in liver cancer have not been fully elucidated.
Ubiquitin-like with PHD and ring finger domains 2 (UHRF2) is a member of the UHRF family, containing five identifiable functional domains: ubiquitin-like (UBL), tandem Tudor domain (TTD), plant homeodomain (PHD), SET and RING-associated (SRA), and a novel RING finger domain9. This complex domain architecture endows UHRF2 with diverse cellular roles. As an ubiquitin E3 ligase, UHRF2 can ubiquitinate PCNP and acts as an important cell cycle by interacting with multiple cell cycle proteins such as CDKs, p53, pRB, and PCNA10. Additionally, UHRF2 can promote DNA damage repair by reducing RING domain-mediated levels of p21. Previous studies have shown that UHRF2 is involved in tumor development and progression in several human cancers, such as esophageal squamous cell carcinoma, lung cancer, and colorectal cancer11. In a recent study, it was found that UHRF2 is significantly increased in HCC tissues compared to normal tissues, and high levels of UHRF2 are significantly associated with the malignant features of HCC12.
ErbB3 is a member of the receptor tyrosine kinase EGFR family and plays an important role in cancer as a homolog of EGFR (ErbB1) and HER2 (ErbB2)13. Overexpression of ErbB3 has been reported in multiple cancers, including breast, gastric, ovarian, prostate, bladder, colon cancers, head and neck squamous cell carcinoma, and melanoma14. Activation of ErbB3 and its downstream signaling pathways, notably PI3K/AKT and Ras/Raf, contributes to tumor development and progression15. Intriguingly, UHRF2 has been suggested to upregulate the ErbB3/Ras/Raf signaling axis, promoting HCC progression14. However, the relationship between UHRF2 and ErbB3 remains elusive, and whether UHRF2 influences this pathway through regulation of PCNP has not been clarified.
In this study, we aimed to elucidate the role of PCNP in HCC, focusing on its regulatory impact on UHRF2 and downstream signaling pathways. We explored the subcellular localization of PCNP, assessed its correlation with clinical features and prognosis, and investigated its role in cell proliferation, invasion, and tumor formation both in vitro and in vivo. Our findings suggest that PCNP drives HCC progression via the UHRF2–ErbB3/Ras/Raf signaling cascade, identifying a novel axis with potential therapeutic implications.
Results
PCNP is upregulated in HCC and correlates with poor prognosis
Increased Expression of PCNP in LIHC (Liver Hepatocellular Carcinoma) To investigate the difference in PCNP expression levels between tumor and adjacent tissues, we employed Singerbox3.0 to analyze RNA-seq data, matched TCGA tumor tissue with GTEx data. PCNP mRNA levels were significantly upregulated in multiple cancer types in tumor vs. matched adjacent tissues across multiple cancers (p < 0.01), including LIHC (Fig. 1A).
Prognostic significance of PCNP expression in pan-cancer analysis and LIHC. A Pan-cancer mRNA expression profiling. PCNP expression is significantly upregulated in tumor tissues versus adjacent normal controls across multiple malignancies (including LIHC, lymphoma, and pancreatic carcinoma) based on RNA-seq data from TCGA and GTEx, analyzed via Sangerbox 3.0. B Overall survival (OS) in LIHC. Kaplan–Meier curves demonstrate significantly shorter OS in patients with high PCNP expression (HR = 1.7, 95% CI 1.2–2.4, *p* = 0.0028). C Disease-specific survival (DSS) in LIHC. Elevated PCNP expression correlates with poorer DSS (HR = 1.59, 95% CI 1.2–2.1, *p* = 0.0033).
Subsequently, the prognostic value of PCNP in liver hepatocellular carcinoma (LIHC) was evaluated using Sangerbox 3.0. The analysis demonstrated that elevated PCNP expression was significantly associated with: Shorter overall survival (OS) (HR 1.7, 95% CI p = 0.0033; Fig. 1B); Poorer disease-specific survival (DSS) (HR 1.59, p = 0.0033; Fig. 1C). These findings suggest that PCNP may be post-transcriptionally upregulated in LIHC and may function as a prognostic indicator.
