Abstract
CXC chemokine ligand 8 (CXCL8) has emerged as a critical mediator in tumorigenesis. This study aims to elucidate the role of CXCL8 in the initiation and progression of papillary thyroid carcinoma (PTC). Comprehensive bioinformatic analyses were conducted to assess CXCL8 expression across pan-cancer datasets, clinical characteristics, biological functions, the tumor immune microenvironment, and single-cell RNA-sequencing data. The expression of CXCL8 and its association with clinicopathological features were validated using qRT-PCR and immunohistochemical analysis of tissue microarrays. The data revealed a significantly elevated expression of CXCL8 in PTC tissues compared to Paired non-cancerous thyroid tissues(PNTs), with CXCL8 expression showing a strong positive correlation with lymph node metastasis. These findings suggest that CXCL8 may function as a key molecular contributor to lymph node metastasis in PTC. CXCL8 is likely to promote tumor cell proliferation, migration, and invasion via the PI3K-Akt signaling pathway. Moreover, CXCL8 expression is closely associated with tumor-infiltrating immune cells, particularly exhibiting high expression levels in dendritic cells. In patients with low CXCL8 expression, anti-CTLA-4 immunotherapy may offer superior clinical benefit compared to anti-PD-1 treatment. Collectively, these findings indicate that CXCL8 represents a promising biomarker involved in the pathobiology of PTC and constitutes a potential therapeutic target, providing novel insights into immunotherapeutic strategies for PTC.
Similar content being viewed by others
Papillary thyroid carcinoma (PTC) is a prevalent endocrine malignancy characterized by a steadily rising global incidence and a complex pathogenesis involving dysregulation of multiple molecular signaling pathways1,2. According to the Global Agency for Research on Cancer (GLOBOCAN), the incidence of PTC continues to rise, particularly among women, who exhibit approximately threefold higher incidence rates than men. Notably, the average 5-year survival rate for PTC approaches 98%3,4. Differentiated thyroid carcinomas, including papillary and follicular subtypes, are generally associated with favorable prognoses, whereas undifferentiated thyroid cancers are marked by aggressive behavior and high mortality rates5. In recent years, advances in molecular biology and genomics have spurred increasing research into the key molecular drivers and mechanistic underpinnings of PTC tumorigenesis. However, despite substantial advances in diagnosis and therapeutic strategies, disease recurrence and metastasis continue to pose significant clinical challenges in PTC management. Therefore, elucidating novel molecular biomarkers and therapeutic targets holds considerable promise for improving the clinical outcomes of patients with PTC.
CXCL8 (also known as interleukin-8) is a multifunctional pro-inflammatory chemokine initially identified as a neutrophil chemoattractant, which has recently been recognized as a key contributor to tumorigenesis6. Aberrant expression of CXCL8 has been documented in various malignancies, including cervical and gastric cancers, and is strongly associated with tumor invasion, metastasis, and clinical prognosis7,8. However, the expression profile and functional role of CXCL8 in thyroid cancer remain incompletely understood. Although preliminary evidence indicates a potential link between CXCL8 and thyroid tumorigenesis, its underlying mechanisms and clinical implications warrant further investigation9. Furthermore, the extent to which CXCL8 modulates thyroid cancer progression through regulation of the immune microenvironment remains a compelling area for in-depth study.
At present, substantial knowledge gaps persist regarding the role of CXCL8 in PTC. First, the regulatory mechanisms underlying CXCL8 expression in PTC remain elusive, and its function within the tumor microenvironment has not been systematically characterized10. Second, the association between CXCL8 expression and clinicopathological parameters—including tumor stage, nodal status, histological subtype, and anatomical location—has not been comprehensively evaluated. In addition, its potential utility as a diagnostic biomarker or prognostic indicator remains to be validated in large-scale clinical cohorts11. These unresolved questions limit current understanding of CXCL8 biology in PTC and hinder the advancement of its translational and clinical applications.
This study aimed to comprehensively characterize the expression landscape of CXCL8 in PTC, investigate its clinical relevance, and elucidate its potential molecular mechanisms12. Through integrated analyses of CXCL8 expression with patient prognosis, immune infiltration patterns, and clinicopathological characteristics, this study seeks to identify novel molecular markers and therapeutic targets for PTC. Furthermore, we aim to delineate the immunomodulatory role of CXCL8 in PTC, thereby providing a theoretical foundation for the development of personalized therapeutic strategies targeting the tumor immune microenvironment. Ultimately, these findings are expected to offer novel insights into improving clinical outcomes and quality of life for patients with PTC.
Materials and methods
Public database analysis
The RNA sequencing data and related clinical information of patients with PTC were downloaded from the TCGA (https://cancergenome.nih.gov/) database.We downloaded the transcriptome profiling (HTseq-FPKM) gene expression quantitative data of 404 PTC (including 355 tumors and 49 para-cancerous tissues) from TCGA database.The list of immune-related genes was downloaded from the import (https://www.immport.org/shared/home) and InnateDB (https://www.innatedb.com/) databases. This study included patients with PTC. The raw data were summarized and processed using Strawberry perl (v5.30.0), and subsequent analyses were conducted using R software (v4.3.3).Our comprehensive from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) for thyroid papillary carcinoma GSE33630 data set, to verify that the related gene expression.The GSE33630 dataset comprises 11 cases of anaplastic thyroid carcinomas (ATC), 49 cases of PTC, and 45 cases of normal thyroid tissues (N). For subsequent analyses in this study, 49 cases of PTC and 45 cases of N were selected.we downloaded pan-cancer data from the UCSC Xena platform (https://xena.ucsc.edu/) to verify the expression levels of CXCL8 between Paired non-cancerous thyroid tissues(PNTs) and tumor samples.
TCGA-derived PTC transcriptome and clinical data—including age at diagnosis, sex, survival time, survival status, clinical stage, and pathological TNM staging—were processed with Strawberry Perl and analyzed with R.
Associations between CXCL8 expression levels and clinical characteristics (age, clinical stage, T stage, M stage, and N stage) were evaluated using χ² tests or Wilcoxon signed-rank tests. Results were visualized as boxplots and heatmaps.
To elucidate the biological role of CXCL8 in PTC, correlation analyses were performed between CXCL8 and other genes, with thresholds set at |Cor| > 0.6 and P < 0.001. Strongly correlated genes were visualized via circular plots. Differential expression analysis was conducted to compare high and low CXCL8 expression groups, using |logFC| > 1 and FDR < 0.05 as cutoffs. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, and Gene Set Enrichment Analysis (GSEA) were performed to identify associated molecular pathways and biological processes. R packages used included “ggplot2”, “ggpubr”, “ggExtra”, “circlize”, “corrplot”, and “pheatmap”.
To assess the relationship between CXCL8 expression and immune infiltration, CIBERSORT was used to estimate the proportions of 22 immune cell subsets. Infiltration differences between high- and low-CXCL8 groups were displayed as boxplots, while scatter plots and lollipop charts illustrated correlations. To evaluate immunotherapeutic potential, associations between CXCL8 expression and immune checkpoint genes and drug targets were analyzed. Immunotherapy scores for anti-CTLA4 and anti-PD-1 responses were derived from TCIA (https://tcia.at), and response differences between CXCL8 expression groups were compared. R packages applied included “reshape2”, “ggpubr”, “vioplot”, and “ggExtra”.
Single-cell RNA sequencing data (GSE241184) from lymph node metastases of PTC patients were obtained from the GEO database. Data integration was performed using the “harmony” R package. Dimensionality reduction, clustering, and cell-type annotation were conducted using “Seurat” and “SingleR”. Violin plots were generated to visualize gene expression within annotated cell types. Screening thresholds were set at P < 0.05 and |logFC| > 1.
Clinical tissue sample collection
To validate bioinformatics findings, we collected clinical tissue samples from 49 patients with PTC who underwent thyroidectomy at Gansu Provincial Hospital (Lanzhou, China), with the following inclusion criteria: (1)pathologically confirmed PTC post-thyroidectomy; (2)no prior anti-tumor therapy (radiotherapy, chemotherapy, or targeted therapy) before surgery; (3)informed consent obtained from patients or their legal guardians.
From these 49 patients, two subsets of samples were used for different experiments:
7 paired tissue samples (7 PTC tissues + 7 PNTs): Tissue (0.5–1 cm in diameter) was excised from the center of PTC tissues; PNTs (of similar size) were obtained from areas > 2 cm from the edge of the tumor. Samples were rinsed with normal saline, rapidly cryopreserved in liquid nitrogen, and confirmed by hematoxylin-eosin (HE) staining by a professional pathologist. These samples were used for high-throughput RNA sequencing.
49 paired tissue samples (49 PTC tissues + 49 PNTs): These samples were used for tissue microarray construction (for immunohistochemical analysis) and qRT-PCR validation (a subset of this cohort, as detailed in “RNA extraction and quantitative real-time polymerase chain reaction”).
The experimental protocol was approved by the Institutional Review Board of Gansu Provincial People’s Hospital (Approval Number: 2022 − 182), and all procedures complied with the Declaration of Helsinki (1964) and its later amendments.
High-throughput RNA sequencing
Fourteen tissue samples from 7 patients from Gansu Provincial People’s Hospital were collected in this study. Tissue 0.5 to 1 cm in diameter was removed from the center of the patient’s PTC tissue. PNTs of similar size was obtained from the patient more than 2 cm from the edge of the thyroid papillary carcinoma tissue. Samples were collected, rinsed with normal saline, and rapidly cryopreserved in liquid nitrogen. The tissues were stained with HE and confirmed by a professional pathologist. The patient had not received preoperative radiotherapy or chemotherapy. Total RNA extraction was provided by Cloud Sequencing Biotechnology (Shanghai, China). RNA concentration was measured for each sample using a NanoDrop ND-1000 instrument (Thermo Fisher Scientific), considering an OD 260/280 value of 1.8 to 2.1 to ensure good RNA purity. CloudSeqInc. (Shanghai, China) provided the high-throughput RNA sequencing service. rRNA was removed from total RNA using the GenSeq®rRNA Removal Kit (GenSeq, Inc.) kit. After RNA removal, RNA sequencing libraries were constructed using the GenSeq® Low-Input Whole RNA Library Preparation Kit (GenSeq, Inc.). The constructed sequencing libraries were quality-controlled and quantified using a Bioanalyzer 2100 system (Agilent Technologies), and high-throughput sequencing of the libraries was controlled using an Illumina NovaSeq 6000 instrument with a paire-end length of 150 bp.
RNA extraction and the quantitative real-time polymerase chain reaction (qRT-PCR)
To validate the CXCL8 expression pattern identified by bioinformatics analysis, we selected 10 paired samples (10 PTC tissues + 10 PNTs from the aforementioned cohort of 49 PTC patients (inclusion criteria: same as “Clinical Tissue Sample Collection”; selection method: simple random sampling to avoid selection bias).
Total RNA was extracted from these 20 tissues (10 pairs) using the TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions.The concentration and purity of the extracted total RNA were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA). The RNA concentrations typically ranged from 178 to 489 ng/µL.Only RNA samples with an A260/A280 ratio between 1.8 and 2.1, indicating high purity without protein or other contamination, were used for subsequent experiments. For reverse transcription, 1 µg of total RNA from each qualified sample was used as input for cDNA synthesis using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Bio Co., Ltd., Japan) according to the manufacturer’s instructions.