PCNP protein is upregulated in HCC tissues and correlates with clinical parameters
To confirm the expression pattern of PCNP at the protein level, we performed immunohistochemical staining using a tissue microarray containing 87 HCC tissues and 80 adjacent non-tumor tissues. As shown in Fig. 2A, B, both nuclear and cytoplasmic PCNP expression were markedly higher in tumor tissues. Western blotting further confirmed elevated PCNP levels in HCC cell lines compared to normal hepatic L-02 cells (Fig. 2C).
Immunohistochemical staining of PCNP in HCC tissues than the adjacent tissues. A Representative IHC stain in HCC tissue array. Images of three representative patients were taken under amplification of 2 or 40. B The immunoreactive score (IRS) in cytoplasmic or nucleus fractions were calculated as intensity of the staining reaction multiplied by the percentage of positive cells. C relative PCNP expression levels in HCC cell lines were measured by performing RT-qPCR using cDNA microarray, including SNU449, Hep3B, SK-HEP-1, PLC-PRF-5, RBE, Huh7 and non-tumor cell line L-02. D The prognostic value of PCNP protein levels in cytoplasmic or nucleus faction on LIHC were analyzed.
Clinicopathological analysis (Table 1) revealed that cytoplasmic PCNP expression showed no significant correlation with pathological grade (p = 0.788) but was significantly associated with T stage (r = 0.245, p = 0.022), clinical stage (r = 0.245, p = 0.022), and recurrence (r = 0.228, p = 0.042). In contrast, nuclear PCNP levels (Table 2)correlated exclusively with pathological grade (r = 0.292, p = 0.006), with no significant associations for T stage (p = 0.755) or clinical stage (p = 0.755). Kaplan–Meier survival analysis revealed that patients with high PCNP expression, either in the nucleus or the cytoplasm, exhibited significantly shorter overall survival and disease-free survival compared with those with low PCNP expression (Fig. 2D). These findings indicate that elevated PCNP levels in both compartments are strong predictors of poor prognosis in HCC.
PCNP knockdown suppresses malignant behaviors in HCC cells
To explore the functional role of PCNP, we performed PCNP knockdown in Huh-7 and SK-HEP-1 cells using shRNA (Fig. 3A). PCNP depletion caused G0/G1 phase arrest (Fig. 3B) and significantly reduced cell viability from day 1 to 5 (Fig. 3C). Moreover, PCNP knockdown inhibited cell invasion, colony formation, and anchorage-independent growth in soft agar (Fig. 4A–C), confirming its pro-oncogenic role.
Knockdown of PCNP decreased cell proliferation in Huh-7 and SK-HEP-1. A After PCNP knockdown, PCNP protein levels were analyzed by performing western blot. *P<0.05, vs. shScram group. B after PCNP knockdown, cell cycle distribution was analyzed by performing PI staining followed by flow cytometry. *P < 0.05, vs. shScram group. C Cell viability was measured from day 1 to 5 after PCNP knockdown. *P < 0.05, vs. shScram group.
The effects of PCNP on malignant behaviors in LIHC. To evaluate the effects of PCNP knockdown on malignant behaviors, including invasion, colony formation and tumor formation in soft agar, Transwell assay (A), colony formation in plate (B) and tumor formation assay (C) were performed. *P < 0.05, vs. shScram group.
PCNP knockdown reduces MAPK pathway activation without inducing apoptosis
Given conflicting roles of PCNP in other cancers16,17, we evaluated MAPK pathway activity following PCNP knockdown. Western blotting showed reduced expression of ERK, JNK, and p38 MAPK proteins (Fig. 5). Apoptotic markers such as cleaved caspase-3, cleaved PARP, Bax, and Bad showed minimal changes (Supplementary Fig. 1), and Annexin V/PI staining showed no significant increase in apoptosis (Supplementary Fig. 2), suggesting PCNP supports cell survival, potentially by regulating stress pathways rather than directly promoting apoptosis.
PCNP transcriptionally regulates UHRF2, which correlates with malignancy
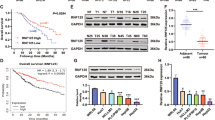
UHRF2, an E3 ubiquitin ligase known to target PCNP, is implicated in HCC progression18,19. GEPIA analysis revealed a strong positive correlation between PCNP and UHRF2 mRNA in HCC tissues (R = 0.72, p < 0.001; Fig. 6A). RT-qPCR further confirmed high UHRF2 expression in Huh-7 cells (Fig. 6B). Although UHRF2 mRNA was slightly downregulated in tumor versus normal tissue (Fig. 6C, D), its high expression was associated with poorer overall survival, mirroring the trends observed for PCNP.