Real-time fluorescence quantitative PCR was conducted using TB Green Premix Ex Taq II (Takara Bio Co., Ltd., Japan) on a Thermo Scientific 7500 Real-Time PCR System (Thermo Fisher Scientific, USA). The reaction conditions were: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. A melt curve analysis was performed post-amplification to confirm the specificity of the PCR products. The cycle threshold (Ct) values were obtained for both CXCL8 and the reference gene β-actin.
The comparative ΔΔCt method was used to analyze the qRT-PCR data. For each sample, the ΔCt = Ct(CXCL8)–Ct(β−actin).For statistical comparison between PTC tissues and their PNTs, the non-parametric Wilcoxon signed-rank test was applied directly to the ΔCt values.The relative gene expression fold change for visualization was calculated as 2^–ΔΔCt, where ΔΔCt = ΔCt(PTC)–ΔCt(PNT).
Each sample was tested in triplicate, and the average Ct value was used for calculations.
The primers for CXCL8 and β-actin were synthesized by Beijing Qingke Biotechnology Co., Ltd. (Beijing, China), and the primer sequences are shown in Table 1.
Immunohistochemical analysis of tissue microarray
The primary antibody used was anti - CXCL8(Clone EPR19824;Proteintech Group, Inc., Chicago, IL, USA; Cat#17038-1-AP; RRID: AB_2126217;dilution 1:200).The secondary antibody was a HRP-conjugated goat anti-rabbit IgG antibody(Cell Signaling Technology, Danvers, MA, USA; Cat#7074;RRID: AB_2099233;dilution 1:3000.The chromogenic substrate was 3, 3’ - diaminobenzidine (DAB)(Dako, Agilent Technologies, Cat. #K3468, RRID: AB_2894767)and prepared according to the manufacturer’s instructions.Tissue sections were deparaffinized twice in xylene (20 min each) and rehydrated through a graded ethanol series (100%, 95%, 80%, and 60%; 3 min each). Following three rinses with distilled water (1 min each), antigen retrieval was performed by submerging sections in retrieval solution and heating them in a microwave at medium power for 10 min. The sections were then cooled in buffer for 35 min. Prior to immunostaining, endogenous peroxidase activity was quenched by incubating sections in 3% H₂O₂ (room temperature, 10 min), followed by three washes with 1X TBST (1 min each). Non-specific binding was blocked with blocking buffer (1 h, room temperature), after which sections were incubated with primary antibody (1.5 h, room temperature), washed with TBST, and treated with peroxidase-conjugated secondary antibody (30 min, room temperature). Color development was achieved using DAB (2–5 min), and sections were counterstained with hematoxylin, rinsed with TBST, and immersed in distilled water (5 min). Finally, sections were dehydrated in graded ethanol (60%, 80%, 90%, and 100%; 5 min each), cleared in xylene (twice, 5 min each), mounted with neutral gum, and examined under a microscope.The slides were scanned on the tissue microarray scanner to quantitatively analyze the expression level of CXCL8.The immunohistochemical staining was evaluated independently by two experienced pathologists who were blinded to the clinical data. The staining intensity was scored as follows: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The proportion of positively stained tumor cells was scored as: 0 (< 5%), 1 (5%−25%), 2 (26%−50%), 3 (51%−75%), and 4 (> 75%). The final immunoreactivity score (IRS) for each case was calculated by multiplying the intensity score by the proportion score, resulting in a range from 0 to 12. An IRS score ≥ 4 was defined as high expression, and < 4 was defined as low expression for statistical analysis.
Statistical analysis
Statistical analyses were conducted using SPSS version 28.0 (IBM Corp., USA) and GraphPad Prism version 10.0 (GraphPad Software, USA). Data conforming to a normal distribution were expressed as mean ± standard deviation (mean ± SD).
For comparisons of CXCL8 mRNA expression levels between paired PTC tissues and PNTs, the non-parametric Wilcoxon signed-rank test was employed due to the sample size and potential deviations from normality assumptions.
For the correlation analysis between CXCL8 expression and clinicopathological features, the following specific handling was applied to the age variable: age was treated as a continuous variable and presented as median [interquartile range (IQR)]. This approach was adopted to avoid information loss and potential bias caused by arbitrary dichotomization (e.g., dividing age into ≤ 55 years/>55 years), which could oversimplify the age distribution and obscure its nuanced relationship with CXCL8 expression. For other categorical clinicopathological features (e.g., gender, lymph node status, Hashimoto’s thyroiditis), correlations with CXCL8 expression were analyzed using Pearson’s χ² test; when any expected cell count was < 5, Fisher’s exact test was applied instead.
For other categorical clinicopathological features (e.g., gender, lymph node status, Hashimoto’s thyroiditis), correlations with CXCL8 expression were analyzed using Pearson’s χ² test; when any expected cell count was < 5, Fisher’s exact test was applied instead to ensure statistical validity under sparse data conditions.
All experiments were independently repeated three times, and only data consistent across two or more repetitions were subjected to statistical evaluation to ensure reproducibility and minimize random error. A P-value < 0.05 was considered statistically significant, and all tests were two-tailed unless otherwise specified.
Results
Analysis and verification of CXCL8 expression in tumors
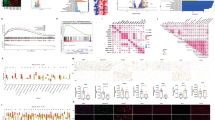
Analysis of pan-cancer data from the UCSC Xena platform revealed elevated CXCL8 expression in tumor samples from multiple cancer types, including Thyroid Cancer (THCA), Bile Duct Cancer (CHOL), Colon Cancer (COAD), Esophageal Cancer (ESCA), Head and Neck Cancer (HNSC), Kidney Papillary Cell Carcinoma (KIRP), Rectal Cance (READ), Stomach Cancer (STAD), and Endometrioid Cancer (UCEC). In contrast, CXCL8 expression was significantly higher in normal samples than in tumor tissues only in Kidney Chromophobe (KICH)(Fig. 1 A).
We next focused on PTC by downloading transcriptome data (HTSeq-FPKM) and corresponding clinical information from the TCGA database, comprising 355 PTC and 49 normal thyroid samples. CXCL8 expression was significantly upregulated in PTC compared to normal thyroid tissue (P < 0.001, Fig. 1B). This finding was corroborated by paired differential analysis (P < 0.001, Fig. 1 C) and further validated in the independent GSE33630 dataset (P < 0.05, Fig. 1D).
To experimentally confirm these observations, we performed RNA sequencing on 7 paired PTC and normal tissues from Gansu Provincial People’s Hospital. Differential expression analysis (threshold: FC > 1.5, |log2FC| > 0.585, P < 0.05) identified 4904 significantly differentially expressed genes (DEGs), including 3035 upregulated and 1869 downregulated genes. Notably, CXCL8 was among the most significantly upregulated genes (log2FC = 3.03, P < 0.001, Fig. 1E), supporting its potential role as a biomarker in PTC.
Unsupervised clustering based on the DEGs clearly separated PTC and normal thyroid samples into two distinct groups, reflecting substantial transcriptome-level heterogeneity (Fig. 1 F). CXCL8 exhibited a consistent high-expression pattern in PTC samples, aligning with the volcano plot results. Minor deviations in gene expression patterns in certain samples (e.g., Cancer-3, Normal-2) suggested underlying molecular heterogeneity within the tumor cohort.
Expression Analysis of CXCL8 in PTC and Various Cancers.(A) Perform a pan-cancer expression analysis of CXCL8 between normal and tumor samples based on UCSC Xena data.(B) In the TCGA database, the expression of CXCL8 in thyroid papillary carcinoma tumor samples is higher than in normal samples (P < 0.001)(C) The expression of CXCL8 in PTC samples from the TCGA database is higher than in adjacent normal tissues (P < 0.001).(D)In the GSE33630 dataset, the expression of CXCL8 is higher in PTC samples compared to normal samples (P < 0.05).(E)The volcano plot shows that CXCL8 is upregulated in tumors based on RNA sequencing data from 7 pairs of PTC and normal thyroid tissues (from Gansu Provincial People’s Hospital)(P < 0.001).(F) The heatmap shows some upregulated and downregulated genes, with samples clustered into the PTC group and the normal group (from Gansu Provincial People’s Hospital)(P < 0.05).
Correlation of CXCL8 with other clinical features
Through the analysis, it can be seen that CXCL8 and T stage, N stage is statistically significant, and there is no statistical significance between CXCL8 and clinical characteristics including age, gender, clinical stage and M stage(Fig. 2).
(A) Heatmap illustrating the distribution of key clinicopathological features (including T stage and N stage) between the CXCL8 high- and low-expression groups.(B-G)Boxplots showing the relationship between CXCL8 expression levels and other clinical parameters: age, gender, M stage, and clinical stage. Statistical significance was observed for T stage and N stage (P < 0.05), while no significant associations were found for age, gender, M stage, or clinical stage.
Biological function of CXCL8 in PTC
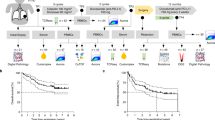
Based on RNA sequencing data from the TCGA database, a co-expression analysis identified 42 genes that were strongly and positively correlated with CXCL8 expression. Among these, 11 genes with the highest correlation coefficients were selected to construct a gene-gene interaction network (Fig. 3). Differential expression analysis between the CXCL8 high-expression and low-expression groups revealed a total of 922 DEGs, including 274 upregulated and 648 downregulated genes (Fig. 3A-B). GO and KEGG pathway enrichment analyses demonstrated that these DEGs were primarily associated with extracellular matrix organization and cytokine–cytokine receptor interaction pathways (Fig. 3C–E). Moreover, GSEA further confirmed that the cytokine–cytokine receptor interaction pathway was significantly enriched in patients with high CXCL8 expression (Fig. 3 F).
(A)Circos plots show the co-expression network of CXCL8 with 11 genes in PTC samples. Each part of the circle represents a gene, and its width indicates the total amount of co-occurrence connecting a gene to another gene. The width of each link represents the total expression time of the linked gene.(B)The heat map shows the top 50 down-regulated genes and top 50 up-regulated genes found in the high expression group.(C-D) Results from GO analysis.(E)Results from KEGG enrichment analysis.(F)GSEA analysis revealed upregulated and downregulated pathways associated with CXCL8 expression.
Relationship between CXCL8 expression and tumor immune microenvironment
Based on “CIBERSORT”, the difference of immune cell infiltration levels between the CXCL8 high expression group and the low expression group was analyzed, which confirmed that the expression of CXCL8 was significantly correlated with tumor-infiltrating immune cells(Fig. 4A-B). We further analyzed the correlation between CXCL8 and immune cells and showed that the infiltration levels of activated dendritic cells, neutrophils, resting dendritic cells(DCs), macrophages M0 and activated mast cells were positively correlated with CXCL8(Fig. 4C-G). However, the infiltration levels of CD8+ T cells, plasma cells, macrophage M1, and γδ T cells were negatively correlated with CXCL8(Fig. 4H-K).The scores of StromalScore, ImmuneScore, and ESTIMATEScore were significantly different between the CXCL8 high and low expression groups, and the CXCL8 high expression group was significantly higher than the CXCL8 low expression group in all three scores(Fig. 4L).