UHRF2 is positively correlated with PCNP in LIHC. A correlation analysis of UHRF2 and PCNP was analyzed and presents positive correlation (R = 0.72, P value = 0). B Relative expression levels of UHRF2 in HCC cell lines. C Differential UHRF2 expression levels between tumor and adjacent tissues in the GTEX databsets. Red code represents tumor tissues data. Green code represents adjacent tissues. D Correlation of UHRF2 expression level with prognosis values in LIHC in the GEPIA database. E After PCNP knockdown or UHRF2 knockdown, PCNP or UHRF2 protein expression was measured by performing western blot. *P < 0.05, vs. shScram-1; #P < 0.05, vs. shScram-2.
Functional experiments showed that knockdown of PCNP significantly downregulated UHRF2 mRNA, whereas knockdown of UHRF2 had little effect on PCNP expression (Fig. 6E), suggesting a unidirectional, transcriptional regulatory relationship.
PCNP activates ErbB3/Ras/Raf pathway via UHRF2
Previous studies have implicated UHRF2 in the activation of the ErbB3/Ras/Raf pathway in HCC12. Consistent with this, PCNP overexpression increased UHRF2 protein levels, along with ErbB3, Ras, and phosphorylated Raf1 (p-Raf1). These effects were reversed by UHRF2 knockdown (Fig. 7), supporting the hypothesis that PCNP indirectly activates the ErbB3/Ras/Raf signaling axis via upregulation of UHRF2.
PCNP knockdown inhibits tumor formation in vivo
To assess the tumorigenic role of PCNP in vivo, we established subcutaneous xenografts using Huh-7 cells stably expressing either shScrambled or shPCNP. Tumor growth was significantly reduced in the shPCNP group compared to control (Fig. 8A). Immunohistochemistry confirmed reduced Ki-67 expression (Fig. 8B) and decreased UHRF2 protein levels (Fig. 8C) in the PCNP knockdown tumors, validating in vitro findings and further supporting that PCNP promotes tumorigenesis through UHRF2 upregulation.
Knockdown of PCNP expression repressed tumor formation in nude mice. A After 30-day maintenance, tumor tissues were surgically excised and tumor growth curve was calculated. *P < 0.05, vs. shScram group. B IHC analyses of Ki-67 in tumors of shScram and shPCNP groups. *P < 0.05, vs. shScram group. C PCNP and UHRF2 protein levels in xenograft tumors of mice.
Discussion
In this study, we identified PCNP as a novel oncogenic factor in LIHC. PCNP expression was significantly upregulated in LIHC tissues compared with adjacent non-tumorous tissues, and this elevation was associated with enhanced proliferation, invasion, and tumorigenicity both in vitro and in vivo. Importantly, tissue array analysis revealed cytoplasmic accumulation of PCNP in HCC samples, which correlated with higher recurrence and poorer survival. This mislocalization implies that PCNP may exert distinct functions depending on its subcellular distribution, with cytoplasmic retention potentially constraining nuclear activities while promoting oncogenic signaling through UHRF2 and ErbB3. Consistently, we demonstrated a positive regulatory relationship between PCNP and UHRF2, leading to activation of the ErbB3/Ras/Raf pathway. Together, these findings highlight both the clinical relevance of PCNP localization and the central role of the PCNP–UHRF2–ErbB3 axis in HCC progression, suggesting its value as a prognostic marker and therapeutic target.
Previous studies have implicated PCNP in various cancers, including neuroblastoma, lung adenocarcinoma, and ovarian cancer20,21. Consistent with those reports, our findings further substantiate the oncogenic role of PCNP. While TCGA data show a significant upregulated in PCNP mRNA expression between tumor tissues and normal liver tissues, immunohistochemical analysis of tissue microarrays confirmed markedly elevated PCNP protein expression in LIHC. These results suggest that PCNP contributes to tumor regulation through both transcriptional and post-transcriptional mechanisms.
We also examined the subcellular localization of PCNP and its clinical significance. Cytoplasmic PCNP expression significantly correlated with advanced T stage, clinical stage, and recurrence. The distinct clinical correlations of cytoplasmic versus nuclear PCNP highlight subcellular localization as a key determinant of its oncogenic function. Cytoplasmic accumulation—likely resulting from failed ubiquitin-mediated nuclear import—promotes tumor progression by activating membrane-proximal signaling (e.g., ErbB3/Ras). Nuclear PCNP, while critical for transcriptional regulation, primarily reflects differentiation status through its association with pathological grade22. Its dynamic turnover and localization may be essential for fine-tuning its oncogenic functions. This aligns with previous studies showing PCNP involvement in tumorigenesis through cell cycle regulation and survival pathways23,24,25.
Our data indicate that PCNP knockdown suppressed malignant phenotypes in HCC cells, including proliferation, colony formation, and invasion. Interestingly, PCNP depletion only mildly altered pro-apoptotic proteins such as cleaved caspase-3, PARP, Bax, and Bad, suggesting that its role may be more related to enhancing tumor cell survival and proliferation rather than inducing apoptosis. This may also imply a potential contribution of PCNP to chemoresistance, a hypothesis worth further investigation.
UHRF2 has been reported to promote the proliferation, migration, and invasion of HCC cells26. Functionally, UHRF2 acts as a nuclear E3 ubiquitin ligase, regulating cell cycle progression through interactions with key cell cycle regulators27. In breast and lung cancers, UHRF2 has been shown to ubiquitinate cyclin D1 and cyclin E1, thereby inducing G0/G1 phase cell cycle arrest, and is considered a potential tumor suppressor28,29. Conversely, UHRF2 has also been implicated as an oncogene in several cancers. For instance, Li et al. reported that UHRF2 is upregulated in colorectal cancer and positively correlates with poor prognosis30. In gastric cancer, UHRF2 promotes cell migration and invasion by modulating epithelial-mesenchymal transition (EMT)31. These seemingly contradictory roles may stem from tissue-specific expression patterns and the context-dependent complexity of UHRF2 function32. Our findings support a model in which PCNP-driven UHRF2 expression facilitates LIHC progression via activation of the ErbB3/Ras/Raf cascade. The observed unidirectional regulation—where PCNP knockdown reduces UHRF2 expression but not vice versa—suggests a potential transcriptional control mechanism.
Bioinformatic evidence further supports this model. STRING analysis (combined score: 0.898) predicts that the RING domain of UHRF2 (its E3 ubiquitin ligase catalytic core) interacts with PEST motifs in PCNP (regions associated with proteolytic degradation) (Supplementary Fig. 3). This aligns with experimental annotations indicating that “UHRF2 ubiquitinates PCNP, leading to its degradation” (STRING: ENSP00000276893), and with the known rapid nuclear turnover of PCNP via the ubiquitin–proteasome system. Moreover, our observation of cytoplasmic PCNP accumulation in advanced HCC (Table 1) may reflect impaired nuclear import or failed degradation, further supporting the proposed mechanism.
In addition, although PCNP enhanced phosphorylation of ErbB3 and downstream Ras/Raf signaling, ERK activation remained largely unchanged. This observation indicates that PCNP selectively engages specific branches of the pathway rather than broadly amplifying MAPK cascades. The stable ERK activity suggests compensatory mechanisms or feedback regulation that limit its responsiveness, highlighting the pathway selectivity of PCNP-driven signaling. Presenting the ratio of phosphorylated to total protein further supports this interpretation and provides a more accurate representation of pathway activation.
Although direct transcriptional regulation has not yet been experimentally confirmed, this model offers a plausible explanation for PCNP-UHRF2 dysregulation in hepatocarcinogenesis: cytoplasmic accumulation of PCNP may evade UHRF2-mediated degradation and aberrantly activate oncogenic signaling pathways such as ErbB3/Ras. To validate this hypothesis, future studies—including chromatin immunoprecipitation (ChIP), luciferase reporter assays, and rescue experiments—are warranted to determine whether PCNP directly binds to and activates the UHRF2 promoter.
In conclusion, our study reveals that PCNP is significantly upregulated in HCC and is associated with adverse clinical features and poor prognosis. We uncover a novel regulatory axis in which PCNP transcriptionally upregulates UHRF2, thereby activating the ErbB3/Ras/Raf signaling pathway and promoting HCC malignancy. While the specific functions of nuclear versus cytoplasmic PCNP remain to be clarified, our findings provide compelling evidence for PCNP as a prognostic biomarker and potential therapeutic target in HCC.