We then explored the correlation between CXCL8 and immune checkpoints using the expression levels of CXCL8 and 27 immune checkpoint-related genes. CXCL8 was significantly correlated with all 27 immune checkpoint-related genes (P < 0.001), and all of them were positively correlated.(Fig. 4M)(STable 1). We also obtained treatment scores for anti-CTLA4 and anti-PD-1 inhibitors, with higher treatment scores indicating better treatment effect. The analysis of immunotherapy response scores revealed that patients with low CXCL8 expression had significantly higher scores for anti-CTLA4 treatment (median [IQR]: 1.58 [1.21–1.95]) compared to those with high CXCL8 expression (median [IQR]: 0.85 [0.62–1.14]; Mann-Whitney U test, P = 0.0073).Statistical significance was found only in the CTLA4-positive -PD-1-negative fraction, indicating that anti-CTLA4 therapy is a better choice than anti-PD-1 therapy for patients with low CXCL8 expression(Fig. 4 N).
(A)Correlation between CXCL8 and immune infiltration.(B)The lollipop plot shows the correlation between immune infiltration and CXCL8 expression.(C-G)Scatter plots show the correlation between CXCL8 expression levels and immune cells: the infiltration levels of activated DCs, neutrophils, resting DCs, macrophages M0, and activated mast cells were positively correlated with CXCL8.(H-K)Scatter plots show the correlation between CXCL8 expression levels and immune cells: infiltration levels of CD8+ T cells, plasma cells, macrophage M1, and γδ T cells were negatively correlated with CXCL8.(L)Immunoscore of CXCL8 in high and low expression groups.(M)Correlation between CXCL8 and immune checkpoint related genes. Twenty-seven immune checkpoints were positively correlated with CXCL8.(N)CXCL8 immunotherapy scores of anti-CTLA4 and anti-PD-1 inhibitors in high and low expression groups.
Single cell date analysis with CXCL8
We obtained raw single-cell RNA sequencing data of one PTC sample and one normal thyroid sample from the GEO database GSE241184. After multiple rounds of cluster analysis and dimensionality reduction, 21 cell clusters were finally identified and labeled as different cell types (Fig. 5A-C). The data showed that CXCL8 was highly expressed in DCs. We also performed a systematic analysis of CXCL8 expression levels in various cell types (Fig. 5D-E).
The expression of CXCL8 mRNA in PTC tissues
In order to verify the expression of CXCL8 in PTC tissues, we extracted mRNA from the tissues for qRT-PCR analysis.The results showed that compared with the PNTs, the relative expression level of CXCL8 mRNA in PTC tissues was significantly higher than the control tissues, with a significant statistical difference (P = 0.0035)(Fig. 6). The results are shown in the figure. This result is consistent with the results obtained from the analysis of GEO and TCGA data in bioinformatics, indicating that CXCL8 is abnormally highly expressed in PTC tissues.
CXCL8 expression and clinicopathological characteristics of PTC patients
To validate CXCL8 expression at the protein level and its clinical significance in PTC, we performed IHC analysis on a tissue microarray constructed from 49 paired PTC and PNT tissues.
In PTC tissues (Fig. 7A-B), HE staining clearly revealed characteristic pathological features of PTC, including papillary architecture, nuclear overlapping, ground-glass nuclei, and nuclear grooves. Corresponding IHC staining (Fig. 7B) demonstrated strong positive staining for the CXCL8 protein, which was localized in the cytoplasm of PTC cells, appearing intense and widely distributed throughout the tumor regions. In contrast, H&E staining of adjacent normal thyroid tissues (Fig. 7C-D) showed preserved normal thyroid follicular structure with uniformly sized follicles containing colloid. In these normal tissues, IHC staining for CXCL8 (Fig. 7D) was negative or showed only very weak positivity, forming a sharp contrast with the PTC tissues.
Based on this IHC analysis, we concluded that CXCL8 protein is significantly upregulated in PTC tissues. We subsequently investigated the correlation between CXCL8 protein expression levels and various clinicopathological parameters. Statistical analysis revealed that high CXCL8 expression was significantly associated with lymph node metastasis (χ2 = 16.034, P < 0.001). Specifically, all patients with central lymph node metastasis (N1a, n = 15) or lateral (± central) lymph node metastasis (N1b, n = 11) exhibited high CXCL8 expression, whereas only 52.2% (12/23) of patients without lymph node metastasis (N0) showed high CXCL8 expression (Table 2).
Furthermore, a significant inverse correlation was observed between CXCL8 expression and the presence of Hashimoto’s thyroiditis (χ2 = 7.646, P = 0.006). Among patients with Hashimoto’s thyroiditis (n = 10), only 40.0% (4/10) showed high CXCL8 expression, compared to 87.2% (34/39) in patients without Hashimoto’s thyroiditis.
No statistically significant correlations were found between CXCL8 expression and other clinicopathological features, including patient gender, age, tumor diameter, tumor location, recurrence risk stratification, capsular invasion, extrathyroidal extension, or multifocality (all P > 0.05; Table 2).
These findings indicate that CXCL8 protein is specifically and highly expressed in PTC tissues, and its elevated expression is closely associated with lymph node metastasis. Furthermore, its expression may be influenced by the presence of Hashimoto’s thyroiditis, providing further evidence for the potential role of CXCL8 in PTC progression and metastasis.