Beyond mechanistic insights, our findings also hold translational elevance. According to Hoshida et al.’s molecular classification of HCC, distinct subclasses are associated with differences in prognosis and therapeutic responsiveness. The observed PCNP–UHRF2 axis, particularly its cytoplasmic and nuclear compartmentalization, could intersect with these subclasses, potentially informing patient stratification. For example, high PCNP expression may identify tumors with aggressive phenotypes and shorter survival, aligning with the poor-prognosis signatures defined by Hoshida’s33 classification. These associations raise the possibility that PCNP could be explored not only as a biomarker but also as a candidate target within subclass-specific therapeutic strategies. Nevertheless, further validation in prospective cohorts will be required to substantiate these translational applications.
Further investigation into its role in drug resistance and subcellular dynamics may pave the way for more effective HCC interventions.
Materials and methods
Tissue microarrays and IHC testing of patient samples
Tissue microarrays (TMAs) were constructed for 87 liver cancer patients using three 2 mm tissue cores and bought from Shanghai Outdo Biotech Company. Sections were cut into 4 μm thickness, deparaffinized, rehydrated and treated with a peroxidase block. IHC staining for PCNP followed the manufacturer’s protocol. Sections were incubated overnight at 4 °C with anti-PCNP antibody (ab97909, Abcam, 1:100), followed by 30 min incubation with EnVision HRP-conjugated secondary antibody (DAKO, Denmark). Staining was visualized using 3,3′-diaminobenzidine (DAB) substrate.
PCNP expression was assessed in both nuclear and cytoplasmic compartments. Immunoreactivity was evaluated using an immunoreactive score (IRS), calculated as the product of staining intensity and percentage of positive cells. Based on IRS values, PCNP expression was categorized as low, medium, or high. Representative micrographs indicating that medium to high PCNP expression was observed exclusively in malignant tissues, while adjacent normal tissues exhibited low expression levels.
Cell culture
The human hepatocellular carcinoma (HCC) cell lines Huh-7 and SK-HEP-1 were purchased from Feiouer Cell Bank (Chengdu, Sichuan, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS). All cells were cultured at 37 °C in a humidified incubator with 5% CO₂.
Bioinformatics analysis using GEPIA and Sangerbox
To evaluate the transcriptional landscape of LIHC, we employed a combination of public database analysis and online bioinformatics platforms. Differential expression analysis and survival analysis were conducted using GEPIA3.0 (http://sangerbox.com/home.html), based on TCGA-LIHC and GTEx datasets. Gene expression was displayed as log₂(TPM + 1), and statistical significance was determined using ANOVA and Spearman correlation. DSS and OS curves were generated using the “Survival Analysis” module, with median expression levels as grouping thresholds. Statistical significance was determined using the log-rank test, and hazard ratios were provided with 95% confidence intervals.
To estimate the potential regulatory association between PCNP and UHRF2, Spearman correlation analysis was performed using GEPIA2, based on TCGA-LIHC tumor and adjacent normal tissue data.
All data used were normalized and statistically analyzed according to the platform’s default pipelines, ensuring reproducibility and public accessibility.
Cell transfection
Cells were seeded in six-well plates at approximately 80% confluence and transfected with lentivirus for either overexpression or knockdown of PCNP or UHRF2. The lentivirus expression vector (GV358) for PCNP overexpression was constructed by GeneChem (Shanghai, China). Lentivirus encoding short hairpin RNAs (shRNA) targeting UHRF2 or PCNP were purchased from Invitrogen (USA) and GenePhama (China). Viral transduction was performed at a multiplicity of infection (MOI) of 20. After 4 h of incubation, the medium was replaced with fresh complete medium containing 10% fetal bovine serum (FBS). Stable cell lines were established using lentiviral vectors provided by Shanghai Heyuan (Shanghai, China), followed by selection with 2 µg/mL puromycin. Transfection efficiency was confirmed by western blot analysis.