Immunohistochemical analysis reveals significantly elevated CXCL8 protein expression in PTC tissues relative to PNTs.(A) HE staining of PTC tissues. (B) CXCL8 staining was positive in PTC tissues. (C) HE staining of PNTs. (D) CXCL8 staining was negative in PNTs.All images were acquired at 200× magnification. Scale bars: 100 μm.
Discussion
PTC is the most common pathological type of thyroid malignancy, particularly prevalent in women13,14. PTC typically grows slowly and has a favorable prognosis, with a key characteristic being its ability to invade adjacent structures, such as lymphatic vessels. Approximately 10% of patients may present with metastatic disease at the time of diagnosis15. In recent years, numerous PTC-associated risk genes have been identified, which significantly affect the proliferation, migration, and invasion of PTC cells.
Chemokine ligand 8 (CXCL8), also known as interleukin-8 (IL-8), is a member of the CXC family of chemokines16,17. This widely expressed cytokine plays a critical role in mediating cellular inflammatory responses and regulating autoimmune diseases within the tumor microenvironment. CXCL8 is upregulated in various types of malignant tumors18. During tumor progression, CXCL8 promotes the proliferation, migration, invasion, angiogenesis, and epithelial-to-mesenchymal transition (EMT) of tumor cells through autocrine or paracrine signaling mechanisms7,19.
Pan-cancer expression analysis of CXCL8 revealed higher expression levels in tumor samples of several cancers, including CHOL, COAD, ESCA, HNSC, KIRP, READ, STAD, and UCEC. Additionally, data from the TIMER2.0 database showed that CXCL8 expression in thyroid cancer samples was significantly higher than in PNTs. Bioinformatics analysis further confirmed that CXCL8 levels were generally elevated in tumor tissues compared to PNTs. To validate these findings, we performed qRT-PCR experiments, which demonstrated significantly increased CXCL8 mRNA expression in PTC tissues compared to PNTs.To investigate the relationship between CXCL8 expression and clinical-pathological features, we conducted tissue microarray experiments using samples from 49 PTC patients. The results revealed that CXCL8 expression was significantly associated with lymph node metastasis and Hashimoto’s thyroiditis, but not with gender, age, tumor size, tumor location, recurrence risk, capsule invasion, or extrathyroidal invasion. The increased concentration of CXCL8 detected in cancerous tissues contributes to enhanced cancer cell proliferation and migration. A notable association between CXCL8 expression and lymph node metastasis suggests that CXCL8 could serve as a potential biomarker for both tumor progression and patient survival in PTC. Gu et al. showed that CXCL8 expression was significantly correlated with clinical stage, distant metastasis, histological type, and grade in cervical cancer, with high CXCL8 expression serving as an independent poor prognostic marker for cervical cancer patients20. Hashimoto’s thyroiditis, a common organ-specific autoimmune disease, is characterized by the presence of various cytokines, including IL-1α, IL-1β, IL-2, IL-3, and IL-8, which amplify the inflammatory response via nitric oxide (NO) and prostaglandins21.
In our study, the Circos diagram illustrated the co-expression network of CXCL8 and 11 genes in PTC samples. We conducted GO and KEGG pathway analyses of differential proteins, leading us to hypothesize the following mechanisms: CXCL8 exhibits significant co-expression with CXCL1, CXCL2, and CXCL3, binding to CXCR1 or CXCR2, triggering multiple G-protein-mediated signaling cascades, including the PLC-PKC, PI3K-Akt, Src, and FAK pathways. These pathways promote tumor cell proliferation, survival, angiogenesis, EMT, migration, and invasion18,22. In our single-cell data, the gene CXCL8 was found to be specifically highly expressed in Dendritic cells. AKT1 in the PI3K-AKT pathway was also highly expressed in Dendritic cells, which provided a basis for the malignant metastasis of CXCL8 in PTC, and our KEGG analysis results also supported CXCL8 as a potential regulator of PI3K-AKT pathway.The PI3K-Akt signaling pathway, crucial for cell membrane receptor signaling, plays an essential role in inflammatory and immune responses23. Zhao et al. demonstrated that blocking the PI3K-Akt pathway eliminated IL-8-induced oxaliplatin resistance in gastric cancer derived from cancer-associated fibroblasts (CAFs)23. Furthermore, CXCL8/CXCR1 and/or CXCL8/CXCR2 signaling induces PI3K expression by activating a cascade that leads to Akt phosphorylation, promoting cell differentiation, proliferation, and survival24. Another important pathway is the MAPK cascade, where CXCL8 activates the RAF/MAPK/ERK pathway25. The FAK/IL-8 axis promotes gastric cancer cell proliferation and migration7. PELI1, a significant transcriptional target, cooperates with CXCL8 in activating the NF-κB pathway, facilitating tumor metastasis, and driving cell proliferation and migration26. High mRNA expression of GPER1, EGFR, and CXCR1 in PTC correlates significantly with lymph node metastasis (LNM), which may help predict LNM risk23.