Western blot
Total protein was isolated by using SoniConvert® Tissue Cell Convertor (DocSense, Chengdu, China) and Animal Tissue/cells/bacteria total protein isolation kit (Cat. No.: PP003; DocSense, Chengdu, China) by following manufacturer’s instruction. Protein concentrations were measured using the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), 20 µg/ml of total protein was fractionated by 10% SDS-PAGE gels. Membrane was blocked in 5% bovine serum albumin (BSA) in PBS for 1 h at room temperature and then incubated overnight at 4 °C with the following primary antibodies (1:1000 dilution): anti-PCNP (ab97909), anti-ERK1/2 (ab184699), anti-phospho-ERK1/2 (ab214036), anti-p38 (ab170099), anti-phospho-p38 (ab195049), anti-JNK1 (ab179461), anti-phospho-JNK1 (ab124956), anti-UHRF2 (ab273667), anti-ErbB3 (ab32121), anti-Ras (ab52939), anti-Raf1 (ab181115), anti-phospho-Raf1 (ab173539), and anti-β-actin (ab8226).Protein bands were visualized and quantified using the C-DiGit Blot Scanner (LI‐COR, Lincoln, NE, USA). All experiments were performed in triplicate, and results are presented as mean fold changes.
RT-qPCR
Total RNA was extracted from cultured cells using the Animal Tissue/Cells Total RNA Isolation Kit (Cat. No.: RP003; DocSense, Chengdu, China). NA quality and integrity were assessed via UV spectrophotometry and formaldehyde denaturing agarose gel electrophoresis. cDNA was synthesized from 1 µg of total RNA using the GoldScript cDNA Kit (Invitrogen) according to the manufacturer’s instructions. Quantitative PCR was performed with SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA) in a 20 µL reaction volume. The amplification protocol included initial denaturation at 95 °C for 35 s, followed by 35 cycles of 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 1 min.
Cell cycle analysis via PI staining
For cell cycle analysis by PI staining, cells were harvested and washed three times with ice-cold PBS, then fixed overnight at 4 °C in 70% ethanol. After fixation, cells were washed again with PBS and incubated with RNase A (100 µg/mL) and propidium iodide (PI, 40 µg/mL; Beyotime, China) in the dark for 15 min. DNA content was analyzed using a 3-laser Navios flow cytometer (Beckman Coulter, Brea, CA, USA) to determine cell cycle distribution.
Cell viability assay
To evaluate cell viability, 2 × 103 cells were plated in each 96-well plates. For each well, 10 µL tetrazolium salt WST-8 (Cell counting Kit-8; Keygen, Nanjing, China) was added to each well for extra 2 h incubation at 37℃. Optical density was measured at a wavelength of 450 nm (OD450) using a microplate reader (Synergy 2 Multi-Mode Microplate Reader; BioTek, Winooski, VT, USA).
Invasion assay
Cells were trypsinized with 0.25% Trypsin and washed three times with ice-cold PBS. A total of 1 × 104 cells were seeded into the upper chambers of transwell inserts with 8 μm pore polycarbonate membranes (Corning Incorporated, Corning, NY, USA) pre-coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Each experiment was performed in triplicate. After 48 h of incubation, non-invading cells on the upper membrane surface were removed, and invading cells on the lower membrane surface were fixed and stained with crystal violet (Beyotime Institute of Biotechnology, Beijing, China). The number of invading cells was counted under a microscope at 100× magnification in four randomly selected fields.
Colony formation assay
Cells were trypsinized, resuspended in PBS, and counted. Subsequently, 1,000 cells were seeded into each well of 6-well plates and cultured for 2–3 weeks. When visible colonies appeared, the plates were gently washed twice with PBS. Colonies were then fixed with 4% paraformaldehyde for 10 min and stained with 0.25% crystal violet for 20 min.
Soft agar colony formation assay
Cells were resuspended in 0.3% soft agar in RPMI-1640 medium supplemented with 10% FBS, and then overlaid onto a base layer of 0.6% solidified agar in RPMI-1640 with 10% FBS in 6-well plates Plates were incubated at 37 °C for 2 weeks. Colonies larger than 50 μm in diameter were counted under a microscope.
Animal experiment
Five-week-oldmale BALB/c nude mice were purchased from Sichuan Dashuo Laboratory Animal Center (Sichuan, China). All procedures were approved by the Medical Ethics Committee of the Chengdu University of Traditional Chinese Medicine and conducted in accordance with institutional guidelines for animal care and use. The study was reported in accordance with the ARRIVE guidelines (https://arriveguidelines.org). For each group (n = 4), 2 × 106 cells were collected for each injection and injected subcutaneously into the left flank of the mice for each group (n = 4). From 10th day after injection, tumor size was measured every five days, and calculated using the following formula: TV (mm3) = a2 × b × 0.52, where a and b were the shortest and longest diameters, respectively. After 30-day later, nude mice were sacrificed and tumor specimens were fixed in 4% paraformaldehyde and analyzed for detecting positive rate of Ki67.
Mice underwent isoflurane anesthesia (4% induction, 2% maintenance via nose cone). Euthanasia was initiated upon reaching predefined humane endpoints (tumor volume > 1.5 cm³, > 20% weight loss, or ulceration) using cervical dislocation by certified personnel.
Annexin V-FITC/PI double staining
Apoptosis was measured using Annexin V-FITC/PI apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. The cells were washed with ice-cold PBS and pelleted. After removing supernatant, pellet was resuspended in 1×binding buffer and stained with 5 µL fluorescein isothiocyanate-labeled Annexin V at 4℃ for 15 min in the dark. Then 10 µL of PI was added at 4℃ for 5 min in the dark. Then cells were analyzed by flow cytometry using 3 laser Navios flow cytometers (Beckman Coulter, Brea, CA, USA).
Statistical analysis
In this study, all experiments were repeat three times independently. Student’s t test was used to determine the statistical significance. All data in this study was presented as the mean ± s.e.m. P < 0.05 was considered as significant.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Di Bisceglie, A. M. Epidemiology and clinical presentation of hepatocellular carcinoma. J. Vasc. Interv. Radiol. 13 (9 Pt 2), S169–S171 (2002).
Brown, Z. J. et al. Management of hepatocellular carcinoma: a review. JAMA Surg. 158 (4), 410–420 (2023).
McGlynn, K. A., Petrick, J. L. & El-Serag, H. B. Epidemiology of hepatocellular carcinoma.Hepatology 73 (Suppl 1):4–13. (2021).
Bruix, J., Boix, L., Sala, M. & Llovet, J. M. Focus on hepatocellular carcinoma. Cancer Cell. 5 (3), 215–219 (2004).
Khan, N. H., Chen, H. J. & Fan, Y. Biology of PEST-containing nuclear protein: a potential molecular target for cancer research. Front. Oncol. 12, 784597 (2022).
Rogers, S., Wells, R. & Rechsteiner, M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234 (4774), 364–368 (1986).
Wu, D. D., Gao, Y. R. & Li, T. PEST-containing nuclear protein mediates the proliferation, migration, and invasion of human neuroblastoma cells through MAPK and PI3K/AKT/mTOR signaling pathways. BMC Cancer. 18 (1), 499 (2018).
Dong, P., Fu, H. & Chen, L. PCNP promotes ovarian cancer progression by accelerating β-catenin nuclear accumulation and triggering EMT transition. J. Cell. Mol. Med. 24 (14), 8221–8235 (2020).
Motnenko, A., Liang, C. C. & Yang, D. Identification of UHRF2 as a novel DNA interstrand crosslink sensor protein. PLoS Genet. 14 (10), e1007643 (2018).
Afzal, A., Sarfraz, M. & Li, G. L. Taking a holistic view of PEST-containing nuclear protein (PCNP) in cancer biology. Cancer Med. 8 (14), 6335–6343 (2019).
Jin, C., Xiong, D. & Li, H. R. Loss of UHRF2 is associated with Non-small cell lung carcinoma progression. J. Cancer. 9 (17), 2994–3005 (2018).
Sun, J., Wu, K. & Chen, S. UHRF2 promotes hepatocellular carcinoma progression by upregulating ErbB3/Ras/Raf signaling pathway. Int. J. Med. Sci. 18 (14), 3097–3105 (2021).
Kiavue, N., Cabel, L. & Melaabi, S. ERBB3 mutations in cancer: biological aspects, prevalence and therapeutics. Oncogene 39 (3), 487–502 (2020).
Sithanandam, G. & Anderson, L. M. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 15 (7), 413–448 (2008).
Roskoski, R. Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 79, 34–74 (2014).
Chen, Y. G., Liu, H. X. & Hong, Y. PCNP is a novel regulator of proliferation, migration, and invasion in human thyroid cancer. Int. J. Biol. Sci. 18 (9), 3605–3620 (2022).
Keshet, Y. & Seger, R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 661, 3–38 (2010).