The tumor immune microenvironment becomes active through paracrine, autocrine, and endocrine communication, generating dynamic signals that drive cancer progression27,28. It is widely accepted that dysfunction in the tumor-immune ecosystem impairs immune surveillance29. Using the “CIBERSORT” tool, we performed differential analysis of immune cell infiltration levels between high- and low-expression CXCL8 groups, confirming a significant correlation between CXCL8 and tumor-infiltrating immune cells. The infiltration levels of activated dendritic cells, neutrophils, resting dendritic cells, M0 macrophages, and activated mast cells positively correlated with CXCL8, while CD8+ T cells, plasma cells, M1 macrophages, and γδ T cells negatively correlated with CXCL8. CXCL8 promotes the migration of CD4+ T cells, leading to immune imbalance and mediating pro-inflammatory functions6,30. In triple-negative breast cancer, CXCL8 inhibits the infiltration of CD4+ and CD8+ T cells and reshapes the tumor immune microenvironment by upregulating CD274 expression31. In endometriosis, CXCL8 secretion from endometrial stromal cells impairs the cytotoxic activity of NK cells, contributing to local immune dysfunction19. M0 macrophages promote tumor cell proliferation, migration, and invasion, with CXCL8 being a key regulator of these processes. Zhao et al. found that CXCL8 regulates the proliferation and polarization of M0 macrophages in cervical cancer progression32. DCs initiate adaptive immunity and regulate inflammation by producing inflammatory chemokines33,34. Infected DCs produce high levels of CXCL8, further stimulating neutrophil activity, contributing to granuloma formation in cat scratch disease35. Mast cells, as multifunctional immune cells, contribute to both innate and acquired immunity. Activated mast cells release various pro-inflammatory mediators, including CXCL8, which may drive mast cell migration35,36.CXCL8 and related molecules (CXC family) primarily exert chemotactic effects on neutrophils. By recruiting macrophages, neutrophils, and T cells to the tumor site, CXCL8 plays a pivotal role in tumor invasiveness and can inhibit anti-tumor immune responses37,38.
Based on the immunosuppressive role of CXCL8 in PTC, targeting the CXCL8 pathway39,40 in combination with PD-1 blockade enhances the tumor immune response and inhibits tumor progression41. CXCL8 is primarily secreted by macrophages, contributing to the immunosuppressive tumor microenvironment by inducing PD-L1+ macrophages in gastric cancer42. Danlos et al. found that mesothelioma patients with primary resistance to PD-1-targeted immunotherapy exhibited elevated CXCL8 levels in both blood and tumors38. Blockade of CTLA4 has been shown to reduce immunosuppressive cells such as myeloid-derived suppressor cells (MDSC) and M2 macrophages, enhancing T-cell activation and presenting a new therapeutic target for PTC43,44.
Conclusions
We detected the expression of CXCL8 in PTC tissues by bioinformatics, qRT-PCR and tissue microarray experiments. The results indicated that CXCL8 plays an important role as a potential biomarker in the biological functions of PTC and can be used as a new target for drug development, providing a possible new direction for the immunotherapy of PTC.
Data availability
The datasets used and/or analysed duringthe current study available from the corresponding author on reasonable request.All data generated or analysed during this study are included in this published article.
References
Filetti, S. et al. Thyroid cancer: ESMO clinical practice guidelines for diagnosis, Tre atment and follow-up†. Ann. Oncol. 30, 1856–1883 (2019).
Mete, O. et al. Correction: Consensus statement: recommendations on actionable Bi Omarker testing for thyroid cancer management. Endocr. Pathol. 35, 309–310 (2024).
Schlumberger, M. & Leboulleux, S. Current practice in patients with differentiated Th Yroid cancer. Nat. Rev. Endocrinol. 17, 176–188 (2021).
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Oh, J. M. & Ahn, B. C. Molecular mechanisms of radioactive iodine refractoriness in differ entiated thyroid cancer: impaired sodium iodide symporter (NIS) expression owing to altered signaling pathway activity and intracellular localization of NIS. Theranostics 1 (2), 1234–1256 (2022).
Asokan, S. & Bandapalli, O. R. CXCL8 signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 1302, 25–39 (2021).
Ma, Y. et al. FAK/IL-8 axis promotes the proliferation and migration of gastric Canc Er cells. Gastric Cancer. 26, 528–541 (2023).
Yin, Y. et al. CXCL8 May serve as a potential biomarker for predicting the prognosi s and immune response in cervical cancer. Discov Onc. 15, 601 (2024).
Li, F. et al. Elucidating the role of pyroptosis in papillary thyroid cancer: prognostic, immunological, and therapeutic perspectives. Cancer Cell. Int. 24, 45 (2024).
Oshlack, A., Robinson, M. D. & Young, M. D. From RNA-seq reads to differential expression results. Genome Biol. 11, 220 (2010).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Sosa, M. S. et al. Subtype-specific overexpression of the Rac-GEF P-REX1 in breast cancer is associated with promoter hypomethylation. Sci. Rep. 10, 12345 (2020).
Lee, J. H. et al. Sonographic and cytopathologic correlation of papillary thyroid carci Noma variants. J. Ultrasound Med. 34, 1–15 (2015).
Luo, X. et al. FOXP4-AS1 inhibits papillary thyroid carcinoma proliferation and M igration through the Akt signaling pathway. Front. Oncol. 12, 900836 (2022).
Limaiem, F., Rehman, A. & Mazzoni, T. Papillary Thyroid Carcinoma. In StatPearls (StatPearls Publishing, 2025).
Cambier, S., Gouwy, M. & Proost, P. The chemokines CXCL8 and CXCL12: Molecu Lar and functional properties, role in disease and efforts towards Pharmacological Inte rvention. Cell. Mol. Immunol. 20, 217–251 (2023).
Russo, R. C., Garcia, C. C., Teixeira, M. M. & Amaral, F. A. The CXCL8/IL-8 Che mokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol. 10, 593–619 (2014).
Liu, Q. et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 31, 61–71 (2016).
Walle, T. et al. Radiotherapy orchestrates natural killer cell dependent antitumor imm Une responses through CXCL8. Sci. Adv. 8, eabh4050 (2022).
Yan, R., Shuai, H., Luo, X., Wang, X. & Guan, B. The clinical and prognostic Valu e of CXCL8 in cervical carcinoma patients: immunohistochemical analysis. Biosci. Re P. 37, BSR20171021 (2017).
Khan, F. A., Al-Jameil, N., Khan, M. F., Al-Rashid, M. & Tabassum, H. Thyroid dy sfunction: an autoimmune aspect. Int. J. Clin. Exp. Med. 8, 6677–6681 (2015).
Gu, L., Yao, Y. & Chen, Z. An inter-correlation among chemokine (C-X-C motif) Lig and (CXCL) 1, CXCL2 and CXCL8, and their diversified potential as biomarkers for tumor features and survival profiles in non-small cell lung cancer patients. Transl. Cancer Res. 10, 748–758 (2021).
Zhao, Z. et al. Calcipotriol abrogates cancer-associated fibroblast-derived IL-8-mediate d oxaliplatin resistance in gastric cancer cells via blocking PI3K/Akt signaling. Acta Pharmacol. Sin. 44, 178–188 (2023).
Korbecki, J. et al. CXCR2 Regulation of Expression, Signal Transduction, and Involvement in Cancer. IJMS 23, 2168 (2022).
Cahyanur, R. et al. CXCL8, MMP1, MMP2, and FN1 Gene Expression and Tumor Extension in Nasopharyngeal Cancer Patients: A Cross-sectional Study. Gene Expressi on 55, (2023).
Zhang, L. et al. Identification of PANoptosis-based signature for predicting the progn osis and immunotherapy response in AML. Heliyon 10, e40267 (2024).
Bejarano, L., Jordāo, M. J. C. & Joyce, J. A. Therapeutic targeting of the tumor M icroenvironment. Cancer Discov. 11, 933–959 (2021).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Chen, C., Wang, Z., Ding, Y. & Qin, Y. Tumor microenvironment-mediated immune evasion in hepatocellular carcinoma. Front. Immunol. 14, 1133308 (2023).
Mei, J. et al. Suppression of autophagy and HCK signaling promotes PTGS2high FC GR3 – NK cell differentiation triggered by ectopic endometrial stromal cells. Autopha Gy. 14, 1376–1397 (2018).
Zhao, X. et al. CXCL8 modulates M0 macrophage proliferation and polarization to i nfluence tumor progression in cervical cancer. Sci Rep 15, 790 (2025).
Vermi, W. et al. Role of dendritic cell-derived CXCL13 in the pathogenesis of Barto Nella Henselae B-rich granuloma. Blood 107, 454–462 (2006).
Wang, Y. et al. Elevated expression of miR-142-3p is related to the pro-inflammatory function of monocyte-derived dendritic cells in SLE. Arthritis Res. Ther. 18, 263 (2016).
Marciscano, A. E. & Anandasabapathy, N. The role of dendritic cells in cancer and anti-tumor immunity. Semin Immunol. 52, 101481 (2021).
Zhang, G. et al. Distribution change of mast cells in human nasal polyps. Anat. Rec (Hoboken). 295, 758–763 (2012).
Solimando, A. G., Desantis, V. & Ribatti, D. Mast cells and interleukins. Int. J. Mol. Sci. 23, 14004 (2022).
David, J. M., Dominguez, C., Hamilton, D. H. & Palena, C. The IL-8/IL-8R axis: A double agent in tumor immune resistance. Vaccines (Basel). 4, 22 (2016).
Danlos, F. X. et al. Genomic instability and protumoral inflammation are associated with primary resistance to Anti-PD-1 + Antiangiogenesis in malignant pleural Mesot helioma. Cancer Discov. 13, 858–879 (2023).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Huang, R. et al. Targeting cancer-associated adipocyte-derived CXCL8 inhibits triple- negative breast cancer progression and enhances the efficacy of anti-PD-1 Immunothe Rapy. Cell. Death Dis. 14, 703 (2023).
Lin, C. et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut 68, 1764–1773 (2019).
Li, S. et al. Molecular subtypes of oral squamous cell carcinoma based on immun osuppression genes using a deep learning approach. Front. Cell. Dev. Biol. 9, 68724 5 (2021).
Wang, Y. et al. Mitochondrial pyruvate dehydrogenase phosphatase metabolism disorder in malignant tumors. Oncol. Res. 33 (8), 1861–1874 (2025).
Acknowledgements
The authors thank the The First Clinical Medical College of Lanzhou University and Gansu Province People’s Hospital for data support and guidance.
Funding
This work was funded by the National Natural Science Foundation of China (No. 82260475); National research project cultivation plan key project of Gansu Provincial Hospital (No. 19SYPYA-13); Gansu Province health industry science and technology innovation major research project (No.GSWSZD2024-02); The 2022 Master-Doctor-Postdoctoral program of NHC Key Laboratory of Diagnosis and Therapy of Gastrointestinal Tumor/TheNon-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (NHCDP2022021); Research Fund project of Gansu Provincial Hospital(21GSSYB-19); The Natural Science Foundation of Gansu Province, China(25JRRA285);Youth Science and Technology Fund of Gansu Province(24JRRA612).
Author information
Authors and Affiliations
Contributions
FL and WZ were the co-first authors.FL, WZ and XW contributed to the conception and design. FL, WZ and XG contributed to the development of the methodology.WZ, XG and QX contributed to material preparation and data collection.FL, WZ and XG contributed to the writing and revision of the manuscript. ZL, QZ and RL contributed to the study supervision. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee of Gansu Province People’s Hospital (Approval Number:2022 − 182) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, F., Zhang, W., Gao, X. et al. Identification of CXCL8 as a potential gene associated with lymph node metastasis in papillary thyroid carcinoma through bioinformatics analysis. Sci Rep 15, 41771 (2025). https://doi.org/10.1038/s41598-025-25686-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-25686-x