Mori, T., Li, Y. & Hata, H. NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP, a PEST-containing nuclear protein. FEBS Lett. 557 (1–3), 209–214 (2004).
Zhang, C., Zhao, Y. & Zhao, N. NIRF Optical/PET Dual-Modal imaging of hepatocellular carcinoma using heptamethine carbocyanine dye. Contrast Media Mol. Imaging 2018, 4979746 (2018).
Wu, D. D. et al. PEST-containing nuclear protein mediates the proliferation, migration, and invasion of human neuroblastoma cells through MAPK and PI3K/AKT/mTOR signaling pathways. BMC Cancer 18, 499. https://doi.org/10.1186/s12885-018-4391-9 (2018).
Han, S. W. et al. Gene expression network analysis of lymph node involvement in colon cancer identifies AHSA2, CDK10, and CWC22 as possible prognostic markers. Sci. Rep. 10, 7170 (2020).
As a nuclear. Protein rich in PEST sequence, PCNP is strictly controlled by the ubiquitination- proteasome degradation pathway, and is also a rapidly degraded protein sequence with a short half-life.
Wu, D. D. et al. PEST-containing nuclear protein mediates the proliferation, migration, and invasion of human neuroblastoma cells through MAPK and PI3K/AKT/mTOR signaling pathways. BMC Cancer 18, 499 (2018).
Wang, D. Y. et al. PEST-containing nuclear protein regulates cell proliferation, migration, and invasion in lung adenocarcinoma. Oncogenesis 8, 22 (2019).
Dong, P. et al. PCNP promotes ovarian cancer progression by accelerating β-catenin nuclear accumulation and triggering EMT transition. J. Cell. Mol. Med. 24, 8221–8235 (2020).
Sun, J. et al. UHRF2 promotes hepatocellular carcinoma progression by upregulating ErbB3/Ras/Raf signaling pathway. Int. J. Med. Sci. 26, 3097—3105 (2021).
Afzal, A. et al. Taking a holistic view of PEST-containing nuclear protein (PCNP) in cancer biology. Cancer Med. 8, 6335–6343 (2019).
Motnenko, A. et al. Identification of UHRF2 as a novel DNA interstrand crosslink sensor protein. PLoS Genet. 14, e1007643 (2018).
He, X. et al. Let-7a elevates p21(WAF1) levels by targeting of NIRF and suppresses the growth of A549 lung cancer cells. FEBS Lett. 583, 3501–3507 (2009).
Li, L. et al. UHRF2 promotes intestinal tumorigenesis through stabilization of TCF4 mediated Wnt/beta-catenin signaling. Int. J. Cancer 147, 2239–2252 (2020).
Lai, M. et al. Multidimensional proteomics reveals a role of UHRF2 in the regulation of epithelial-mesenchymal transition (EMT). Mol. Cell. Proteom. 15, 2263–2278 (2016).
Jiang, K. C. et al. Regulation of PEST-containing nuclear proteins in cancer cells: implications for cancer biology and therapy. Front. Oncol. 15, 1548886 (2025).
Goossens, N., Sun, X. & Hoshida, Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat. Oncol. 2 (4), 371–379. https://doi.org/10.2217/hep.15.26 (2015).
Acknowledgements
The author would like to thanks Tao Hong for language editing.
Funding
The Technical Innovation Research and Development Project of Scientific and Technological Office of Chengdu(2021-YF05-01726-SN).
Key Science and Technology Project of Zigong Scientific and Technological Office(2024-YGY-02-03).
Author information
Authors and Affiliations
Contributions
JLL and YTY designed the experiments. XYZ and XYZ built xenograft nude mice models and performed pathological analysis. JLL, ZYZ and XYZ are responsible cell culture, morphological analysis, and data collection and performed statistical analysis. JLL and XYZ wrote the manuscript. All authors read and approved the final manuscript.Jinlong Luo and Zhiyi Zhou contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All the animal experiments were conducted according to the Medical Ethics Committee of the Chengdu University of Traditional Chinese Medicine and performed according to the ethical guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, J., Zhou, Z., Zhao, X. et al. PCNP promotes hepatocellular carcinoma progression by upregulating UHRF2 to activate ErbB3/Ras/Raf pathway. Sci Rep 15, 41312 (2025). https://doi.org/10.1038/s41598-025-25127-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-25127-9